Abstract
Glyphosate is a non-selective organophosphate herbicide that is widely used in agriculture, but its effects on soil microbial communities are highly variable and often contradictory, especially for high dose applications. We applied glyphosate at two rates: the recommended rate of 50 mg active ingredient kg−1 soil and 10-fold this rate to simulate multiple glyphosate applications during a growing season. After 6 months, we investigated the effects on the composition of soil microbial community, the catabolic activity and the genetic diversity of the bacterial community using phospholipid fatty acids (PLFAs), community level catabolic profiles (CLCPs), and 16S rRNA denaturing gradient gel electrophoresis (DGGE). Microbial biomass carbon (Cmic) was reduced by 45%, and the numbers of the cultivable bacteria and fungi were decreased by 84 and 63%, respectively, under the higher glyphosate application rate. According to the PLFA analysis, the fungal biomass was reduced by 29% under both application rates. However, the CLCPs showed that the catabolic activity of the gram-negative (G−) bacterial community was significantly increased under the high glyphosate application rate. Furthermore, the DGGE analysis indicated that the bacterial community in the soil that had received the high glyphosate application rate was dominated by G− bacteria. Real-time PCR results suggested that copies of the glyphosate tolerance gene (EPSPS) increased significantly in the treatment with the high glyphosate application rate. Our results indicated that fungi were impaired through glyphosate while G− bacteria played an important role in the tolerance of microbiota to glyphosate applications.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Extensive use of herbicides is a serious matter of environmental concern because of potential adverse effects on ecosystems (Busse et al. 2001; Pizarro et al. 2016). Glyphosate ([N-phosphonomethyl] glycine), one of the most commonly used herbicides worldwide, is applied to control a broad spectrum of weeds in agriculture, horticulture, silviculture, and urban landscapes (Candela et al. 2007; Duke 2017). It inhibits the activity of 5-enoylpyruvylshikimate-3-phosphate synthase (EPSPS), an enzyme involved in the shikimate pathway. This enzyme is essential for the synthesis of aromatic amino acids, which are necessary for plant survival (Duke and Powles 2008). The shikimate synthesis pathway is also found in microorganisms, and it would be assumed that the microbial growth and activity of susceptible species may be disrupted by glyphosate (Nguyen et al. 2016). In vitro experiments demonstrated that glyphosate can inhibit microbial growth of environmental isolates (Bonnet et al. 2007) and gut microbiota at concentrations greater than 75 mg L−1 (Shehata et al. 2013). However, results of studies investigating glyphosate impacts on soil microbial communities are highly variable and often contradictory (Newman et al. 2016a; Nguyen et al. 2016).
Soil microbes are vital to maintain soil ecosystem functions including nutrient cycling and bioremediation (Rick and Thomas 2001; Li et al. 2018). Despite the fact that glyphosate was considered to have a low mammalian and ecological toxicity (Busse et al. 2001), recently, concerns have been raised by some scientists that the increasing use of glyphosate might affect non-target organisms in the soil (Bai and Ogbourne 2016). Many studies investigated the effects of glyphosate on soil microbial properties, such as microbial biomass, enzyme activity, and respiration, applying broad-scale or integrative methods (Hosseini Bai et al. 2014; Imfeld and Vuilleumier 2012; Newman et al. 2016b). A recent meta-analyses indicated that the effect of glyphosate on soil microbial communities is not consistent but depends on many different factors, including, for example, the concentration and formulation of glyphosate in an application, soil pH and exposure time (Nguyen et al. 2016). For example, glyphosate concentration > 200 mg kg−1 increased soil microbial biomass under short-term exposure (< 100 days) and low soil pH (< 5.5) conditions, while lower glyphosate concentrations decreased the microbial biomass in the longer term (> 100 days) and at neutral pH (Nguyen et al. 2016). Although these studies provided insight into the response of soil microbial communities to glyphosate application, its effects on community-level dynamics and microbial metabolic activity are still poorly understood.
Previous studies on microbial communities showed different responses to glyphosate applications. In general, bacterial community composition was shifted adaptively toward more glyphosate-tolerant species over time (Jacobsen and Hjelmsø 2014; Lancaster et al. 2010). Glyphosate use led to a decrease in most bacterial and all fungal biomass in rhizosphere and favored more glyphosate-tolerant bacteria through shifting the composition of bacterial communities (Newman et al. 2016a). The shifts in the microbial community composition might subsequently affect microbial functions involving carbon (C), nitrogen (N), and phosphorus (P) cycling (Hosseini Bai et al. 2014). Therefore, it is necessary to determine whether microbial groups are impaired by different glyphosate application rates.
Previous studies observed a glyphosate-induced increase in soil microbial activity and functional diversity 15 days after glyphosate application, while it was inconsistent with the response of the microbial community 30 days after glyphosate treatment (Iker et al. 2009). Most previous studies were short-term studies, and the potential long-term (here defined as studies > 100 days) effects are not yet comprehensively documented (Newman et al. 2016a, b). To identify endpoint effects of glyphosate use, application rates used are often much higher than recommended label rates in contrast to agronomic studies, which usually investigate application rates relevant to agricultural practices (Nguyen et al. 2016). Our objective was to evaluate the long-term effects of glyphosate applied at different application rates on soil microbial communities. We used the recommended glyphosate application rate and 10-fold higher rate. Specifically, the following hypotheses were tested: (1) soil bacterial community responds to glyphosate application, and, in turn, increases the catabolic activity of Gram-negative bacteria; (2) glyphosate application impairs soil fungal community, ultimately, decreasing the fungi to bacteria ratio (F/B); and (3) the increase in the abundance of the glyphosate tolerance gene EPSPS depends on the glyphosate application rate.
Materials and methods
Experimental design and soil sampling
This experiment was carried out in Nanjing (118° 46′ E, 32° 03′ N), Jiangsu Province, China. This area belongs to the semitropical zone with an annual mean temperature of 15.3 °C and a mean annual rainfall of 1106.5 mm. Loam soil was collected at a depth of 0–20 cm from an agriculturally used site that has not received any agrichemicals. After removing the plant residues by sieving the soil to < 2.0 mm, the soil sample was divided into samples of 10 kg. Each sample was loosely packed into a 20-cm high pot with 20 cm diameter. The pots were cultivated with grass (Cynodon dactylon L. Pars) and randomly placed with a distance of about 2 m from each other. At the end of the experiment, the erect stems of the grass reached heights of approximately 10 cm with deep root systems. Three treatments with four replicates were established in a completely randomized design: a control without glyphosate (GLY0), the recommended glyphosate application rate (GLY1) with 50 mg commercial product kg−1 soil (equal to 1.33 kg active principle per hectare) (Busse et al. 2001; Weaver et al. 2007), and a high glyphosate application rate (GLY10) of 500 mg commercial product kg−1 soil. Applying glyphosate several times during one growing season, sometimes at higher than recommended application rates as an attempt to control glyphosate-tolerant weeds in glyphosate-resistant crops, is common practice. This can lead to glyphosate accumulation in soils, a situation we have mimicked with our high glyphosate application rate treatment. Glyphosate (commercial product) was dissolved into 50 mL distilled water and evenly sprayed to the surface of the soils to simulate agricultural application and create a decreasing stress gradient from the soil surface to the bottom of the pots. The control soil samples (GLY0) received the same amount of distilled water. The glyphosate application was a single application. The half-life of glyphosate is about 20 days (provided by manufacturer).
After 6 months (April–September) soil samples were collected in 0 to10 cm depth and sieved to 2 mm. Soil carbon (C), nitrogen (N), and phosphorus (P) concentrations, phospholipid fatty acids (PLFAs), and community level catabolic profiles (CLCPs) were then immediately measured in the soil samples. The measurement techniques are described below. Sub-samples were stored in sterile plastic bags at − 20 °C for 16S rRNA gene analysis. Physico-chemical properties of all soils were measured with the methods described in a previous study (Wang et al. 2006).
Soil microbial biomass C, soil organic C, total N, and total P concentrations
Soil microbial biomass C (Cmic) was measured by the chloroform fumigation-extraction method (Vance et al. 1987). In brief, each replicate of the three treatments (equivalent to 50 g oven-dried soil weight) was divided into two parts and labeled as CF (chloroform fumigated) and NF (non-fumigated). Only the CF sub-sample was exposed to CHCl3 vapor at 20 °C for 24 h in the dark. To elute the water-soluble oxidized C, 250 mL of 0.5 M K2SO4 was added to all sub-samples prior to filtering. Extractable organic C was measured using an organic C autoanalyzer (Shimadzu TOC 5000A, Japan). The following equation was used to calculate Cmic (μg g−1) = 2.64 Ec, where Ec is the difference in organic C concentration between CF and NF.
The concentration of total organic C (Corg), total N (Ntot), and total P (Ptot) was determined using air-dried soil samples with PerkinElmer 240c analyzer (PerkinElmer Co., Ltd., USA), Foss Heraeus CHN-O-rapid elements analyzer (Foss Heraeus Co., Ltd., Germany), and Jarell-Ash ICP quantometer (Jarell-Ash Co., Ltd., USA), respectively.
PLFA analysis
The analysis of PLFAs was conducted according to White and Findlay (1988). In brief, total lipids were extracted from 5 g freeze-dried soil using a mixture of chloroform, methanol, and citrate buffer (V/V = 1:2:0.8). Then the PLFAs were separated using silica solid phase extraction columns (Supelco, Inc., Bellefonte, PA, USA) followed by trans-esterification with a mild alkaline to produce fatty acid methyl esters (FAMEs). The FAMEs were then separated and identified using gas chromatography (N6890, Agilent, USA). In order to identify PLFAs, chromatographic retention times and mass spectra were compared with those of the standard qualitative bacterial acid methyl ester mix (Supelco UK, Poole, Dorset, UK) that ranged from C11 to C20 using GC 6890 Plus analyzer (Hewlett-Packard Co. USA).
In this study, the sum of 15:0, i15:0, a15:0, i16:0, i17:0 cy17:0, 18:1ω9t, 18:1ω9c and cy19:0 was considered to represent bacterial biomass (Frostegård et al. 1993, Liang et al. 2008, Tong et al. 2007). The sum of i15:0, a15:0, i16:0 and i17:0 was considered to represent gram-positive (G+) bacterial biomass, and the sum of cy17:0, cy19:0, 18:1ω9t and 18:1ω9c was considered to represent gram-negative (G−) bacteria (Tong et al. 2007). The polyenoic and unsaturated 18:2ω6 was used as indicator of fungal biomass (Frostegård et al. 1993), and the ratio of 18:2ω6 to bacterial PLFAs was used as estimate of the ratio of fungal to bacterial biomass in the soil (Frostegård and Bååth 1996).
CLCPs of soil G− and G+ bacteria
Community level catabolic profiles (CLCPs) of soil microbial communities were measured with Biolog GP and GN microtiter plates (Biolog Inc. USA) (Garland and Mills 1991). Fresh soil equivalent to 25 g of dry weight was added to 250 mL of 0.1 M potassium phosphate buffer (pH 7.0) and shaken at 200 rpm for 1 min. After settling for 5 min at 0 °C, the supernatant was diluted 100 times using phosphate buffer (pH 7.0) to equalize the turbidity of the eluted solutions from all soil samples (A595 < 0.1). Then, 125 μL of the aliquot was inoculated to wells of preheated plates and incubated at 30 °C for 1 week. During incubation, absorbance values in each well were read at 595 nm every 12 h with Sunrise Plate Reader (TECAN, Switzerland).
Average well color development (AWCD) was used to assess the overall catabolic activity of the microbial community. It was calculated from the mean of all net absorbance values of the 95 wells containing substrates.
Quantification of cultivable bacteria, phosphobacteria, and fungi
Fresh soil (approximate dry weight of 10 g) was dissolved in 100 mL of 0.1 M sodium phosphate buffer (pH 7.0) by agitating the slurry for 30 min. The suspension was diluted 10-fold for three times in succession, and then 100 μL of the diluted solution was incubated to (1) trypcasein soy agar (TSA) medium to quantify the amount of bacteria (Wu et al. 2001), (2) modified standard SRSM1 enrichment medium of Sundara Rao and Sinha specific for the isolation of phosphate-solubilizing microorganisms (Vazquez et al. 2000), and (3) fungi agar medium to enumerate the amount of fungi (Toledohernández et al. 2007). The density of microbes was assessed by visually quantifying colony formation unities (CFU) per milliliter of sample (CFU mL−1) after 5 days of cultivation at 28 °C in the dark.
Soil DNA extraction and 16S rRNA-DGGE fingerprinting
Soil microbial DNA was extracted with the method of SDS-CTAB lysis (Zhou et al. 1996). In brief, crude DNA samples were size-fractionated by agarose gel electrophoresis to reduce the possibility of chimera formation and to remove inhibitors such as heavy metals and humic acids. DNA of about 23 kb was recovered using Geneclean UNIQ-10 Spin kit (Sangon, China) according to the manufacturer’s instruction. For 16S rRNA gene nested PCR, the eubacterial primers 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-AGAGTTTGATCCTGGCTCAG-3′) were used in the first round, and 1 μL of the resulting PCR product was used as template in the second round with the primers GC40-341f (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCACGGGGGGCCTACGGGAGG-3′) and 907r (5′-CCGTCAATTCCTTTGTTT-3′). The 16S rRNA gene was amplified in 50 μL of reaction system, containing 2.5 U Taq DNA polymerase (Tiangen, China), 5 μL ExTaq buffer, 3 μL MgCl2, 1 μL of 10 mmol L−1 dNTPs, 1 μL of 10 μmol L−1 of the primer, and 1 μL of a template DNA. PCR reactions were performed with the following cycling conditions: 5 min at 94 °C; 30 cycles of 30 s at 94 °C, 30 s at 55 °C, and 30 s at 72 °C.
Denaturing gradient gel electrophoresis (DGGE) was performed using D-code™ Universal Mutation Detection System (Bio-Rad, USA). PCR products were loaded into 6% (w/v) polyacrylamide gel, which consisted of a linear denaturing gradient ranging from 40% denaturant at the top of the gel to 65% denaturant at the bottom (100% denaturant is defined as 7 M urea and 40% (v/v) deionized formamide). Electrophoresis was conducted at 130 V and 60 °C for 330 min. The gel was incubated in 0.5 μg mL−1 ethidium bromide solution for 30 min, rinsed in water for 20 min, photographed with GelDoc 2000 gel photo system (BioRad, USA), and analyzed with BioRad’s Quantity One software package. In order to detect the diversity of soil bacteria at the molecular level, the mutual and specific bands were excised and suspended in 20 μL water overnight at 4 °C. Approximately 5 μL of the supernatants was used for re-amplification with the primers 341f (5′-CCTACGGGAGGCAGCAG-3′) and 518r (5′- ATTACCGCGGCTGCTGG-3′). The PCR products were purified using AXYGEN PCR purification kit (AXYGEN, China) and sequenced.
The 16S rRNA sequences were identified using the BLAST algorithm against the GenBank database. Sequences in the GenBank database with more than 98% similarity to the 16S rRNA sequence were assigned to the same phylotype. All identified sequences were aligned with their closest relatives.
Quantification of EPSPS gene abundance
The class II EPSPS gene was amplified with the primers (5′-CGTAAGTAAGTGAAAGTAACAATACAGC-3′) and (5′-CGCTTCTTCGGACAATGACAGAC-3′) (Tian et al. 2010) from the microbial genomic DNA as mentioned above. The amplified fragment was cloned into the vector pCR2-TOPO, and transferred into Escherichia coli TOPO10. The plasmid DNA were extracted and purified with a commercial plasmid kit (AXYGEN, China). Then, 101–108 copies of the recombinant EPSPS gene were used to generate a standard curve. Real-time quantitative PCR was carried out with EvaGreen reagents (Biotium, USA) according to the manufacturer’s standard protocol. Amplification reactions were performed using ABI Prism 7000 sequence detector (Applied Biosystems, USA), and the data were analyzed by Applied SDS software version 1.1 (Applied Biosystems, USA).
Statistical analyses
Statistical analyses were performed with SPSS version 16.0 (SPSS, Chicago, IL, USA). One-way ANOVA (n = 3, p < 0.05) of the completely randomized design with glyphosate as factor was performed to test significant differences. Before performing the ANOVA, the normality of distribution and homogeneity of the variance was tested, and the data were log-transformed if the assumption of homogeneity was violated. Principal component analysis (PCA) was carried out to test the effect of glyphosate application on soil bacterial communities measured by CLCP and PLFA analysis. The DGGE bands were transformed into a 1–0 binary matrix based on the presence or absence of DGGE bands. Then, the binary matrix was also used to generate cluster dendrogram with the GenStat 10.0 software package to illustrate the soil bacterial community variance. The statistical significance was determined at the 5% level.
Results
Soil properties and microbial biomass C
There was no significant difference between the three treatments in soil pH, Corg, Ntot, and the other basic soil parameters analyzed (Table 1). Compared to the Ptot concentrations in the control GLY0 and the treatment GLY1, the concentration of Ptot was increased by 118% in the treatment GLY10, accompanied by a significant decrease in Corg/Ptot (Table 2). On average, Cmic and Cmic/Corg were significantly reduced by 45 and 47% in the treatment GLY10, respectively, compared to those of the control.
Community structure and diversity of soil bacteria
The changes in the composition of microbial PLFAs, as an indicator of active soil microbial biomass and community composition, are summarized in Table 3. One of the G+ bacteria’s PLFA (a15:0) decreased by 21% in the treatment GLY10 compared to the control GLY0 (Fig. 1). However, no significant difference was found in the total microbial PLFA and bacterial biomarkers. PCA was conducted to visualize the differences between the PLFAs that contributed to the difference induced by the glyphosate treatment (Fig. 2). The first two components explained 67% of the total variance, and eight PLFAs were strongly correlated (r ≥ 0.6, r is the absolute value of correlation coefficient) to PC1. Four of these PLFAs (i15:0, a15:0, i16:0 and cy17:0) were bacterial signatures, and the other four (18:2ω6, 20:0, 18:1ω9t and 2-OH 14:0) were strongly correlated to PC2, indicating that these PLFAs played important roles in the response to the glyphosate treatment. The CFU count of total bacteria was reduced significantly by 84% in the treatment GLY1 compared to the control (Table S1).
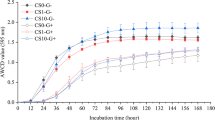
Biolog GN and GP microplates were used to determine the substrate utilizing potential of G− and G+ heterotrophic bacteria, respectively. Changes in the average well color development (AWCD) values reflected the different response of G− and G+ bacteria to the imposed glyphosate stress (Fig. 3). After 72 h of incubation, only the AWCD value of G− bacteria in the treatment GLY10 was higher than for the control, while no difference was detected in the AWCD value of G+ bacteria. The PCA results further illustrate the impacts of glyphosate on G− and G+ bacteria (Fig. S1). Two components, PC1 and PC2, accounted for 52 and 51% of the total variance in Biolog GN and GP microplates, respectively. Only the treatment GLY10 was different to the control in the GN Biolog plates. Moreover, both glyphosate treatments, GLY1 and GLY10, were different from the control in the GP Biolog plates.
DGGE pattern of amplified 16S rRNA fragments was used to examine the effects of glyphosate application on the genetic structures of the soil bacterial community (Fig. S2). A total of 18 distinguished bands from the soil samples and the solution in Biolog plates were excised from DGGE gels and sequenced. Based on the results of blast and phylogenetic analysis, we found that 72% of the identified bands were related to G− bacteria (Table 4). Compared to the control, four bands decreased in intensity at position a, b, d, and e, and one band disappeared at position c (Fig. S2). In addition, one band increased in intensity at position f, and four new bands appeared at position g, h, i, and j on the DGGE gel in the treatments GLY1 and GLY10 compared to the control soil (Fig. S2). We also identified cultivable bacteria from the solution of the Biolog plates to investigate the response of soil bacteria to the glyphosate application. Some cultivable bacteria, such as Burkholderia and Gamma proteobacterium, were detected (Table 4). However, there were only two bands in the Biolog samples, and one band diminished at position i in the treatments GLY1 and GLY10 compared with the bands of the control. The cluster analysis based on DGGE pattern indicated that the community structure of soil bacteria in GLY1 was more similar to the control than to the community of the treatment GLY10 (Fig. 4).
Soil fungal community
There was a significant decrease of 29% in the concentration of fungal PLFA (18:2ω6) in the treatments GLY1 and GLY10 compared to the control (Fig. 1; Table 3). The ratio of fungal to bacterial biomass decreased significantly by 38% in both the treatments GLY1 and GLY10. Moreover, the CFU counting of soil fungi in both the treatments GLY1 and GLY10 was significantly reduced by 77 and 63%, respectively, compared to the control (Table S1).
Abundance of EPSPS gene and enumeration of cultivable phosphobacteria
Real-time PCR was employed to determine the change in copy numbers of the EPSPS gene in the soils treated with glyphosate. The EPSPS gene was detected in all three treatments with different copies, of which the copy number in the treatment GLY10 was approximately 2.6-times higher than those in the other two treatments (Fig. 5). Furthermore, the CFU counting of phosphobacteria revealed significantly higher numbers of phosphobacteria in the treatments GLY1 and GLY10 than in the control (Table S1).
Discussion
Response of the soil bacterial community to glyphosate applications
A long-term effect of glyphosate applications at a recommended and 10-fold higher application rate increased the catabolic activity of G− bacteria but inhibited the fungal community in the soils. This finding was consistent with a previous meta-analysis showing that the glyphosate application rate is a vital factor in determining microbial response (Nguyen et al. 2016). Our results showed that glyphosate application at a 10-fold higher than recommended rate (GLY10) increased the AWCD value rather than the biomass of G− bacteria compared to the control (GLY0), indicating that the G− bacteria activity had a stronger response at the higher dose than at the recommended rate. The soil microbial community could have increased their catabolic activity rather than reproducing themselves to resist environmental stress (Dahlin and Witter 1998). Similarly, a previous study found that the abundance of G− bacteria decreased in the rhizosphere of glyphosate-treated plants (Newman et al. 2016b). The increase in catabolic activity of G− bacteria can be interpreted as self-rescue approach of microbes allowing them to survive under environmental stress caused by glyphosate applications (Wang et al. 2008).
The Cmic was reduced significantly through the glyphosate application in the treatment GLY10 compared to the treatment GLY1 and the control GLY0 (Table 2), suggesting that the high concentration of glyphosate had a negative effect on soil microbial activities, which is consistent with previous findings (Andréa et al. 2003; Stratton and Stewart 1992). The persistence of glyphosate varies across soil properties and environmental conditions (Bai and Ogbourne 2016). For example, the recent literature indicates that the half-life of glyphosate ranges from 10 to 151 days based on a study with a loamy soil in China (Zhang et al. 2015) and a study with a clay soil in Sweden (Bergström et al. 2011), respectively. In our study, the half-life of glyphosate was indicated to be about 20 days. Higher rates of glyphosate application (> 100 mg kg−1) could provide a metabolizable source of C, N, and P (Duke et al. 2012; Haney et al. 2002). In this study, Cmic/Corg was significantly lower in the treatment GLY10 than in the control GLY0 and the treatment GLY1, implying that less substrate C was incorporated into the soil microbial biomass at the high glyphosate application rate. Although the amounts of total and bacterial PLFAs showed no significant differences among the three treatments (Table 3), both PCAs based on the PLFAs (Fig. 2) and the pattern of 16S rRNA DGGE (Fig. S2) confirmed that the soil bacterial community composition, specifically G− bacteria, responded to the glyphosate application, supporting our first hypothesis.
Phylogenetic analysis based on 16S rRNA DGGE pattern showed that 72% of the identified bands belonged to G− bacteria (Table 4), suggesting that G− bacteria might have dominated the whole soil bacterial community and played an important role in responding to the stress of the glyphosate applications. Previous studies indicated that G− bacteria tend to grow well under substrate-rich condition (Chang and Chiu 2015; Margesin et al. 2009), and it was proposed that G− bacteria are more resistant to chemical stress than other microbes because they possess a specific cell wall structure (Berg et al. 2005; Lapygina et al. 2002; Wang et al. 2008). In the present study, some bacterial species, mainly Ralstonia sp., Gamma proteobacterium, Burkholderia sp., and Devosia sp. were detected in the treatments GLY1 and GLY10. These had been shown to be capable of degrading some chemicals, such as mono and dichlorophenols and trichloroethylene (Ayoubi and Harker 1998; Cobos-Vasconcelos et al. 2006; Lee 2007). Our results are consistent with a previous study, which showed that the dominance of some G− bacteria in pesticide-stressed soils decreased the richness of the bacterial community (Wang et al. 2008). Our results confirm the presence of a spectrum of sensitive bacteria among the microbial population depending on the glyphosate application rate as reported in previous study (Nguyen et al. 2016).
Impairment of the soil fungal community through glyphosate applications
In this study, both the concentration of fungal PLFA and numbers of cultivable fungi decreased significantly in the treatment GLY1 and GLY10 compared to the control, indicating that the soil fungal community was impaired at both application rates. This finding is consistent with a previous meta-analysis showing that less resilient microbial population were inhibited by increasing glyphosate concentrations over approximately 10 mg kg−1 (Nguyen et al. 2016), suggesting that the fungal community is more sensitive to stress caused by glyphosate than other microorganisms. Fungi are considered to be more sensitive to chemical stress (Edvantoro et al. 2003), and it was also reported that fungal PLFA biomarkers in the rhizosphere decreased when plants were treated with glyphosate (Newman et al. 2016b). The F/B biomass ratio is often used as indicator of changes in the carbon use efficiency of microbial communities in response to environmental stress (Chen et al. 2013). In the present study, the lower F/B ratio in the treatments GLY1 and GLY10 supports our second hypothesis and suggests that both the recommended and 10-fold higher glyphosate application rate decreased the microbial community’s efficiency to utilize carbon, which is in line with the decreased Cmic/Corg ratio induced by the glyphosate applications.
Glyphosate-induced increase in abundance of EPSPS gene
The EPSPS gene showed high tolerance to glyphosate in certain strains (Sun et al. 2005). In the present study, the copy number of the EPSPS gene in the treatment GLY10 was 2.6 times higher than that in the control GLY0, while there was no significant difference between the treatments GLY0 and GLY1 (Fig. 5), supporting our third hypothesis that the abundance of the glyphosate tolerance EPSPS gene depended on the glyphosate application rate. Some bacterial strains, such as Pseudomonas sp. and Agrobacterium tumefaciens sp., are G− bacteria, whose EPSPS genes can tolerate exposure to glyphosate (Jacob et al. 1988; Mcauliffe et al. 1990; Zhou et al. 2006). For example, previous studies reported that both Pseudomonas sp. and Agrobacterium sp. have the capability to metabolize glyphosate (Jacob et al. 1988; Mcauliffe et al. 1990), and A. tumefaciens sp. strain was successfully used in glyphosate-tolerant crops due to its high glyphosate resistance (Zhou et al. 2006). In addition, the high glyphosate application rate (GLY10) increased the Ptot content of the soil and thereby decreased the ratio of Corg/Ptot compared to the treatments GLY0 and GLY1 (Table 2), suggesting that P assimilation increased through the high glyphosate application (Wang et al. 2008). Correspondingly, the numbers of cultivable phosphobacteria increased 24- and 40-fold in the GLY1 and GLY10 treatments, respectively (Table S1). Although glyphosate application had no effect on the contents of Corg in the soils of this study, the chemical composition of Corg is likely to influence the soil microbial population size (Li et al. 2013). Recently, shifts in fungal community were shown to be linked to the chemical composition of Corg in forest soils (Li et al. 2017a, b). Therefore, some specific microbial communities, such as G− bacteria, phosphobacteria, and fungi, could play important roles in tolerating or resisting glyphosate, but their functions need to be further elucidated in future study.
Conclusion
In our 180-day study, we showed that the soil fungal community was impaired by the recommended glyphosate application rate. In contrast, the high glyphosate application rate had significant effects on the structure and functional and genetic diversity of the soil microbial community. Both, the G+ bacteria and fungal community were inhibited while the catabolic activity of G− bacteria was increased, indicating that these taxa were sensitive and responsive to the environmental change caused by glyphosate applications. The higher application rate increased the dominance of G− bacteria but decreased its richness, suggesting that G− bacteria play an important role in the degradation of glyphosate. Although the recommended glyphosate application rate had no effects on the soil bacterial community, the shifts observed in the community structure and function of the soil microbial community at the recommended glyphosate application rate warrant more detailed investigations. Future research should employ a combination of various approaches, including quantification of functional gene activity at the mRNA level and next-generation sequencing to assess potential ecological effects of glyphosate applications on soil microbial composition and its functions over a longer time period.
Acknowledgements
We thank Jianfang Feng from State Key Laboratory of Pollution Control and Resource Reuse in the School of Environment of Nanjing University, China, for her assistance with the GC-MS analysis. We are very grateful to Prof. Shuijin Hu at the Department of Plant Pathology, North Carolina State University for his helpful comments to revise a previous version of the manuscript.
References
Andréa DMM, Bonanholuchini T, Carlosbazarin L, Papini S, Matallo S, Barifousesavoy M, Tedeschi VL (2003) Influence of repeated applications of glyphosate on its persistence and soil bioactivity. Pesqui Agropecu Bras 38:1329–1335
Ayoubi PJ, Harker AR (1998) Whole-cell kinetics of trichloroethylene degradation by phenol hydroxylase in a Ralstonia eutropha JMP134 derivative. Appl Environ Microbiol 64:4353–4356
Bai SH, Ogbourne SM (2016) Glyphosate: environmental contamination, toxicity and potential risks to human health via food contamination. Environ Sci Pollut Res 23:18988–19001
Berg J, Tom-Petersen A, Nybroe O (2005) Copper amendment of agricultural soil selects for bacterial antibiotic resistance in the field. Lett Appl Microbiol 40:146–151
Bergström L, Börjesson E, Stenström J (2011) Laboratory and lysimeter studies of glyphosate and aminomethylphosphonic acid in a sand and a clay soil. J Environ Qual 40:98–108
Bonnet JL, Bonnemoy F, Dusser M, Bohatier J (2007) Assessment of the potential toxicity of herbicides and their degradation products to nontarget cells using two microorganisms, the bacteria Vibrio fischeri and the ciliate Tetrahymena pyriformis. Environ Toxicol 22:78–91
Busse MD, Ratcliff AW, Shestak CJ, Powers RF (2001) Glyphosate toxicity and the effects of long-term vegetation control on soil microbial communities. Soil Biol Biochem 33:1777–1789
Candela L, Álvarez-Benedí J, De Melo MC, Rao P (2007) Laboratory studies on glyphosate transport in soils of the Maresme area near Barcelona, Spain: transport model parameter estimation. Geoderma 140:8–16
Chang EH, Chiu CY (2015) Changes in soil microbial community structure and activity in a cedar plantation invaded by moso bamboo. Appl Soil Ecol 91:1–7
Chen JH, Liu XY, Zheng JW, Zhang B, Lu HF, Chi ZZ, Pan GX, Li LQ, Zheng JF, Zhang XH (2013) Biochar soil amendment increased bacterial but decreased fungal gene abundance with shifts in community structure in a slightly acid rice paddy from Southwest China. Appl Soil Ecol 71:33–44
Cobos-Vasconcelos DDL, Santoyo-Tepole F, Juárez-Ramírez C, Ruiz-Ordaz N, Galíndez-Mayer CJJ (2006) Cometabolic degradation of chlorophenols by a strain of Burkholderia in fed-batch culture. Enzyme Microb Technol 40:57–60
Dahlin S, Witter E (1998) Can the low microbial biomass C-to-organic C ratio in an acid and a metal contaminated soil be explained by differences in the substrate utilization efficiency and maintenance requirements? Soil Biol Biochem 30:633–641
Duke SO (2017) The history and current status of glyphosate. Pest Manag Sci. https://doi.org/10.1002/ps.4652
Duke SO, Powles SB (2008) Glyphosate: a once-in-a-century herbicide. Pest Manag Sci 64:319–325
Duke SO, Lydon J, Koskinen WC, Moorman TB, Chaney RL, Hammerschmidt R (2012) Glyphosate effects on plant mineral nutrition, crop rhizosphere microbiota, and plant disease in glyphosate-resistant crops. J Agr Food Chem 60:10375–10397
Edvantoro BB, Naidu R, Megharaj M, Singleton I (2003) Changes in microbial properties associated with long-term arsenic and DDT contaminated soils at disused cattle dip sites. Ecotox Environ Safe 55:344–351
Frostegård A, Bååth E (1996) The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol Fert Soils 22:59–65
Frostegård Å, Bååth E, Tunlio A (1993) Shifts in the structure of soil microbial communities in limed forests as revealed by phospholipid fatty acid analysis. Soil Biol Biochem 25:723–730
Garland JL, Mills AL (1991) Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol 57:2351
Haney R, Senseman S, Krutz L, Hons F (2002) Soil carbon and nitrogen mineralization as affected by atrazine and glyphosate. Biol Fert Soils 35:35–40
Hosseini Bai S, Xu Z, Blumfield TJ, Wild CH, Chen C (2014) Soil carbon and nitrogen dynamics in the first year following herbicide and scalping in a revegetation trial in south-east Queensland, Australia. Environ Sci Pollut Res 21:5167–5176
Iker M, Josem B, Isabel A, Lur E, Carlos G (2009) Effects of glyphosate on rhizosphere soil microbial communities under two different plant compositions by cultivation-dependent and -independent methodologies. Soil Biol Biochem 41:505–513
Imfeld G, Vuilleumier S (2012) Measuring the effects of pesticides on bacterial communities in soil: a critical review. Eur J Soil Biol 49:22–30
Jacob GS, Garbow JR, Hallas LE, Kimack NM, Kishore GM, Schaefer J (1988) Metabolism of glyphosate in Pseudomonas sp. strain LBr. Appl Environ Microbiol 54:2953–2958
Jacobsen CS, Hjelmsø MH (2014) Agricultural soils, pesticides and microbial diversity. Curr Opin Biotech 27:15–20
Lancaster SH, Hollister EB, Senseman SA, Gentry TJ (2010) Effects of repeated glyphosate applications on soil microbial community composition and the mineralization of glyphosate. Pest Manag Sci 66:59–64
Lapygina EV, Lysak LV, Zviagintsev DG (2002) Tolerance of soil bacterial complexes to salt shock. Microbiology 71:143–147
Lee SD (2007) Devosia subaequoris sp. nov., isolated from beach sediment. Int J Syst Evol Microbiol 57:2212–2215
Li YF, Zhang JJ, Chang SX, Jiang PK, Zhou GM, Fu SL, Yan ER, Wu JS, Lin L (2013) Long-term management effects on soil organic carbon pools and chemical composition in Moso bamboo (Phyllostachys pubescens) forests in subtropical China. Forest Ecol Manag 303:121–130
Li YC, Li YF, Chang SX, Xu QF, Guo ZY, Gao Q, Qin ZY, Yang YF, Chen JH, Liang X (2017a) Bamboo invasion of broadleaf forests altered soil fungal community closely linked to changes in soil organic C chemical composition and mineral N production. Plant Soil 418:507–521
Li YC, Li YF, Chang SX, Liang X, Qin H, Chen JH, Xu QF (2017b) Linking soil fungal community structure and function to soil organic carbon chemical composition in intensively managed subtropical bamboo forests. Soil Biol Biochem 107:19–31
Li YF, Hu SD, Chen JH, Müller K, Li YC, Fu WJ, Lin ZW, Wang HL (2018) Effects of biochar application in forest ecosystems on soil properties and greenhouse gas emissions: a review. J Soils Sediments 18:546–563
Liang C, Fujinuma R, Balser TC (2008) Comparing PLFA and amino sugars for microbial analysis in an Upper Michigan old growth forest. Soil Biol Biochem 40:2063–2065
Margesin R, Jud M, Tscherko D, Schinner F (2009) Microbial communities and activities in alpine and subalpine soils. FEMS Microbiol Ecol 67:208–218
Mcauliffe KS, Hallas LE, Kulpa CF (1990) Glyphosate degradation by Agrobacterium radiobacter isolated from activated sludge. J Ind Microbiol Biotechnol 6:219–221
Newman MM, Hoilett N, Lorenz N, Dick RP, Liles MR, Ramsier C, Kloepper JW (2016a) Glyphosate effects on soil rhizosphere-associated bacterial communities. Sci Total Environ 543:155–160
Newman MM, Lorenz N, Hoilett N, Lee NR, Dick RP, Liles MR, Ramsier C, Kloepper JW (2016b) Changes in rhizosphere bacterial gene expression following glyphosate treatment. Sci Total Environ 553:32–41
Nguyen DB, Rose MT, Rose TJ, Morris SG, van Zwieten L (2016) Impact of glyphosate on soil microbial biomass and respiration: a meta-analysis. Soil Biol Biochem 92:50–57
Pizarro H, Vera M, Vinocur A, Pérez G, Ferraro M, Helman RM, dos Santos Afonso M (2016) Glyphosate input modifies microbial community structure in clear and turbid freshwater systems. Environ Sci Pollut Res 23:5143–5153
Rick WY, Thomas SM (2001) Microbial nitrogen cycles: physiology, genomics and applications. Curr Opin Microbiol 4:307–312
Shehata AA, Schrödl W, Aldin AA, Hafez HM, Krüger M (2013) The effect of glyphosate on potential pathogens and beneficial members of poultry microbiota in vitro. Curr Microbiol 66:350–358
Stratton GW, Stewart KE (1992) Glyphosate effects on microbial biomass in a coniferous forest soil. Environ Toxicol 7:223–236
Sun YC, Chen YC, Tian ZX, Li FM, Wang XY, Zhang J, Xiao ZL, Lin M, Gilmartin N, Dowling DN (2005) Novel AroA with high tolerance to glyphosate, encoded by a gene of Pseudomonas putida 4G-1 isolated from an extremely polluted environment in China. Appl Environ Microbiol 71:4771–4776
Tian YS, Xiong AS, Xu J, Zhao W, Gao F, Fu XY, Xu H, Zheng JL, Peng RH, Yao QH (2010) Isolation from Ochrobactrum anthropi of a novel class II 5-enopyruvylshikimate-3-phosphate synthase with high tolerance to glyphosate. Appl Environ Microbiol 76:6001–6005
Toledohernández C, Bonesgonzález A, Ortizvázquez OE, Sabat AM, Bayman P (2007) Fungi in the sea fan Gorgonia ventalina: diversity and sampling strategies. Coral Reefs 26:725–730
Tong Z, Bischoff M, Nies L, Applegate B, Turco RF (2007) Impact of fullerene (C60) on a soil microbial community. Environ Sci Technol 41:2985–2991
Vance ED, Brooks PC, Jenkinson DS (1987) An extraction method for measuring soil microbial biomass. Soil Biol Biochem 19:703–707
Vazquez P, Holguin G, Puente ME, Lopezcortez A, Bashan Y (2000) Phosphate-solubilizing microorganisms associated with the rhizosphere of mangroves in a semiarid coastal lagoon. Biol Fert Soils 30:460–468
Wang MC, Gong M, Zang HB, Hua XM, Yao J, Pang YJ, Yang YH (2006) Effect of methamidophos and urea application on microbial communities in soils as determined by microbial biomass and community level physiological profiles. J Environ Sci Health B 41:399–413
Wang MC, Liu YH, Wang Q, Gong M, Hua XM, Pang YJ, Hu S, Yang YH (2008) Impacts of methamidophos on the biochemical, catabolic, and genetic characteristics of soil microbial communities. Soil Biol Biochem 40:778–788
Weaver MA, Krutz LJ, Zablotowicz RM, Reddy KN (2007) Effects of glyphosate on soil microbial communities and its Mississippi soil. Pest Manag Sci 63:388–393
White DC, Findlay RH (1988) Biochemical markers for measurement of predation effects on the biomass, community structure, nutritional status, and metabolic activity of microbial biofilms. Hydrobiologia 159:119–132
Wu VC, Fung DY, Kang DH, Thompson LK (2001) Evaluation of thin agar layer method for recovery of acid-injured foodborne pathogens. J Food Prot 64:1067–1071
Zhang CP, Hu XQ, Luo JY, Wu ZY, Wang L, Li B, Wang YL, Sun GC (2015) Degradation dynamics of glyphosate in different types of citrus orchard soils in China. Molecules 20:1161–1175
Zhou JZ, Bruns MA, Tiedje JM (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322
Zhou M, Xu HL, Wei XL, Ye ZQ, Wei LP, Gong WM, Wang YQ, Zhu Z (2006) Identification of a glyphosate-resistant mutant of rice 5-enolpyruvylshikimate 3-phosphate synthase using a directed evolution strategy. Plant Physiol 140:184–195
Funding
This work was financially supported by the Program for Changjiang Scholars and Innovative Research Team in University (IRT_14R27), the National Natural Science Foundation of China (NSFC) (31372140, 30971871, 40371071), and the Fundamental Research Funds for the Central Universities (No. 020814380002).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible editor: Diane Purchase
Electronic supplementary material
ESM 1
(DOCX 283 kb)
Rights and permissions
About this article
Cite this article
Liu, Y., Li, Y., Hua, X. et al. Glyphosate application increased catabolic activity of gram-negative bacteria but impaired soil fungal community. Environ Sci Pollut Res 25, 14762–14772 (2018). https://doi.org/10.1007/s11356-018-1676-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-018-1676-0