Abstract
The biological control agent (BCA) Pseudomonas fluorescens DR54 was applied to seeds (experiment 1) or roots (experiment 2) of barley growing in microcosms, while noninoculated plants served as controls. The fate of the BCA and its effects on the rhizosphere microbial community was evaluated in microcosms destructively sampled at days 2, 4, 6, and 9 after inoculation. In both experiments the number of P. fluorescens DR54 cells decreased immediately after application as enumerated by immunostaining and microscope direct counting. Substrate-induced respiration (SIR) was taken as a measurement of the active microbial biomass, while indicators of the total microbiota (and main taxonomic groups) were obtained using the phospholipid fatty acid (PLFA) technique. In experiment 1, these parameters were unaffected by the relatively small number of BCA cells applied, whereas in experiment 2, the larger BCA input resulted in an enhanced level of both SIR and PLFAs from Gram-negative bacteria (which included the BCA itself). However, at day 9 after inoculation, treatments with P. fluorescens DR54 and controls were similar in all measured parameters in both experiments. This was also illustrated very clearly by principal component analysis of the PLFA data, which in both experiments were able to discriminate between treatments in the first days after BCA inoculation, thus confirming the sensitivity of this method. Laccase activity has a potential as an indicator of fungal stress, e.g., when challenged with an antifungal BCA. This seemed to be supported in experiment 2, where the activity of this enzyme was enhanced four-fold in the BCA treatment at day 2. Our study shows that under the present conditions, P. fluorescens DR54 disappears from the soil and causes only transient effects on the soil microbiota. It also shows that the PLFA technique is a sensitive and reliable monitoring tool in in situ assessment of BCA nontarget effect on indigenous microorganisms in soil.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Biological control agents (BCAs) have become a promising alternative to chemical pesticides for disease control in crop plants [36]. This development will supposedly accelerate with the increasing pressure by environmental authorities and public opinion to diminish the use of chemical pesticides that are often hazardous to the environment and human health. Numerous bacterial and fungal isolates have been tested in the laboratory for their potential inhibitory effects as BCAs toward a range of plant pathogenic microorganisms [6, 34]. From a crop-protection point of view, there is a wish to isolate (or modify genetically) BCAs that satisfy two main requirements. They should be effective in inhibiting plant pathogens in situ and they should be able to stay in the environment long enough to serve as an effective pathogen control. However, other microorganisms than the target organisms may be affected by the BCA, thereby changing the taxonomic and/or functional structure of the indigenous microbial community in the soil. This, in turn, could be detrimental to biological processes in the soil that are vital to plant growth.
Typically, BCAs produce substances that are toxic to the pathogen, e.g. 2,4-diacetylphloroglucinol [23] or viscosinamide [31], but they may also extrude hydrolytic enzymes [7], siderophores [26], and other substances that may suppress disease. In most cases, BCAs decline in numbers quite quickly when introduced into soil [40, 49] and the microbial community returns to its original size and composition within a short time. Even so, there is a risk that BCAs may change the nontarget microbial community in a soil and thereby affect its functional properties. Hence, there is also a need for developing reliable standard methods that can be used by environmental authorities to assess the risk of new BCAs in future approval procedures.
Possible effects of BCAs on the soil microbiota may be estimated by measuring changes in parameters that are characteristic for functional processes (e.g., cellulase activity, respiration, and nitrification) and/or by monitoring changes in the size and composition of the total microbial community or in distinct groups (e.g., pseudomonads or saprophytic fungi). This may be done by classical cultivation-dependent methods or by DNA fingerprinting methods such as community PCR with TGGE/DGGE analysis [21] or community DNA hybridization [16, 25]. Phospholipid fatty acid (PLFA) analysis can be used to detect changes in the structure of microbial communities [45], and although several fatty acids are specific to microbial main taxa, the PLFA technique is broad-scaled, picturing perturbations in the total microbial community structure after changes in growth conditions [52]. It has, for example, been used to demonstrate effects of heavy metals [9, 12] and substrate amendment [17] on the composition of the microbial community in soil. The PLFA technique has the advantage over DNA fingerprinting techniques that analysis is performed on microbial lipids extracted directly from the soil and is therefore considered a measurement of the in situ situation in the soil.
The aim of this work was to measure possible nontarget effects on the native soil microbial community after release of the antifungal BCA P. fluorescens DR54. The BCA was introduced into the spermosphere or rhizosphere of barley, in each of two experiments, and the effects on the composition and size of the soil microbial community was measured, using techniques that would allow us to describe the in situ situation. The fate of the BCA was monitored using a fluorescent antibody (FA) technique, while the size of the total active microbial community was measured by substrate-induced respiration (SIR). Perturbations in the soil microbial community were measured, with the PLFA technique, as changes in the total soil phospholipid fatty acid profiles as well as actual levels of fatty acids characteristic for specific microbial groups. Fungal response to P. fluorescens DR54 has recently been shown to induce laccase production in Rhizoctonia solani [8]. This is maybe a defense mechanism to overcome lipid peroxidative stress [11] caused by surfactants, such as viscosinamide, in P. fluorescens DR54 [8]. Hence, laccase activity was measured as a possible indicator of fungal defense response when confronted with antagonists producing biosurfactants.
Methods
BCA
The antifungal antagonist P. fluorescens DR54 (wild-type) was used because it is considered a biological control agent (BCA). The strain was originally isolated from sugar-beet rhizosphere in our laboratory [30], and it inhibits growth of the plant-pathogenic fungi Rhizoctonia solani and Pythium ultimum under both in vitro and in situ conditions [31, 42, 43]. The cyclic depsipeptide viscosinamide, produced by P. fluorescens DR54, has biosurfactant properties and is believed to play a role in the growth inhibition, as deduced from changes in morphology and physiology in various microfungi [8, 31, 41].
Experiment 1
Hordeum vulgare (barley, cv. Lamba) seeds were germinated to a root length of <1 mm. Fifty growth units (glass tubes, 4 cm dia.) were filled with 145 g moist soil (a clay loam from a conventionally managed field at Snuppekorsgård in Denmark, Table 1) passed through a 4-mm sieve. Four seeds were placed on the surface of the soil in each growth unit and 20 μL of a P. fluorescens DR54 suspension, containing a total of 5 × 108 cells, was supplied to each seed in half of the growth units. The cells originated from a P. fluorescens DR54 batch culture (agitated, 20°C), grown to early stationary phase in LB medium [38]. They were washed once and resuspended in spent medium, deprived of cells by centrifugation, and filtered though a 0.22-μm polycarbonate filter. The seeds in the remaining growth units received 20 μL spent medium without P. fluorescens DR54 cells to serve as controls. This procedure ensured that effects observed in the P. fluorescens DR54 treatment were caused by the cells and not by compounds dissolved in the spent medium. Immediately after inoculation, the seeds were covered with 5 g of sieved soil, and Milli-Q water (Millipore, Bedford, MA, USA) was added to a final water content of 75% of the water holding capacity. The glass tubes were covered with nontransparent foil and placed in a growth cabinet at a 16/8 h light/dark cycle and a temperature of 15°C. The plants were watered to weight daily. Growth units were harvested destructively (five replicates per treatment) at 2, 4, 6, and 9 days after sowing/inoculation. Roots were collected and shaken so that only closely attached soil (1–3 mm around the roots) was retained and defined as rhizosphere soil.
Experiment 2
Experimental conditions were the same as in experiment 1 regarding experimental design and biological material, growth units, and growth of P. fluorescens DR54. However, the barley seeds were pregerminated to a root length of ∼5 cm before the roots were coated with P. fluorescens DR54 by submerging (30 s) in a suspension of ∼5 × 109 cells mL−1. The cells were washed once and resuspended in 25 mM phosphate-buffered saline (PBS, pH 7.4). Controls were coated with sterile PBS only. The soil used was a sandy loam from a conventionally managed field at Højbakkegård in Denmark (Table 1). Conditions for growth, harvest events, sampling, and analyses were the same as in experiment 1, except that laccase activity was additionally measured.
Immunodetection of P. fluorescens DR54
Root pieces, with ∼300 mg soil adhering, were sampled uniformly from the root system (so that root sections from the entire root system were represented in approximately equal proportions) and placed in test tubes together with 5 mL of 25 mM PBS (pH 7.4) and 0.5 g sterile quartz sand. After 10 min of vortexing and 30 s of sedimentation, 0.75 mL of the suspension was transferred to Eppendorf tubes, supplied with formaldehyde to a final concentration of 1%, and stored at 1°C until immunostaining. Diluted samples were filtered onto poly-L-lysine coated polycarbonate filters (black 0.22 μm, Poretics, Livermore, CA, USA) using a filtration manifold (Carbon14 Agency, Hørsholm, DK). The filtered samples were blocked (20 min) in Milli-Q water with 0.05 M Tris-HCl pH 7.4, 0.15 M NaCl, 2% Tween, and 1 g BSA L−1, washed twice (5 min) in Milli-Q water with 0.05 M Tris-HCl pH 7.4, 0.15 M NaCl, 0.05% Tween, and 0.5 g BSA L−1, and incubated with the primary P. fluorescens DR54 specific antibody (overnight) in a 1:500 dilution in blocking solution with only 0.05% Tween. The P. fluorescens DR54 specific antibody was raised in rabbits and cross-reactions tested (and removed by absorption) against a panel of common soil bacteria following the procedure of [19]. After two washings, the filters were incubated with the secondary FITC-swine-anti-rabbit antibody (DakoCytomation A/S, Glostrup, Denmark) for 2 h and washed twice. Finally, the filtered samples were DAPI (4′,6-diamidino-2-phenylindole) stained (1 mg DAPI L−1, 10 min), washed once in Milli-Q water, air dried, and mounted between a slide and a coverslip in Vectashield antifading mounting oil (Vector Lab, Peterborough, UK). All washing and incubation procedures were performed in six-well culture clusters (Costar, Corning Inc., Corning, NY, USA). Filters (in duplicates of two), from all samples in the P. fluorescens DR54 treatment and the first harvest in the control treatment, were produced as described above and monitored in a Zeiss Axioskop epifluorescence microscope at 1000× magnification. A minimum of 200 labeled cells, or alternatively the number of labeled cells in 30 fields, was counted. Cross-reactions in the experimental soils were practically absent, as checked in control samples not supplied with P. fluorescens DR54.
Substrate-Induced Respiration
In order to estimate the size of the total microbial biomass in the rhizosphere, a substrate-induced respiration (SIR) assay [1] was performed on 300-mg portions of rhizosphere soil carefully removed from the roots using a fine pair of forceps. The soil was transferred to 3-mL Venoject tubes and 3 mg of a mixture of glucose and talcum (4:1, w:w) was incorporated into the soil (as modified after [2]). After capping and incubation for 30 min, the tubes were flushed for 1 min with a technical gas (79% N2 and 21% O2) to remove CO2. The tubes were incubated in the dark and the headspace gas sampled 1.5 and 3 h after flushing and analyzed for its concentration of respired CO2. Analyses were performed on a Hewlett Packard 6890 GC equipped with a Porapak-Q column and a thermal conductivity detector at an oven temperature of 30°C. The concentration of CO2 in the headspace was determined by comparing the obtained areas to a CO2 standard curve.
Laccase Assay
One g of rhizosphere soil was added to 375 μl 1 M Na-acetate buffer (pH 5.6) in 10-ml polypropylene centrifuge tubes (Ole Dick). The tubes were capped, sonicated (1 min), and agitated gently in 3 h at room temperature. After centrifugation (10,000 g, 15 min), 200 μL of the supernatant was transferred to cuvettes and mixed with 750 mL of a 0.1 M Britton and Robinson buffer (pH 5.0) [50] and 50 μL 20 mM ABTS (2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid)–diammonium salt, Sigma). The laccase activity was measured spectrophotometrically as the increase in the absorbance at 405 nm (over 12 h at 25°C) that followed the oxidation of ABTS by laccase. The ABTS substrate used in the assay can also be converted by peroxidases [18], which depend on H2O2 to show any activity. However, including catalase in the assay (to remove H2O2) resulted in a decrease of no more than 40% in the oxidation of ABTS, confirming that most of the enzyme activity measured was caused by laccases.
Phospholipid Fatty Acid Analysis
The procedure for extraction of phospholipid fatty acids (PLFA) was modified after Frostegård et al. [12]. Root pieces with adhered soil were placed in Teflon centrifuge tubes (Oak Ridge, Nalge Nunc Int.) with 6 mL citrate buffer (0.15 M, pH 4.0), agitated for 10 min, and the root pieces removed. Great care was taken to ensure that no root pieces were left in the tubes. The tubes were weighed before and after this procedure to allow measurement of the amount of soil (2–4 g) left in the tubes. The weights were adjusted for the amount of liquid removed together with the root material. The procedure for extraction, purification, and derivatization of fatty acids is described in detail in [40], while analysis equipment and its settings are described in [22]. Identification of fatty acids was performed as described by [12] and by comparison to the retention times of known fatty acid. Nomenclature of fatty acids is described by Tunlid and White [45]. Attempts were made to estimate proportions of the following main taxa of the microbiota in samples by using PLFAs indicative of these groups: Gram-positive bacteria (i15:0, a15:0, i16:0, i17:0, a17:0) [33], Gram-negative bacteria (18:1w7, cy17:0, cy19:0) [51], fungi (18:2w6,9) [10], and actinomycetes (10me18:0) [24].
Statistical Methods
Data are presented as means together with their standard error (SE) of the mean. Principal component analysis was performed on the entire PLFA data set (unless otherwise noted) after log10 transformation using Unscrambler software (CAMO ASA, Norway). P. fluorescens DR54 direct count (log10 transformed), SIR, laccase, and PLFA concentration data were statistically treated using the GLM procedure and Tukey two-way comparison in SYSTAT 9.0.
Results and Discussion
In the present work we studied the behavior of a Pseudomonas BCA under controlled environmental conditions, in order to predict its fate and impact on the indigenous microbial microbiota in a barley rhizosphere. The methods chosen for the study appeared appropriate to enumerate the introduced BCA and to detect the resulting perturbations in size and structure of the microbial community. In contrast to classical cultivation-dependent method or DNA fingerprinting, the immunofluorescence, SIR, and PLFA methods involve no cultivation or amplification steps (which often induce methodological bias) in an effort to obtain a reliable picture of the in situ situation in the soil.
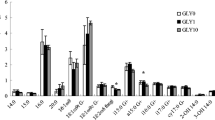
The number of P. fluorescens DR54 detected in the rhizosphere in experiment 1 (Fig. 1a) decreased by ∼50% between days 2 and 4 (P = 0.02) and remained stable until day 9. When P. fluorescens DR54 was coated onto the roots in experiment 2 (Fig. 1b), the initial cell density was ∼100 times higher than in experiment 1, but decreased gradually to only 1 % of initial cell density at the last harvest. In the first experiment the cells were applied only to the seeds, and if root colonization was not efficient, the decline in number of cells may partially be due to a dilution effect because soil from the entire root system was included in the assay. However, it was evident in both experiments that the cells became gradually smaller and lost their integrity during the 9 days, as judged from visual inspection in the microscope. A traditional plating technique would have yielded a lower number of cells and failed detection of any nonculturable cells. Information about this group of cells may be important in case the compound responsible for the antagonism is bound to the cells, like viscosimamide in P. fluorescens DR54 [31], and released predominantly after cell death. However, we cannot preclude from the present data that a low number of viable P. fluorescens DR54 cells could remain in the soil for a longer time than the actual 9 days. The die-off rate was clearly higher in the Højbakkegård soil than in the Snuppekorsgård soil (experiments 2 and 1, respectively), perhaps because of its low clay content, leaving only a few microsites that could protect the inoculant against protozoan grazing [35, 47]. In most cases, when bacteria are introduced into nonsterile soil, their survival rate is poor [49]. This may be due to bad adaptation to the new environment combined with strong competition from the indigenous microorganisms [35]. In a recent work [32], the fate of P. fluorescens DR54 was followed, using a green fluorescent protein (GFP) transformant introduced into the rhizosphere of barley. In accordance with the results in experiment 2, the total number of GFP-positive cells decreased by about two log units within the first 2 weeks and the proportion of culturable cells was even less, especially in the first few days (∼10%). In a long-term experiment with P. fluorescens DR54 [40], CPUs declined to ∼1% within 2 weeks, whereafter the die-off rate seemed more moderate during the next month, leaving about 105 P. fluorescens DR54 cells g−1 dry rhizosphere soil. Similar results have been obtained with the BCA P. fluorescens CHA0 [20] and other fluorescent pseudomonads [27, 48]. Bacillus subtilis BCA survival was also poor in three soils [5], although sterilizing the soil highly enhanced its survival, demonstrating the importance of the interactions with the indigenous soil microbiota.
Recovery of biological control agent Pseudomonas fluorescens DR54 in barley rhizosphere soil. P. fluorescens DR54 was supplied at 5 × 108 cells per seed in experiment 1 (a) or as a dense suspension to 5-cm-long seedling roots in experiment 2 (b). Controls were noninoculated. Total microbial biomass C (measured as SIR), total microbial PLFA, and Gram-negative PLFA (c, e, g and d, f, h in experiments 1 and 2, respectively) were likewise measured (bars indicate SE, n = 5; asterisk indicates significant difference between treatments at the P = 0.05 level).
SIR is supposed to reflect the size of the active microbiota in the soil [1, 28]. When viable bacteria are supplied to soil they contribute directly to the respiration, or more indirectly, by supporting new growth of the resident microbial community by serving as a substrate source. In experiment 2 (Fig. 1d), P. fluorescens DR54 more than doubled the SIR 2 days after introduction, compared to the control (P = 0.001), suggesting that a considerable proportion of the P. fluorescens DR54 cells was actually live and metabolically active 2 days after introduction. However, as the SIR from the P. fluorescens DR54 treatments and the control were similar at day 9, eventual growth of indigenous microorganisms feeding on P. fluorescens DR54 seemed insignificant. This further suggests that all P. fluorescens DR54 cells were not disappearing immediately after dying, but persisted for some time in the soil before lysis. The SIR increased about 20% (P = 0.04) between the first two harvests in experiment 1 (Fig. 1c), but was unaffected by P. fluorescens DR54, probably because the respiration of the small number of introduced cells could not be detected as a difference in SIR between the treatments. Likewise, in an experiment with field-grown wheat [15], coating the seeds with 107 cells of P. putida (genetically modified to produce the antifungal compound phenazine-1-carboxylic acid) had no effect on the SIR, although the population of culturable fungi was affected negatively.
The amount of PLFA from both the total microbial community of the Gram-negative bacteria in experiments 1 (Figs. 1e and g, respectively) and 2 (Figs. 1 f and h , respectively) was not affected by P. fluorescens DR54 in any of the harvests. This was also true for other main microbial groups, such as Gram-positive bacteria, fungi, and actinomycetes (data not shown). An exception was in experiment 2, where the amount of PLFA from Gram-negative bacteria was increased by ∼40% in the start of the experiment (P = 0.04). So with the PLFA method, it was possible to directly detect the introduced P. fluorescens DR54 cells and their decline over time, as was also demonstrated using the immunodetection and SIR methods. Similarly, the most prominent PLFA in a Flavobacterium balustinum was elevated in cucumber rhizosphere following seed coating [46]. The amount of P. fluorescens DR54 applied to the soil in experiment 1 escaped PLFA detection due to the low number of cells.
Using the approach of Anderson and Domsch [2] the measured SIR values (both experiments) were used to estimate the microbial biomass C level to range from 1500 to 4000 μg microbial C g−1 dry soil. The level of total PLFA ranged from 100 to 300 nmol g−1 dry soil, with no consistent difference between the experiments or treatments. From the total PLFA data set we estimated the average number of C atoms in fatty acids to be close to 17. Hence, the PLFA contained from 2.4 to 4.3 μmol fatty-acid C g−1 dry soil, equivalent to 20.4 and 61.2 μg fatty-acid C g−1 dry soil. Thus, the relation between the content of C in microbial biomass and C in PLFAs shows that 1–2% of the microbial biomass C was found in membrane lipids. This is in accordance with previously reported values [4, E. Bååth pers. comm.] and indicates a good correspondence between the SIR and the PLFA estimates.
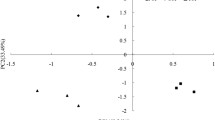
The distribution of the treatments/harvests in the score plots in the PCA of the PLFAs gave information about how the antagonist and natural succession affected the microbial community structure. In experiment 1, scores from the first two harvests ordinated to the left in the score plot (Fig. 2a) along PC1, while the last two harvests were located to the right, a pattern similar for both the control and the P. fluorescens DR54 treatment. In contrast, the P. fluorescens DR54 scores separated from the control scores along the PC2 axis in the first two harvests. In the last two harvests the treatments were placed together, although the two sampling times separated. Hence, succession during time (represented by PC1 = 49%) was the factor responsible for most of the variation in the microbial community. P. fluorescens DR54 (represented by PC2 = 21%) influenced the structure of the microbial community only in the first few days after introduction—parallel to the decline in population size and the possible dilution effect discussed above. The situation in experiment 2 (Fig. 3a) resembled that in experiment 1, although the distance between the two treatments was much larger in the first harvest, reflecting the relatively larger input of P. fluorescens DR54. Also, this distance in harvest 1 and 2 was along PC1, showing that in the first days after inoculation, P. fluorescens DR54 was the main factor contributing to the variation in the structure of the microbial community. The PLFA profiles in the two treatments were apparently quite similar at days 6 and 9, suggesting that the effect of P. fluorescens DR54 was quickly fading out. It is also clear from the loading plot in experiment 2 (Fig. 3b) that P. fluorescens DR54 itself was directly causing a substantial part of the variation. The PLFAs 16:0, 16:1w7, 18:1w7, cy17:0, cy19:0, and 18:0, constituting the absolute main part of the fatty acids found in the membrane of axenically grown P. fluorescens DR54 (and normally found in Gram-negatives [51]), were clustering in the lower left corner of the loading plot—at the same position where the P. fluorescens DR54 score from day 2 was located.
Scatter plot of the score (a) and loading (b) values of the first two principal components in a principal component analysis of the relative molar abundance of fatty acids originating from the phospholipids in the total microbial community in barley rhizosphere. The seeds were either supplied (□) or not (○) with the biological control agent Pseudomonas fluorescens DR54 at 5 × 108 cells. The day after inoculation is shown in the individual markers. Additional plots show score (c) and loading (d) values calculated without the PLFAs found in the P. fluorescens DR54 cells. Each data point represents the average of the coordinate values within each treatment at the individual sampling events (experiment 1, bars indicate SE, n = 5). The circle encompass the following fatty acids in (b): i16:0, i17:0, 10me16:0, 10me17:0, 10me18:0, br18:0, cy17:0, cy19:0, and in (d): i16:0, i17:0, 10me16:0, 10me17:0, 10me18:0, br18:0.
Scatter plot of the score (a) and loading (b) values of the first two principal components in a principal component analysis of the relative molar abundance of fatty acids originating from the phospholipids of the total microbial community in barley rhizosphere. Five-cm-long roots of seedlings were either supplied (□) or not (○) with a dense suspension of the biological control agent Pseudomonas fluorescens DR54. The day after inoculation is shown in the individual markers. Additional plots show score (c) and loading (d) values calculated without the PLFAs found in the P. fluorescens DR54 cells. Each data point represents the average of the coordinate values within each treatment at the individual sampling events (experiment 1, bars indicate SE, n = 5). The circle encompass the following fatty acids in (b) and (d): i15:0, i16:0, i17:0, a15:0, 10me16:0, 10me18:0, br18:0, 16:1w5, 17:1w8.
Using a PCA to depict differences in microbial community structure in the two treatments could reflect that fatty acids from P. fluorescens DR54 itself was present in only one of the treatments. However, excluding the PLFAs from P. fluorescens DR54 from the PCA calculation changed the ordination pattern very little (experiments 1 and 2; Figs. 2c and 3c, respectively), suggesting that differences in the structure of the indigenous microbial community did actually occur. In another study [40], significant but transient effects of P. fluorescens DR54 on microbial community structure were likewise detected with the PLFA technique, whereas with PCR-DGGE and BIOLOG techniques no disturbance was seen. Community changes were observed, using BIOLOG and a fatty acid technique based on whole-cell lipid content, after introduction of a genetically engineered P. chlororaphis BCA, but only for as long as the strain could be detected on selective media [13]. Part of such changes may be due to displacement of bacteria that are closely related to the introduced MCA and occupying the same ecological niche [29, 39]. The action of most BCA are directed toward fungi, and a few studies have focused on nontarget effects on the soil fungal community [3, 14] finding only short-term effects as judged from ARDRA and species diversity index.
Fungi commonly produce laccases [44] which are enzymes that, beside taking part in the conversion of phenolic compounds, may also degrade compounds toxic to the fungus [11] or change the permeability of the cell wall by oxidative cross-linking [37]. Crowe and Olsson [8] demonstrated that laccases were induced in R. solani mycelium in response to stress treatments aimed at the membrane integrity, including the presence of P. fluorescens DR54. In experiment 2, laccase activity in the rhizosphere soil was measured as a possible indicator of fungal stress in vivo when confronted with P. fluorescens DR54. This seemed to be the case in the first harvest after introduction of the BCA as the laccase activity was much higher (P = 0.001) in the P. fluorescens DR54 treatment than in the control (Fig. 4). To minimize the possibility that the laccase activity was from the roots or from the BCA itself, laccase activity from sterile barley roots and P. fluorescens DR54 was measured, without finding any (data not shown). In case the roots were induced only under in vivo growth conditions, laccase activity would probably also have been enhanced in the controls. Hence, we find it plausible that fungi in the rhizosphere reacted to P. fluorescens DR54 by producing laccase. Although our results may point to laccase activity as a way to measure some aspects of fungal stress under in vivo conditions, more studies are needed to establish this more thoroughly.
Laccase activity in rhizosphere soil of barley supplied (DR54) or not (Control) with the biological control agent Pseudomonas fluorescens DR54. The cells were supplied to 5-cm-long roots of the seedlings as a dense suspension (experiment 2, bars indicate SE, n = 5; asterisk indicates significant difference between treatments at the P = 0.05 level).
In summary, our results indicate that introducing the BCA P. fluorescens DR54 into soil represents no significant risk to the indigenous microbiota. Even at high densities (as in experiment 2), the BCA survival rate was poor and the impacts on the other microbial organisms minor. Nevertheless, with principal component analysis it was possible to show that the observed difference in the PLFA profiles represented both the presence (and decline) of the antagonist itself and the perturbations of the resident microbial community. The PLFA profiles in the BCA and control treatments were no longer discernible 6–9 days after inoculation of the BCA and confirmed together with the SIR and laccase data that the effects of the BCA were only short-term. Our results confirm the sensitivity and usefulness of the PLFA technique to detect even small changes in the structure of microbial populations in the environment.
References
JPE Anderson KH Domsch (1978) ArticleTitleA physiological method for the quantitative measurement of microbial biomass in soils Soil Biol Biochem 10 215–221 Occurrence Handle10.1016/0038-0717(78)90099-8
JPE Anderson KH Domsch (1985) ArticleTitleMaintenance carbon requirements of actively-metabolizing populations under in situ conditions Soil Biol Biochem 17 197–203 Occurrence Handle10.1016/0038-0717(85)90115-4
PAHM Bakker DCM Glandorf M Viebahn TWM Ouwens E Smit P Leeflang K Wernar LS Thomashow JE Thomas-Oate LC Loon Particlevan (2002) ArticleTitleEffects of Pseudomonas putida modified to produce phenazine-1-carboxylic acid and 2,4-diacetylphloroglucinol on the microflora of field grown wheat A Van Leeuw J Microb 81 617–624 Occurrence Handle10.1023/A:1020526126283
DL Balkwill FR Leach JT Wilson JF McNabb DC White (1988) ArticleTitleEquivalence of microbial biomass measures based on membrane lipid and cell wall components, adenosine triphosphate, and direct counts in subsurface aquifer sediments Microb Ecol 16 73–84
AJ Bennet C Leifert JM Whipps (2003) ArticleTitleSurvival of the biocontrol agents Coniothyrium minitants and Bacillus subtilis MBI 600 introduced into pasteurised, sterilised and non-sterile soil Soil Biol Biochem 35 1565–1573 Occurrence Handle10.1016/j.soilbio.2003.08.001
GJ Boland LD Kuykendall (1998) Plant–Microbe Interactions and Biological Control Marcel Dekker New York
K Broglie R Broglie N Benhamou I Chet (1993) The role of cell wall degrading enzymes in fungal disease resistance I Chet (Eds) Biotechnology in Plant Disease Control Wiley-Liss New York 139–156
JD Crowe S Olsson (2001) ArticleTitleInduction of laccase activity in Rhizoctonia solani by antagonistic Pseudomonas fluorescens strains and a range of chemical treatments Appl Environ Microbiol 67 2088–2094 Occurrence Handle10.1128/AEM.67.5.2088-2094.2001 Occurrence Handle11319086
F Ekelund S Olsson A Johansen (2003) ArticleTitleChanges in the succession and diversity of protozoan and microbial populations in soil spiked with a range of copper concentrations Soil Biol Biochem 35 1507–1516 Occurrence Handle10.1016/S0038-0717(03)00249-9
TW Federle (1986) Microbial distribution in soil—new techniques F Meguar M Gantar (Eds) Perspectives in Microbial Biology Slovene Society for Microbiology Ljubljana, Slovenia 493–498
J Fernández-Larrea U Stahl (1996) ArticleTitleIsolation and characterization of a laccase gene from Podospora anserina Mol Gen Genet 252 539–551 Occurrence Handle10.1007/s004380050261 Occurrence Handle8914515
Å Frostegård A Tunlid E Bååth (1993) ArticleTitlePhospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals Appl Environ Microbiol 59 3605–3617
JV Gagliardi JS Buyer JS Angle E Russek-Cohen (2001) ArticleTitleStructural and functional analysis of whole-soil microbial communities for risk and efficacy testing following microbial inoculation of wheat roots in diverse soils Soil Biol Biochem 33 25–40 Occurrence Handle10.1016/S0038-0717(00)00110-3
M Girlanda S Perotto Y Mönne-Loccoz R Bergero A Lazzari G Défago P Bonfante AM Luppi (2001) ArticleTitleImpact of biocontrol Pseudomonas fluorescens CHA0 and a genetically modified derivative on the diversity of culturable fungi in the cucumber rhizosphere Appl Environ Microbiol 67 1851–1864 Occurrence Handle10.1128/AEM.67.4.1851-1864.2001 Occurrence Handle11282643
DCM Glandorf P Verheggen T Jansen J-W Jorritsma E Smit E Leeflang K Wernars LS Thomashow E Laureijs JE Thomas-Oates PAHM Bakker LC Loon Particlevan (2001) ArticleTitleEffect of genetically modified Pseudomonas putida WCS358r on the fungal rhizosphere microflora of field-grown wheat Appl Environ Microbiol 67 3371–3378 Occurrence Handle10.1128/AEM.67.8.3371-3378.2001 Occurrence Handle11472906
BS Griffiths K Ritz N Ebblewhite E Paterson K Killham (1998) ArticleTitleRye grass rhizosphere microbial community structure under ambient and elevated CO2 concentrations, with observations on wheat rhizosphere Soil Biol Biochem 30 315–321 Occurrence Handle10.1016/S0038-0717(97)00133-8
BS Griffiths K Ritz N Ebblewhite G Dobson (1999) ArticleTitleSoil microbial community structure: effects of substrate loading rates Soil Biol Biochem 31 145–153 Occurrence Handle10.1016/S0038-0717(98)00117-5
Halliwell, B, Gutteridge, JMC (1999) Free Radicals in Biology and Medicine, 3rd ed. Oxford Scientific Publications
M Hansen L Kragelund O Nybroe J Sørensen (1997) ArticleTitleEarly colonization of barley roots by Pseudomonas fluorescens studied by immunofluorescence technique and confocal laser scanning microscopy FEMS Microbiol Ecol 23 353–360 Occurrence Handle10.1016/S0168-6496(97)00037-8
C Hase F Marscher Y Moënne-Loccoz G Défago (1999) ArticleTitleNutrient deprivation and the subsequent survival of biocontrol Pseudomonas fluorescens CHA0 in soil Soil Biol Biochem 31 1181–1188 Occurrence Handle10.1016/S0038-0717(99)00036-X
H Heuer K Smalla (1997) Application of denaturing gradient gel electrophoresis for studying soil microbial communities JD Elsas Particlevan JT Trevors EMH Wellington (Eds) Modern Soil Microbiology Marcel Dekker New York 353–374
EJ Joner A Johansen AP Loibner MA Cruz Particlede la OH Szolar JM Portal C Leyval (2001) ArticleTitleRhizosphere effects on microbial community structure and dissipation and toxicity of polycyclic aromatic hydrocarbons (PAHs) in spiked soil Environ Sci Technol 35 2773–2777 Occurrence Handle10.1021/es000288s Occurrence Handle11452608
C Keel U Schnider M Maurhofer C Voisard J Laville U Burger P Wirthner D Haas G Défago (1992) ArticleTitleSuppression of root diseases by Pseudomonas fluorescens CHA0: importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol Mol Plant–Microbe Interact 5 4–13
RM Kroppenstedt (1985) Fatty acid and menaquinone analysis of actinomycetes and related organisms M Goodfellow DE Minnikin (Eds) Chemical Methods in Bacterial Systematics Academic Press London 173–199
S Lee JA Fuhrman (1990) ArticleTitleDNA hybridization to compare species composition of natural bacterioplancton assemblages Appl Environ Microbiol 56 739–746 Occurrence Handle2317044
JE Loper JS Buyer (1991) ArticleTitleSiderophore in microbial interactions on plant surfaces Mol Plant–Microbe Interact 4 5–13
PS Lübeck M Hansen J Sørensen (2000) ArticleTitleSimultaneous detection of the establishment of seed-inoculated Pseudomonas fluorescens strain DR54 and native soil bacteria on sugar beet root surfaces using fluorescent antibody and in situ hybridization technique FEMS Microbiol Ecol 33 11–19 Occurrence Handle10.1016/S0168-6496(00)00038-6 Occurrence Handle10922498
H Marstorp E Witter (1999) ArticleTitleExtractable dsDNA and product formation as measures of microbial growth in soil upon substrate addition Soil Biol Biochem 31 1443–1453 Occurrence Handle10.1016/S0038-0717(99)00065-6
DC Naseby JM Lynch (1998) ArticleTitleImpact of wild-type and genetically modified Pseudomonas fluorescens on soil enzyme activity and microbial population structure in the rhizosphere soil Mol Ecol 7 617–625 Occurrence Handle10.1046/j.1365-294x.1998.00367.x
TH Nielsen J Sørensen J Fels HC Pedersen (1998) ArticleTitleSecondary metabolite- and endochitinase-dependent antagonism toward plant-pathogenic microfungi of Pseudomonas fluorescens isolates from sugar beet rhizosphere Appl Environ Microbiol 64 3563–3569 Occurrence Handle9758768
TH Nielsen C Christophersen U Anthoni J Sørensen (1999) ArticleTitleViscosinamide, a new cyclic depsipeptide with surfactant and antifungal properties produced by Pseudomonas fluorescens DR54 J Appl Microbiol 86 80–90 Occurrence Handle10.1046/j.1365-2672.1999.00798.x
B Normander NB Hendriksen O Nybroe (1999) ArticleTitleGreen fluorescent protein-marked Pseudomonas fluorescens: localization, viability, and activity in the natural barley rhizosphere Appl Environ Microbiol 65 4646–4651 Occurrence Handle10508101
WM O’Leary SG Wilkinson (1988) Gram-positive bacteria C Ratledge SG Wilkinson (Eds) Microbial Lipids, vol 1 Academic Press London 117–201
DJ O’Sullivan F O’Gara (1992) ArticleTitleTraits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens Microbiol Rev 56 662–676 Occurrence Handle1480114
J Postma CH Hok-A-Hin JA Veen Particlevan (1990) ArticleTitleRole of microniches in protecting introduced Rhizobium leguminosarum biovar trifolii against competition and predation in soil Appl Environ Microbiol 56 495–502
PL Pusey (1996) Micro-organisms as agents in plant disease control LG Copping (Eds) Crop Protection Agents from Nature: Natural Products and Analogues. Crit Rep Appl Chem, vol 35 The Royal Society of Chemistry Cambridge, UK 426–436
ADM Rayner ZR Watkins JR Beeching (1999) Self-integration—an emerging concept from the fungal mycelium NAR Gow G Robson G Gadd (Eds) The Fungal Colony Cambridge University Press London
J Sambrook EF Fritsch T Maniatis (1989) Molecular Cloning: A Laboratory Manual Cold Spring Harbor Laboratory Press Cold Spring Harbor, NY
L Thirup K Johnsen A Winding (2001) ArticleTitleSuccession of indigenous Pseudomonas spp. and actinomycetes on barley roots affected by the antagonistic strain Pseudomonas fluorescens DR54 and the fungicide imazalil Appl Environ Microbiol 67 1147–1153 Occurrence Handle10.1128/AEM.67.3.1147-1153.2001 Occurrence Handle11229904
L Thirup A Johansen A Winding (2003) ArticleTitleMicrobial succession in the rhizosphere of live and decomposing barley roots as affected by the antagonistic strain Pseudomonas fluorescens DR54-BN14 or the fungicide imazalil FEMS Microb Ecol 43 383–392 Occurrence Handle10.1016/S0168-6496(02)00450-6
C Thrane S Olsson TH Nielsen J Sørensen (1999) ArticleTitleVital fluorescent stains for detection of stress in Pythium ultimum and Rhizoctonia solani challenged with viscosinamide from Pseudomonas fluorescens DR54 FEMS Microbiol Ecol 30 11–23 Occurrence Handle10.1016/S0168-6496(99)00034-3
C Thrane TH Nielsen MN Nielsen J Sørensen S Olsson (2000) ArticleTitleViscosinamide-producing Pseudomonas fluorescens DR54 exerts a biocontrol effect on Pythium ultimum in sugar beet rhizosphere FEMS Microbiol Ecol 33 139–146 Occurrence Handle10.1016/S0168-6496(00)00054-4 Occurrence Handle10967213
C Thrane MN Nielsen J Sørensen S Olsson (2001) ArticleTitlePseudomonas fluorescens DR54 reduces sclerotia formation, biomass development, and disease incidence of Rhizoctonia solani causing damping-off in sugar beet Microb Ecol 42 438–445 Occurrence Handle10.1007/s00248-001-0007-y Occurrence Handle12024268
CF Thurston (1994) ArticleTitleThe structure and function of fungal laccases Microbiology 140 19–26
A Tunlid DC White (1992) Biochemical analysis of biomass, community structure, nutritional status, and metabolic activity of microbial communities in soil G Stotzky JM Bollag (Eds) Soil Biochemistry, vol 7 Marcel Dekker New York 229–262
A Tunlid HAJ Hoitink C Low DC White (1989) ArticleTitleCharacterization of bacteria that suppress Rhizoctonia damping off in bark compost media by analysis of fatty acid biomarkers Appl Environ Microbiol 55 1368–1374
JD Elsas Particlevan AF Dijkstra JM Govaert JA Veen Particlevan (1986) ArticleTitleSurvival of Pseudomonas fluorescens and Bacillus subtilis introduced into two soil of different texture in field microplots FEMS Microbiol Ecol 38 151–160 Occurrence Handle10.1016/0378-1097(86)90046-7
LS Overbeek Particlevan JA Veen Particlevan JD Elsas Particlevan (1995) ArticleTitleInduced reporter gene activity, enhanced stress resistance, and competitive ability of a genetically modified Pseudomonas fluorescens strain released into a field plot planted with wheat Appl Environ Microbiol 63 1965–1973
JA Veen Particlevan LS Overbeek Particlevan JD Elsas Particlevan (1997) ArticleTitleFate and activity of microorganisms introduced into soil Microbiol Mol Rev 61 121–135
JA Wahleithner F Xu M Brown SH Brown EJ Golightly T Halkier S Kauppinen A Pederson P Schneider (1996) ArticleTitleThe identification and characterization of four laccases from the plant-pathogenic fungus Rhizoctonia solani Curr Genet 29 295–403 Occurrence Handle10.1007/s002940050061
SG Wilkinson (1988) Gram-negative bacteria C Ratledge SG Wilkinson (Eds) Microbial Lipids, vol 1 Academic Press London 299–488
L Zelles (1999) ArticleTitleFatty acid patterns of phospholipids and lipopolysaccharides in the characterization of microbial communities in soil: a review Biol Fertil Soils 29 111–129 Occurrence Handle10.1007/s003740050533
Acknowledgments
The authors thank lab technician Ulla Rasmussen for competent help with PLFA analyses and immunostaining procedures. This study was supported by a grant from the Danish Environmental Research Programme (SMP2): “Centre for Effects and Risks of Biotechnology in Agriculture.”
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Johansen, A., Olsson, S. Using Phospholipid Fatty Acid Technique to Study Short-Term Effects of the Biological Control Agent Pseudomonas fluorescens DR54 on the Microbial Microbiota in Barley Rhizosphere. Microb Ecol 49, 272–281 (2005). https://doi.org/10.1007/s00248-004-0135-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-004-0135-2