Abstract
Biological nitrogen fixation is the primary source of new N in terrestrial arctic ecosystems and is fundamental to the long-term productivity of arctic plant communities. Still, relatively little is known about the nitrogen-fixing microbes that inhabit the soils of many dominant vegetation types. Our objective was to determine which diazotrophs are associated with three common, woody, perennial plants in an arctic glacial lowland. Dryas integrifolia, Salix arctica, and Cassiope tetragona plants in soil were collected at Alexandra Fiord, Ellesmere Island, Canada. DNA was extracted from soil and root samples and a 383-bp fragment of the nifH gene amplified by the polymerase chain reaction. Cloned genotypes were screened for similarity by restriction fragment length polymorphism (RFLP) analysis. Nine primary RFLP phylotypes were identified and 42 representative genotypes selected for sequencing. Majority of sequences (33) were type I nitrogenases, whereas the remaining sequences belonged to the divergent, homologous, type IV group. Within the type I nitrogenases, nifH genes from posited members of the Firmicutes were most abundant, and occurred in root and soil samples from all three plant species. nifH genes from posited Pseudomonads were found to be more closely associated with C. tetragona, whereas nifH genes from putative alpha-Proteobacteria were more commonly associated with D. integrifolia and S. arctica. In addition, 12 clones likely representing a unique clade within the type I nitrogenases were identified. To our knowledge, this study is the first to report on the nifH diversity of arctic plant-associated soil microbes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the Canadian High Arctic, low temperature and poor substrate quality frequently limit decomposition and N mineralization, and consequently plant production is often N-limited [3, 18, 22]. In these ecosystems, biological nitrogen fixation is the primary source of new N to plants [2], highlighting the importance of the nitrogen-fixing microbial community (diazotrophs) in the distribution and abundance of High Arctic vegetation.
Biological nitrogen fixation is the reduction of atmospheric dinitrogen to ammonium as catalyzed by the enzyme nitrogenase. nifH is the gene that encodes the iron protein subunit of nitrogenase and it is highly conserved among all diazotrophic groups, making it an ideal molecular marker for these organisms in nature. Good correlation between the nifH phylogeny and the 16S rRNA phylogeny has been found for diazotrophic microbes [24, 25], lending support to the application of nifH as a molecular phylogenetic tool.
Recently, efforts have been made to examine nifH diversity across multiple ecosystems [25]. A principal finding of this analysis was that the distribution of nitrogen-fixing microorganisms is nonrandom and can be predicted on the basis of habitat characteristics [25]. Diazotroph activity is important in many environments that remain un- or underinvestigated. An understanding of the distribution of nitrogen-fixing organisms in nature is a prerequisite step for assessing changes in these communities that are caused by environmental change.
Previously, we reported on the effects of warming and fertilization on diazotroph community structure and activity at Alexandra Fiord, Ellesmere Island, Canada [6]. We found that nitrogenase activity and nifH gene community structure were controlled by different factors; although nitrogenase activity was spatially variable and strongly controlled by the presence of moss, nifH gene community structure was temporally variable in response to warming. As a consequence of their greater nifH diversity, subsurface diazotrophs accounted for the greatest change in nifH community structure with warming. We hypothesized that the structural shifts in diazotroph communities could be driven by plant processes under phenological control [6]. However, little is known about the diazotrophs that inhabit the soils of many of the dominant vegetation types in the Canadian High Arctic. In this study, we examined the diazotroph community associated with three common, woody, perennial arctic plant species in a glacial lowland. Our objective was to determine which diazotrophs are commonly associated with roots, rhizospheres, and bulk soils surrounding these plants, in order to gain information about what organisms may be important in the nitrogen nutrition of these long lived species. Given their potential for change under future environmental conditions, baseline information on the organisms that inhabit arctic soils is critical for predictions of how arctic ecosystems will respond to future warming.
Methods
Plant Collection
Plants were collected in a glacial lowland at Alexandra Fiord, Ellesmere Island, Canada (78°53′N, 75°55′W). Plants were randomly selected within the dwarf-shrub cushion-plant type community, which is dominant in the lowland [15]. On July 13, 2002, four Dryas integrifolia plants were harvested. Whole plants with intact root systems were excavated with a shovel and placed in large plastic bags that were immediately frozen and remained so for the duration of transport to the laboratory. On August 4, 2002, four additional D. integrifolia plants were harvested along with two individuals of S. arctica, and two Cassiope tetragona plants. These eight plants were stored and transported frozen as previously described.
Once in the laboratory, plants were allowed to thaw at room temperature until the soil was friable and three subsamples per plant were collected: “bulk soil,” “roots with aggregates,” and “surface-sterilized roots.” Soil near the plant, but not in direct contact with any roots, was designated as the bulk soil sample. These samples were placed in a sealed plastic bag and homogenized via shaking by hand for 1 min. The entire root system of each plant was then dissected and divided in half. The first half on the root system was shaken by hand to remove soil particles that adhered loosely, and this “roots with aggregates” comprised the second sample. The remainder of the root system was placed into a glass jar of distilled water, sealed with a tight fitting lid, and shaken vigorously by hand for 1 min to remove soil aggregates. The root system was then removed and rinsed several times with distilled water prior to immersion in 5% bleach solution (by volume) for 5 min. After immersion in bleach, the roots were rinsed several times with sterile distilled water and comprised the third subsample of “surface-sterilized roots.”
DNA Extraction and PCR Amplification, Cloning
DNA was extracted from 1-g subsamples of bulk soil by using an UltraClean Soil DNA Isolation kit according to the manufacturer's instructions (MoBio, Carlsbad, CA, USA). DNA was extracted from roots with aggregates and surface-sterilized roots samples using the same kit; however, prior to applying the standard protocol, roots were macerated with a micropestle under aseptic conditions. To ensure that micropestles were DNA-free, they were soaked in 2 M HCL for 48 h, rinsed three times with sterile distilled water, and air-dried prior to sample maceration. When the mass of roots with aggregates exceeded 1 g, root clusters and root aggregates were preferentially selected over larger diameter woody roots. Extracted DNA was stored frozen at −20°C.
A half-nested polymerase chain reaction (PCR) protocol was used to amplify a 383-bp fragment of the nifH gene from a diluted extract using the protocol described by Deslippe et al. [6]. To isolate single genotypes, PCR products were cloned using a TOPO Cloning kit (Invitrogen, Burlington, ON, Canada) following the supplier's instructions. White colonies were grown overnight in 100 μL SOC medium containing 100 mg/L ampicillin at 37°C, then 10-μL aliquots of these colonies were killed by boiling at 100°C for 5 min in a heat block, and diluted 1:4 with sterile, DNA-free water. These dilutions were used as template for the reamplification of nifH genes for restriction fragment length polymorphism (RFLP) analysis, using the half-nested protocol described by Deslippe et al. [6], except that only the final amplification was performed. Nine clones per soil or root fraction were selected for the reamplification of nifH genes, for a total of 324 clones. The purity of the sterile distilled water used in PCR and cloning reactions was continuously monitored by the use of negative controls in each reaction set.
RFLP Analysis
PCR products were screened for similarity using RFLP analysis. Amplified PCR products were digested using the enzymes TaqI and HhaI (Invitrogen) according to the manufacturer's instructions. Resulting fragments were run with 1-kb ladder marker (Invitrogen), on a 2.5% high-resolution gel at 100 mV, stained with ethidium bromide, and visualized on an ultraviolet transilluminator. Gels were photographed digitally using a Gel Print 2000i photodocumentation system (BioPhotonics, Pittsfield, MA, USA). Fragment patterns were analyzed using Gene Profiler 4 (Scanalytics, Rockville, MD, USA). Fragment size was calibrated using the desmile calibration method with log piecewise linear curve fitting. Fragments in all lanes of a gel were matched using a pairwise method at a tolerance of 2%; although fragment patterns from different gels were matched at a tolerance of 5% to compensate for differences among gels. Fragments less than 75 bp were excluded from the analysis to reduce the possibility of including PCR artifacts and primer dimers [14]. Cluster analysis of the fragment patterns was performed in Gene Profiler using the Neighbor module and Dice's similarity index [7] (converted to a distance measure as 1 − Dice's index) [14]. The results were viewed by using Tree View software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). Isolates representative of the different RFLP clades were selected for sequencing.
Statistical Analyses
General Linear Model ANOVAs were used to analyze the effect of sampling date (July vs August) and soil fraction on the nine RFLP types for D. integrifolia. Because of the uneven sampling of the three plant species, the effects of plant species and soil fraction on the RFLP types were determined by using nonparametric Kruskal-Wallis ANOVAs for D. integrifolia, S. arctica, and C. tetragona in August. All ANOVAs were performed via STATISTICA version 6.1 [21].
Contingency tables of RFLP-type by root and soil fraction were constructed for 216 clones from the August sampling date for use in correspondence analysis (CA). Clone counts were weighted by column totals prior to analysis. The goal of correspondence analysis is to reproduce the distances between points in a contingency table in a lower dimensional space. Correspondence analyses were run by using the Multivariate Exploratory Techniques module in STATISTICA 6.1 [21].
Sequencing and Phylogenetic Analysis
PCR was used to amplify a 581-bp fragment of pCR 2.1 TOPO vector (Invitrogen) containing our nifH insert from diluted, heat-killed clones. The primers M13F (5′-GTAAAACGACGGCCAG) and M13R (5′-CAGGAAACAGCTATGAC) annealed at the 2977–2992 and 176–197 positions of the vector, respectively. Both primers were synthesized by Invitrogen. PCR cocktails consisted of genomic DNA (approximately 80 ng), 0.2 mM dNTPs, 0.4 μM primers, 10× PCR Buffer, 2 mM MgCl2, and 0.72 U of Platinum Taq DNA polymerase (Invitrogen) in a final volume of 30 μL. PCRs were performed with a thermocycler program consisting of an initial denaturing temperature of 94°C for 4 min followed by 30 cycles of: denaturing at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min. A final extension period of 10 min at 72°C completed the program. A PTC-100 Programmable Thermal Controller (MJ Research Inc., Waltham, MA, USA) was used for all amplifications. PCR products were cleaned by ethanol precipitation before sequencing. The sequencing reaction employing dye-labeled dideoxy nucleotides was performed by using the CEQ DTCS Sequencing Kit (Beckman Coulter, Fullerton, CA, USA) with 1.6 μmol forward primer M13F (Invitrogen) and 30 ng reamplified clone DNA. The use of the M13 primer allowed sequencing to begin within the vector so that the entire nifH insert could be read. A CEQ8000 automated capillary electrophoresis sequencer (Beckman-Coulter) was used to generate all sequences. Sequences were visually checked for quality and completeness using Sequencher software 4.2.2 (Gene Codes, Ann Arbor, MI, USA). When necessary, a second sequencing reaction using the reverse primer, M13R, was performed, and the results were used to clarify ambiguous positions. All sequences were compared to known sequences in GenBank (http://www.ncbi.nlm.nih.gov) by using the BLAST nucleotide–nucleotide search function.
To ensure correct the alignment of nifH sequences, we downloaded the fer4_NifHPfam seed alignment (http://www.sanger.ac.uk/Software/Pfam/data/jtml/seed/PF00142.shtml) (see [1]) and used it to align protein translations of our 31 clone sequences and 38 selected representative nifH, anfH, and vnfH gene sequences obtained from GenBank. Protein alignment sequences were assembled in MacVector 7.2 (Accelrys, San Diego, CA, USA) and aligned using the ClustalW function. This alignment was visually checked and manually corrected for errors, and used to create a second alignment of DNA sequences. Phylogenetic analysis was performed using PAUP 4b10 (Sinauer Associates, Sunderland, MA, USA), a single starting tree was inferred using Neighbor Joining and then refined using Maximum Likelihood. To determine the model of evolution that best fit our data, the Akaki information criterion (AIC) was implemented using Modeltest 3.7 [16]. The General Time Reversible model with invariable sites and a gamma distribution (GTR+I+γ) was selected. Branch swapping used the tree bisection–reconnection (TBR) algorithm.
Results
Thirty-six clone libraries, each with 36 clones, were constructed for the root and soil fractions of D. integrifolia, S. arctica, and C. tetragona plants. Nine clones were randomly selected from each clone library to be screened for similarity using RFLP analysis (total, 324 clones). Ninety-two percent of these clones (297 of 324) formed nine RFLP phylotypes. RFLP phylotypes one through nine will be abbreviated as R1–R9 hereafter.
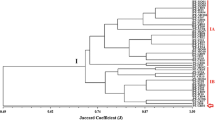
The correspondence analysis of plant samples and RFLP phylotypes shown in Fig. 1 extracted two principal dimensions: dimension 1, which accounts for 62.5% of the inertia (eigenvalue, 0.08280), and dimension 2, which accounts for 38.5% of the inertia (eigenvalue, 0.05179). The total chi-square value for the analysis was 24.2 with 14.9 and 9.3 contributed by dimension 1 and dimension 2, respectively (df = 14, p = 0.0432). In Fig. 1, we see that R6 was most closely associated with samples from C. tetragona (ellipse A), which is supported by the results of Kruskal–Wallis ANOVA that revealed that this pattern was driven predominantly by the root aggregate samples (p = 0.03; df = 2, χ2 = 7). R5 and R9 were most frequently associated with the root and soil fractions of D. integrifolia (ellipse B). R5, which was the largest of the phylotypes, contained 63 clones (Table 1); clones belonging to this phylotype occurred significantly more often in samples of D. integrifolia taken in August than in July (p = 0.04). Ellipse C highlights the association of S. arctica samples with R1, R2, and R7. The large distance among the phylotypes in ellipse C is attributable to differences in the frequency of phylotypes among soil and root fractions of S. arctica. Although R1 and R7 were both frequently associated with the root aggregates and bulk soils of this species, R2 occurred predominantly in the surface-sterilized root samples of S. arctica. In August, R3 and R4 were common to all three plant species studied, and frequently occurred in the bulk soil samples.
Clones representative of the nine phylotypes were selected for sequencing. Forty-two nifH genes were sequenced in total. In phylogenetic analysis, the majority (33) clustered within type I nitrogenases, which comprise Mo-containing nifH and a number of vnfH genes [4, 25]. Figure 2 shows that four primary clades occurred within the type I group. The first clade contained 11 sequences belonging to R2, R1, R9, and R8, and represented nifH sequences similar to those of alpha- and beta-Proteobacteria. Clones 7-4, 11-2, and 1C-4 clustered most closely with the nifH gene of Azospirillum lipoferum in phylogenetic analysis (Fig. 2) and returned the nifH genes of uncultivated alpha-Proteobacteria bacterium A2 and A5 from a Douglas-fir forest soil, and uncultured bacterium clone GYMC-22B from a nickel mine spoils in New Caledonia, as their most similar GenBank taxa in BLAST searches (Table 2). Clones 2C-3, 2C-4, 2C-1, and 4C-9 clustered with the nifH gene of Azorhizobium caulinodans in phylogenetic analyses (Fig. 2) and were most similar to Rhizobium-type (Rhizobium, Bradyrhizobium; Table 2) nifH gene sequences. Clones 5-9, J11-6, and 12-3 clustered together, and returned nifH clones Langqian-7 from the Tibetan Plateau, China, and NIW9-4 associated with dead aboveground biomass of Spartina alterniflora as their most similar taxa in GenBank. Branching from this group was clone J12-7, the only representative of RFLP phylotype 8, which was infrequent and detected mainly in July samples. Clone J12-7 was most similar to the nifH gene of the beta-Proteobacterium Herbaspirillum sp. B501 associated with rice (Oryza officinalis) seed from Japan, and to the nifH gene of Azoarcus tolulyticus, also a member of the beta-Proteobacteria (Fig. 2).
Maximum likelihood tree based on phylogenetic analysis of a 383-bp DNA fragment of nifH. Bootstrap values occur at nodes that received a minimum of 50% node support and are based on 1000 replicate Neighbor Joining trees. Taxa are followed by accession numbers in parentheses; G: GenBank; U: Universal Protein Knowledgebase. All sequence data are from samples collected on August 4, 2002; unless specified by “July” then samples were collected on July 13, 2002. Root: surface-sterilized roots; agg.: root aggregates; bulk: bulk soil; Dryas: Dryas integrifolia; Salix: Salix arctica; Cassiope: Cassiope tetragona.
The second clade within the type I nitrogenases contained six nifH gene sequences belonging to R3 and Gram-positive, high G + C members of the Firmicutes as well as cyanobacterial sequences. Within the Firmicutes, six clones grouped tightly together in 100% of the bootstrap replicates. Clones 2A-4, 2A-5, 5C-9, 3-7, J7-5, and J7-1 grouped with the nifH gene of Paenibacillus durus in phylogenetic analyses, and returned nifH genes of P. polymyxa strain DSM356 and uncultivated clone TP60021 from a Massachusetts pasture as their most similar taxa.
The third clade of the type I nitrogenases contained four clones from the present study and nifH gene sequences from several members of the gamma-Proteobacteria. Of the four clones, three belonged to R6, whereas one (clone 1B-2) belonged to RFLP phylotype 5. Our sequences were of two distinct types. Two clones of the first type (clones 10-5 and J1-8) clustered with the nifH gene of Pseudomonas stutzeri in phylogenetic analyses and were most similar to the nifH sequence of Pseudomonas sp. DC obtained from a rice inoculum in BLAST searches. The other two clones, 1B-2 and 12-7, had moderate sequence identity with the nifH gene of Azoarcus communis (Table 2).
The fourth clade contained 12 clones from the present study only, which are most similar to the nifH genes of uncultivated organisms from extreme environments. These sequences were 386 nucleotides in length in contrast to the all other sequences that amplified a 383-bp product. Many of the sequences belonging to clade 4 were most similar to nifH genes from uncultivated organisms from the Qing Zang Plateau of the Tibetan region of China (clones Qinglin-5, and Yushu-15; Table 1). None of our sequenced clones clustered with the type II or the type III nitrogenases (Fig. 2).
The remaining nine clones formed R5 in the RFLP analysis and returned a bchL gene of the purple photosynthetic bacterium Rhodopseudomonas palustris as their phylogenetic closest taxon in BLAST searches with approximately 85% sequence identity. Table 2 describes the RFLP phylotypes and taxonomic affiliations of the nifH gene sequences derived from organisms associated with the three plant species studied.
Discussion
Our phylogenetic analysis revealed that the majority of the nifH gene sequences recovered from root and soil samples of D. integrifolia, Salix arctica, and C. tetragona clustered within the type I nitrogenases, which comprise Mo-containing nifH and a number of vnfH genes [4, 25]. Clones comprised nine RFLP phylotypes and all, except R5, belonged to the type I group. In general, RFLP and phylogenetic analysis agreed; however, in two cases, members of RFLP phylotypes were split among clades in our phylogenetic analysis. Sequenced clones from R2 were most similar to Rhizobium-type nitrogenases (Rhizobium sp., Bradyrhizobium sp.; 84–90% similarity) as well as to Yushu-15, a nifH clone from the Tibetan Plateau, China (∼92% similar). In phylogenetic analysis, members of R2 were split depending on the number of nucleotides amplified. Sequences that amplified a 383-bp fragment (the “Rhizobium-like” sequences) grouped among the nifH genes of the alpha-Proteobacteria, whereas sequences that amplified a 386-bp product (the “Tibetan clone-like” sequences) grouped within clade four (Fig. 2).
A second instance where the RFLP and phylogenetic analyses disagreed was the inclusion of clone 1B-2 in clade 3. We were surprised to find that clone 1B-2 was most similar to the nifH gene of A. communis because of its placement in R5. All other sequences belonging to R5 appear to be type IV “nif-like” genes most similar to a chlorophyllide reductase gene. Clone 1B-2 was unique in our analysis in that it has only a single restriction fragment for both enzymes assayed. In silico digests of the sequence with HhaI revealed a single restriction site at 326 bp but because of our exclusion of fragments smaller than 75 bp in length, only the larger fragment was recorded. Similarly, the sequence had five restriction sites in in silico assays with TaqI, but all except the first 185-bp fragment were too small for inclusion in our analysis. Although R5 clones produced two restricted fragments for each enzyme, one of these was 187 bp in length, which was indistinguishable from a fragment of 185 bp with a match tolerance of 2%. Because Dice's index uses the number of common bands to calculate the similarity between clones, and because no other clone in our dataset was similar to clone 1B-2, our RFLP analysis falsely grouped 1B-2 with R5.
Clones 1B-2 and 12-7 were included within clade 3 (the gamma-Proteobacteria) despite the fact that they share moderate sequence identity with the nifH gene of a member of the beta-Proteobacteria A. communis (Table 2). These sequences are likely derived from members of the “authentic” Azoarcus branch described by Hurek et al. [10]; the monophyletic grouping of Azoarcus-like nifH sequences within the gamma-Proteobacteria is in agreement with the 16S ribosomal DNA phylogeny. Both 1B-2 and 12-7 amplified a 386-bp product, and were the only sequences outside of clade 4 to do so. The inclusion of Azotobacter vinelandii alternative vanadium nitrogenase gene vnfH in clade 3 illustrates that we were unable to distinguish among vnfH and nifH within the type I nitrogenases, which is consistent with the observation of Kessler et al. [11] that nifH phylogenies do not indicate metal type.
Correspondence analysis revealed differences in the distribution of diazotrophs among plants and their soil and root fractions. Figure 1 shows that C. tetragona was most strongly associated with members of R6 that comprise nifH genes from posited gamma-Proteobacteria. In the phylogenetic analysis, sequences from R6 formed a monophyletic group with P. stutzeri. We anticipated that we would find nifH genes from Pseudomonads in our samples. Pseudomonas sp. strain GR20-5 was isolated in pure culture from the rhizosphere of D. integrifolia growing in the southern region of Ellesmere Island [13]. This strain was characterized as a psychrotrophic diazotroph and an aggressive root colonizer [13]. P. stutzeri is a ubiquitous organism that is known to occupy diverse environments, including hydrocarbon-contaminated Antarctic soils [8].
D. integrifolia was most strongly associated with R9 and R5 (Fig. 1). Sequenced clones from R9 revealed that this phylotype is composed of nifH genes most similar to those of uncultivated alpha-Proteobacteria in soils and litter of a Douglas-fir forest in the Cascade Mountains of Oregon, USA. All sequenced clones belonging to R5 were moderately similar to a bacteriochlorophyllide reductase gene (bchL) from the organism R. palustris. BchL is a nifH homolog, and the gene sequences that formed R5 likely belong to the so-called type IV “nitrogenases,” which have been described as a highly divergent and loosely coherent group of nif-like genes that include sequences from the Archaea as well as homologous chlorophyllide reductase genes [4, 25]. The gene bchL encodes the ATP-binding iron–sulfur protein protochlorophyllide reductase, a residue of the photosynthetic reaction center complex of bacteriochlorophyll. In their article describing the complete genome sequence of R. palustris, Larimer et al. [12] note that the reaction center proteins of this organism are most closely related to the unusual photosynthetic bacterium Bradyrhizobium sp. strain ORS278, which forms nitrogen-fixing nodules on the stems of a tropical legume that grows in waterlogged soils. Many of the type IV genes are deeply branching and belong to organisms that are not known to perform nitrogen fixation and that are not photosynthetic. These sequences may represent relic proteins that are ancestral to both the bacteriochlorophyll and the nitrogenase genes [17]. If actively transcribed, products of these sequences are not likely involved in nitrogen fixation.
D. integrifolia was uniquely associated with the smallest phylotype, R8. All but a single nifH clone of this phylotype were derived from samples taken in the July sampling period, and R8 was not included in our correspondence analysis of plants from the August sampling period. A sequenced clone from R8 returned the organism Herbaspirillum sp. strain B501 as its phylogenetic nearest taxon (Table 2). In phylogenetic analyses, this clone was grouped among members of the beta-Proteobacteria, and R8 is thought to be composed of nifH genes from beta-Proteobacteria. Despite the small number of clones comprising R8, this phylotype appeared to be fairly common among D. integrifolia plants, with clones from R8 occurring in three of the four plants sampled in the July sampling period. The paucity of R8 types in the August sampling period suggests a decline in this group of beta-Proteobacteria later in the growing season. However, because of the small number of R8, no such change was detected in our ANOVAs.
S. arctica was most closely associated with three RFLP phylotypes (R1, R7, and R2), which all appear to be composed of nifH gene sequences from Proteobacteria. Clones that formed R1 occurred in the bulk soil and root aggregate samples. Sequenced clones of R1 were most similar to nifH genes from uncultured organisms, especially clone Langqian-7, a nifH clone from the Tibetan Plateau, China. Clones belonging to R7 were common in the root aggregate samples of S. arctica. A sequenced clone from R7 returned nifH clone CB893H27 from a marine surface sample at Chesapeake Bay, USA, as its phylogenetic nearest taxon (Table 2). Unlike clones from R1 and R7, clones from R2 were strongly associated with the surface sterilized root samples of S. arctica. One of the sequenced clones of R1 and some of the sequenced clones in R2 were most similar to Rhizobium-type nifH genes (Table 2). The remaining clones in R2 and R7 amplified a nifH gene sequence that was 386 nucleotides in length, in contrast to most other sequences that amplified a 383-bp product. In phylogenetic analysis, these grouped with the clones that formed R4.
Common throughout soil and root samples of all three plant species studied were clones belonging to phylotypes R4 and R3. All sequenced clones belonging to R4 were 386 nucleotides in length, and formed a monophyletic group with the 386-bp nifH clones from R2 and R7 in our phylogenetic analysis (Fig. 2). The 386-bp sequences differed from other type I sequences by the insertion of a threonine residue at the 111th codon position (330–333 bp). Figure 2 shows that these clones formed clade 4 within the type I nitrogenase genes. Sequences belonging to clade 4 were most similar to nifH genes from uncultivated organisms from extreme environments. These include a nifH gene obtained from a rhizosphere sample of a plant grown on nickel mine spoil [9]; however, majority of the sequences were most similar to nifH genes obtained from soil samples obtained in the Qing Zang Plateau of the Tibetan region of China (clones Qinglin-5, and Yushu-15; Table 2). This area has a mean elevation of over 4000 m and a mean January temperature of −50°C, suggesting that many of the organisms common to these high arctic plant rhizosphere soils are well adapted to extreme cold. However, the similarity of our clones with the Tibetan sequences was moderate, on the order of 84–85% (Table 2). When the nifH sequences of the Tibetan clones Qinglin-5 and Yushu-15 were included in our phylogenetic analysis they did not form a monophyletic group with our sequences (data not shown), indicating that this clade may represent a novel subcluster within the type I nitrogenases.
R3, which was nearly equal in size to R5 (Table 1), contained sequences with high similarity (96–98%) to the nifH gene of Paenibacillus polymyxa (Table 2). In phylogenetic analysis, sequenced clones belonging to R3 clustered tightly together and formed a monophyletic group with the nifH gene of P. durus. However, given the divergence of the R3 group from the Paenibacillus nifH sequence in the phylogenetic analysis (Fig. 2), our sequences likely represent nifH genes derived from a Firmicutes group, which is most similar to Paenibacillus but not a member of that genus. In our analysis, the Firmicutes nifH group was intermediate between nifH gene sequences from the cyanobacteria and those of the genus Frankia (Fig. 2), indicating the similarity of cyanobacterial nifH sequences to some members of the Firmicutes. Several other studies have reported close phylogenetic placement of these two groups (see [10, 11]).
Despite observing structures that appeared to be root nodules on D. integrifolia, we obtained no Frankia nifH gene sequences from any of our samples. Frankia sp., which is believed to form actinorhizal symbioses with members of the genus Dryas (clade III; [23]), has nodules with nonseptate vesicles and exhibits intercellular penetration of root hair cells. No isolates of this group have been obtained in pure culture [5, 23]. Although we are not able to rule out the possibility that our extraction protocol was ineffective at extracting Frankia DNA from the posited Dryas nodules, this seems unlikely as we ground root tissue to facilitate extraction of nodular DNA. Moreover, Frankia has been found in free-living form in northern forest soils, particularly those that are not extremely acidic and that have deciduous plant species [19, 20] and these are characteristics of our own study site. We were surprised to find no Frankia-type nifH genes in any soil fraction in this study, especially given that R3, which contains nifH gene sequences of posited Firmicutes, was the second largest phylotype.
Previously, we observed a poor relationship between nifH community structure and nitrogenase activity at this site. We found that as a consequence of their greater nifH diversity, subsurface diazotrophs accounted for the greatest change in nifH community structure due to warming, although surface-dwelling cyanobacteria were the most important contributors to nitrogenase activity. We also documented seasonal change in the belowground nifH community [6]. In the present study, we investigated nifH diversity belowground, and found that 21% of the “nifH” gene sequences probably did not encode functional nitrogenases at all, but rather nifH homologs belonging to the divergent type IV group. Moreover, R5, comprising type IV sequences, was the only phylotype found to be significantly more common in August than July, suggesting that these nifH homologs could be partly responsible for the seasonal succession we previously observed. NifH homologs complicate the issue of gene diversity and functional redundancy within ecosystems. NifH diversity was likely overestimated in our own and other previous studies that employed PCR, rather than sequencing-based approaches, in assessing nifH diversity.
We sought to determine which diazotrophs are commonly associated with roots, rhizospheres, and bulk soils of D. integrifolia, S. arctica, and C. tetragona, in order to identify the organisms that are important in the nitrogen nutrition of these long-lived plants. In general, nifH gene communities were composed of nondominant types, with the largest group accounting for only 21% of the total (Table 1). Figure 1 reveals that RFLP phylotypes were differentially associated with the three plant species and, in the case of S. arctica, these differences were sometimes driven by the soil or root fraction from which the clones were derived, suggesting that plants may select for different diazotrophic communities. It appears that members of the Firmicutes, which had nifH genes most similar to those of the genus Paenibacillus, were common throughout the samples and may be widespread throughout the dwarf-shrub cushion-plant community at Alexandra Fiord (Fig. 1). In this study, nifH genes from posited Pseudomonads were found to be more closely associated with C. tetragona than the other two plants species, whereas nifH genes from putative Proteobacteria, especially alpha-Proteobacteria, were more commonly associated with D. integrifolia and S. arctica.
Many of the nifH sequences generated in this study had relatively low sequence identity with known nifH sequences, which suggests a degree of uniqueness of this particular site but more likely reflects the limited investigation of nifH gene diversity in polar plant communities generally. nifH sequences generated in this study were often similar to nifH genes from other uncultivated organisms from environmental samples (Table 2). An interesting finding was the degree of similarity between nifH genes from the present study and those from other extremely cold environments including the Tibetan plateau in China. The finding that similar groups of functional genes exist in geographically isolated, but physically similar, locations lends support to the hypothesis that the distribution of nitrogen-fixing organisms can be predicted on the basis of habitat characteristics [25]. However, judging from the divergence of many of our sequences from known taxa, these predictions are still constrained by the need for baseline data on functional nitrogenase genes in un- or underinvestigated environments. There exists a need to predict and gauge biological response to environmental change in arctic latitudes. To our knowledge, this study is the first report on the nifH diversity of arctic plant-associated soil microbes.
References
Bateman, A, Cerruti, L, Durbin, R, Etwiller, L, Eddy, SR, Griffiths-Jones, S, Howe, KL, Marshall, M, Sonnhammer, ELL (2002) The Pfam protein families database. Nucleic Acids Res 30: 276–280
Chapin, FS, Jefferies, RL, Reynolds, JF, Shaver, GR, Svoboda, J (1992) Arctic plant physiological ecology: a challenge for the future. In: Chapin, FS, Jefferies, RL, Reynolds, JF, Shaver, GR (Eds.) Arctic Ecosystems in a Changing Climate: An Ecophysiological Perspective. Academic Press, San Diego, pp 3–8
Chapin, FS, Shaver, GR, Kedrowski, RA (1986) Environmental controls over carbon, nitrogen and phosphorus fractions in Eriophorum vaginatum in Alaskan tussock tundra. J Ecol 74: 167–195
Chein, Y-T, Zinder, SH (1996) Cloning, functional organization, transcript studies, and phylogenetic analysis of the complete nitrogenase structural genes (nifHDK2) and associated genes in the archeaon Methanosarcina barkeri 227. J Bacteriol 178: 143–148
Clawson, ML, Bourret, A, Benson, DR (2003) Assessing the phylogeny of Frankia-actinorhizal plant nitrogen-fixing root nodule symbioses with Frankia 16S rDNA and glutamine synthetase gene sequences. Mol Phylogenet Evol 31: 131–138
Deslippe, JR, Egger, KN, Henry, GHR (2005) Impacts of warming and fertilization on nitrogen-fixing microbial communities in the Canadian High Arctic. FEMS Microbiol Ecol 53: 41–50
Dice, LR (1945) Measures of the amount of ecologic association between species. Ecology 26: 297–302
Eckford, R, Cook, FD, Saul, D, Aislabie, J, Foght, J (2002) Free-living heterotrophic nitrogen-fixing bacteria isolated from fuel-contaminated Antarctic soils. Appl Environ Microbiol 68: 5181–5185
Hery, M, Phillippot, L, Meriaux, E, Poly, F, Le Roux, X, Navarro, E (2005) Nickel mine spoils revegetation attempts: effects of pioneer plants on two functional bacterial communities in the N-cycle. Environ Microbiol 7: 486–498
Hurek, T, Egener, T, Reinhold-Hurek, B (1997) Divergence in nitrogenases of Azoarcus spp., Proteobacteria of the β subclass. J Bacteriol 179: 4172–4178
Kessler, PS, McLarnan, J, Leigh, JA (1997) Nitrogenase phylogeny and the molybdenum dependence of nitrogen fixation in Methanococcus maripaludis. J Bacteriol 179: 541–543
Larimer, FW, Chain, P, Hauser, L, Lamerdin, J, Malfatti, S, Do, L, Land, ML, Pelletier, DA, Beatty, JT, Lang, AS, Tabita, FR, Gibson, JL, Hanson, TE, Bobst, C, Torres y Torres, JL, Peres, C, Harrison, FH, Gibson, J, Harwood, CS (2004) Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat Biotechnol 22: 55–61
Lifshitz, R, Kloepper, FW, Scher, FM, Tipping, EM, LaLiberte, M (1986) Nitrogen fixing Pseudomonads isolated from roots of plants grown in the Canadian High Arctic. Appl Environ Microbiol 51: 251–255
Mah, K, Tackaberry, LE, Egger KN, Massicotte, HB (2001) The impacts of broadcast burning on the diversity of ectomycorrhizal fungi associated with hybrid spruce seedlings in central British Columbia. Can J For Res 31: 224–235
Muc, M, Freedman, B, Svoboda, J (1989) Vascular plant communities of a polar oasis at Alexandra Fiord (79°N), Ellesmere Island. Can J Bot 67: 1126–1136
Posada, D, Crandall, KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14(9): 817–818
Raymond, J, Siefert, JL, Staples, CR, Blankenship, RE (2004) The natural history of nitrogen fixation. Mol Biol Evol 21: 541–554
Shaver, GR, Chapin, FS (1986) Effect of fertilizer on production and biomass of tussock tundra, Alaska, USA. Arct Antarct Alp Res18: 261–268
Smolander, A (1990) Frankia populations in soils under different tree species—with special emphasis on soils under Betula pendula. Plant Soil 121: 1–10
Smolander, A, Sundman, V (1987) Frankia in acid soils of forests devoid of actinorhizal plants. Physiol Plant 70: 297–303
StatSoft, Inc. (2002) STATISTICA (data analysis software system), version 6. www.statsoft.com
Van Wijk, MT, Clemmensen, KE, Shaver, GR, Williams, M, Callaghan, TV, Chapin, FS, Cornelissen, JHC, Gough, L, Hobbie, SE, Jonasson, S, Lee, JA, Michelsen, A, Press, MC, Richardson, SJ, Rueth, H (2004) Long-term ecosystem level experiments at Toolik Lake, Alaska, and at Abisko, Northern Sweden: generalizations and differences in ecosystem and plant type responses to global change. Glob Chang Biol 10: 105–123
Wall, LG (2000) The actinorhizal symbiosis. J Plant Growth Reg 19: 167–182
Young, J (1992) Phylogenetic classification of nitrogen-fixing organisms. In: Stacey, G, Burris, RH, Evans, HJ (Eds.) Biological Nitrogen Fixation. Chapman & Hall, New York, pp 43–87
Zehr, JP, Jenkins, BD, Short, SM, Steward, GF (2003) Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ Microbiol 5: 539–554
Acknowledgments
We thank Dr. Greg Henry for logistic support at the Alexandra Fiord site. We thank Mark Thompson of the UNBC sequencing facility for technical service and advice concerning phylogenetic analyses. This research was supported by funding from the Natural Sciences & Engineering Research Council of Canada to K.N.E., with additional logistic support from the Northern Scientific Training Program (Department of Indian and Northern Affairs Canada), the Polar Continental Shelf Project, and the Royal Canadian Mounted Police.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Deslippe, J.R., Egger, K.N. Molecular Diversity of nifH Genes from Bacteria Associated with High Arctic Dwarf Shrubs. Microb Ecol 51, 516–525 (2006). https://doi.org/10.1007/s00248-006-9070-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-006-9070-8