Abstract
Although mercury contamination of fish is a widespread phenomenon, its regional evaluation is hindered by the reluctance of permitting agencies to grant collection permits, problems in securing adequate freezer space, and time to process whole, large fish or filets. We evaluated mercury concentrations in 210 filet biopsies from 65 sites in 12 western states relative to whole-body mercury concentration in the same fish. We found a highly significant relationship (r2 = 0.96) between biopsy and whole-fish mercury concentrations for 13 piscivorous and nonpiscivorous fish species. We concluded that relative to conventional fish-tissue sampling and analysis procedures for whole fish or filets, the biopsy procedure for mercury in fish tissue is nonlethal, less cumbersome, more likely to be permitted by fisheries agencies, and a precise and accurate means for determining both filet and whole-fish mercury concentrations.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
We provide a model for predicting whole-fish mercury concentrations based on a nonlethal muscle biopsy procedure (Pearson 2000). We believe this technique is an improvement over conventional sample collection and preparation procedures.
Contamination of fish tissue with mercury is a long-standing concern (Tsubaki and Irukayama 1977; Wiener and Spry 1996). Contamination usually is assessed in one of two ways: either by analyzing filets relative to human health exposure or analyzing whole fish relative to fish and wildlife exposure. Either approach is fraught with problems because both require collecting and killing fish, usually freezing fish, homogenizing the entire fish or an entire filet, and subsampling before mercury analysis by conventional atomic absorption. Fisheries agencies are becoming more reluctant to issue collection permits, especially in areas where threatened or endangered species might be encountered. Shipping whole fish from the field to the analytical laboratory is costly. Homogenization of whole fish or fish filets is both storage (freezer space) and labor intensive. Sample preparation involves cutting, heavy duty grinding, dilution, cleaning of all equipment between samples, and subsampling of the homogenate. The possibility of contamination and/or error exists at all sample-preparation stages.
A partial solution to these problems is to capture live fish, biopsy their muscle tissue (Williamson 1992), and return them alive to the water body. Waddell and May (1995) used Williamson’s biopsy protocol to characterize selenium content in the muscle tissue (1 to 2 cm below the dorsal fin) of live razorback suckers (Xyrauchen texanus). Because the biopsies were limited in mass, they were analyzed by gamma-ray spectrometry. Muscle tissue samples taken adjacent to the biopsies for comparison were prepared and analyzed by a more conventional and less-expensive method (freeze dried; homogenized; subsample prepared with nitric, perchloric, and hydrochloric acids; and analyzed for selenium by hydride generation atomic-absorption spectrometry [AAS]). They determined from these analyses that there was no difference between biopsy and whole filet. Also, they concluded that selenium concentrations in biopsies from the dorsal fin and posterior muscle areas were virtually the same, whereas those from the anterior muscle areas were slightly lower.
Pearson (2000) evaluated mercury concentrations in biopsies (anterior, dorsal, and posterior) against mercury concentrations in the whole filets of 11 walleye (Sander vitreus) and 18 northern pike (Esox lucius). He concluded that the mercury concentrations in biopsies and filets from the same fish were not significantly different. He concluded further that dorsal muscle biopsies were slightly more accurate predictors of filet mercury concentrations than biopsies from the anterior or posterior areas of the filet. The dorsal sampling area was reaffirmed to be best by Cizdziel et al. (2002) during their evaluation of the cold-vapor AAS method for measuring mercury in fish tissue against the newer combustion AAS (CAAS) method (United States Environmental Protection Agency [USEPA] 1998). They concluded that the two mercury measurement methods provide statistically equivalent results.
Thus, studies indicate a consistent relationship between biopsy and filet mercury concentrations (Pearson 2000) and between filet and whole-fish mercury concentrations (Goldstein et al. 1995). Baker et al. (2004) determined that biopsies of northern pike are not lethal and pose no greater risk to the fish than routine “tagging.” However, there are no published results directly relating biopsy mercury concentrations to whole-fish mercury concentrations. We are interested in regional-scale surveys of whole-fish mercury concentrations as indicators of contamination and potential risk to piscivorous wildlife. However, we want to avoid the problems of whole-fish processing, so we evaluated the nonlethal biopsy procedure (Pearson 2000) coupled with direct mercury analysis (USEPA 1998) as a means to predict whole-fish mercury concentrations.
Approach
Probability Sampling Design and Sample Collection
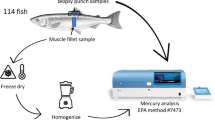
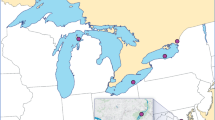
Stream and river sampling sites were drawn from Arizona, California, Colorado, Idaho, Montana, Nevada, North Dakota, Oregon, South Dakota, Utah, Washington, and Wyoming, on a probability basis, from all lotic waters appearing on the 1:100,000-scale Digital Line Graph database of the United States Geological Survey (1989). Sample site selection followed a procedure that recognized the continuous nature of lotic systems, controlled for spatial distribution, and considered their variable spatial density (Herlihy et al. 2000). We collected fish from streams and rivers according to wadeable and nonwadeable electrofishing protocols (Peck et al. 2003a, 2003b). Field crews were directed to collect 1 to 5 large fish (>120 mm long) of various sizes for each species encountered. From June through September 2001, we collected 210 fish from 13 species at 65 sites in 12 western USA, states (Fig. 1). Fish were measured, wrapped in aluminum foil, double plastic bagged, labeled, and iced for shipment to the analytical laboratory the next day. The fish originally were collected for conventional whole-body mercury concentration analysis. The opportunity to compare biopsy mercury concentrations with whole-body mercury concentrations presented itself only after fish had been frozen 4 to 6 months after the summer 2001 field season. Thus, the potentially nonlethal biopsy sampling was done in the laboratory, on previously frozen fish, rather than in the field on live fish as might be done in practice.
Laboratory Procedures
We wanted to know the stability of biopsy samples during freezer storage, i.e., did potential moisture loss change mercury concentrations in the biopsies We took several biopsy plugs from one nonsample northern pikeminnow (Ptychocheilus oregonensis) and placed individual biopsies into three types of storage container for freezing. The storage containers were (1) Packard LSC 20-mL polyethylene scintillation vials, (2) I-Chem brown borosilicate 40-mL vials, and (3) I-Chem clear borosilicate 40-mL vials. Containers of each type were acid soaked (2% HCl) overnight and then rinsed five times with deionized water and air dried before biopsy samples were placed into them for freezing at −20°C. Biopsies from each container type were removed at various intervals during 100 days and analyzed for mercury by the CAAS method (USEPA 1998).
Fish samples received at the analytical laboratory were frozen at −20°C and stored until analyzed. At the time of analysis, fish were removed from the freezer, reweighed, remeasured, and allowed to partially thaw. Biopsies were removed from partially thawed fish in a manner used on live fish (Pearson 2000). The protocol consisted of (1) scraping a few scales from the anterior end of and 1 to 2 cm below the dorsal fin on the left side; (2) inserting a disposable 6-mm diameter biopsy punch (Biopunch, Fray Products, Buffalo, NY), with a slight twisting motion, to cut through the skin and into the tissue; (3) tilting or bending the punch slightly once it was inserted to full depth (8 mm) to break off the end of the tissue sample; (4) removing the punch carefully to retain the sample; (5) removing the sample with the self-contained punch removal tool or a scalpel (alternate procedure was to blow sample out with a pipette bulb); (6) trimming skin from the end of the plug; and (7) placing the plug into a sterile 20-mL scintillation vial or other suitable container for refreezing at −20°C until analysis. Pearson (2000) used 5-mm punches but indicated that inadequate amounts of sample were sometimes obtained for conventional mercury analysis. Therefore, after some experimentation we chose 6-mm biopsy punches. Biopsies were sliced in half lengthwise at the time of analysis and run as duplicates. Results were reported as means of the duplicates.
The remainder of the whole fish was then ‘‘chunked’’ with stainless steel knives and blended with an approximate 1:1 ratio of deionized water by weight until the mixture appeared to be completely homogeneous. The exact amount of water added was recorded and used to adjust the amount of mercury in the ‘‘as-received’’ fish samples at the time of analysis. All cutting utensils, cutting surfaces, and blenders were cleaned with hot soapy water and rinsed three times with deionized water between samples to prevent cross contamination. Subsamples were removed immediately from the whole-fish sample to prevent separation of lipids, and the subsamples were refrozen at −20°C in sterilized 40-mL glass, screw-cap vials until analysis. At the time of analysis, homogenate samples were thawed, remixed with two or three replicate aliquots removed, and analyzed. Results were reported as means of the two or three replicate analyses.
Both biopsy and whole-fish samples were analyzed using CAAS (Milestone DMA80 direct Mercury Analyzer, Milestone, Monroe, CT) according to USEPA (1998). After each thermal decomposition analysis of fish tissue, ash was removed from the sample weigh boats, the boats were soaked in deionized water for 30 minutes and heated to 550°C for 1 hour, then cooled before their next use. A major advantage of the CAAS method is that it requires a small sample (approx. 0.25 g) and no sample preparation (direct mercury analysis).
Quality Assurance
An instrument detection limit (IDL) of 0.05 ng mercury was determined by replicate analysis of acidified, aqueous mercury standard solutions to assess precision. This corresponds to an IDL of 0.0003 μg mercury/g for a nominal fish-tissue sample of 0.25 g. However, tissue analysis is more complex than standards, and the method detection limit (MDL) for fish tissue is expected to be >0.0003 μg mercury/g.
The thermal decomposition-amalgamation method for mercury analysis combines the release of mercury from the matrix and analysis in a single step. Therefore, doing an MDL study by spiking mercury (in acidified, aqueous solution) will not give the best estimate of MDL because the mercury added is already in a ‘‘released’’ form. In addition, it is nearly impossible to obtain mercury-free fish tissue. Thus, we used the method of Taylor (1987) to estimate the MDL for tissue samples. This method is essentially the same as the Environmental Monitoring and Assessment Program (EMAP) protocol for MDL determination (USEPA 1997) except a sample is used rather than fortifying a clean matrix. The SD for 202 measurements of Standard Reference Material (SRM) 2976 made during the previous year’s (2000) fish-tissue analyses was used to estimate the MDL for tissue samples (0.02 μg mercury/g). Also, analyses included 103 tissue samples run in duplicate. The relative SD (RSD) of the duplicates ranged from 0.01% to 25.8% (mean 3.66%). In addition, 215 replicates of dogfish reference material DORM-2 and 202 replicates of SRM 2976 mussel tissue were analyzed with the project samples. Both groups met the precision objective of 15% RSD or MDL, whichever was larger.
Accuracy was assessed by analysis of the reference materials DORM-2 (high level) and SRM 2976 (low level) as calibration checks during sample analysis. DORM-2 has a certified total mercury concentration of 4.64 μg mercury/g. Only 1 of the 215 replicate results slightly exceeded the criteria of 85% to 115% (average = 101% ± 4.4%). The certified total mercury value for SRM 2976 is 0.0610 μg mercury/g. The average recovery from 202 samples of this reference material was 115.1% ± 13.3%, which is an acceptable result because the certified value is within a factor of 5 of the estimated MDL for this method. Each day of analysis, reagent blanks (2% nitric acid) were analyzed after the standards were analyzed to ensure there was no carryover of mercury before environmental samples were analyzed. No samples were analyzed until acceptable blanks were obtained (corresponds to approximately 0.0004 μg mercury/g or approximately 0.0001 μg mercury in a typical 0.25-g environmental sample).
Fish-Tissue Data Analysis
We used all of the fish in our database (n = 210) to describe the relationship between biopsy and whole-body mercury concentration except for two pikeminnows that were outliers relative to the rest of the database. Thus, 208 individual fish comprised the total sample size for our primary analysis.
Biopsy and whole-body mercury concentrations can be viewed as two similar measurements that both have natural variation and/or measurement error. Thus, we considered using a geometric mean functional relationship (GMFR; Ricker 1973) rather than ordinary least squares (OLS) regression to relate the two quantities. GMFR clearly is an appropriate tool where the functional relationship itself (for example, whether two measurements are equivalent) is of primary interest (Schmitt and Finger 1987).
Our goal was not to identify a functional relationship per se. Instead, we sought to predict one measurement (whole-body mercury concentration) from the other (biopsy mercury concentration). Sokal and Rohlf (1981) argued that OLS is the preferred tool for making predictions, in part because GMFR does not supply confidence limits for regression parameters or predictions. Moreover, in our prediction setting, the value of ‘‘x’’ (biopsy mercury concentration) is best viewed as a fixed realization of a random variable (Sprent and Dolby 1980). Thus, having observed ‘‘x’’ and lacking an informative measurement error model, we can only take ‘‘x’’ at face value and use it to predict ‘‘y’’ (whole-body mercury concentration). In this situation, the maximum likelihood estimates of regression parameters are OLS estimates (Sprent and Dolby, 1980; Draper and Smith 1998). For these reasons, we employed OLS rather than GMFR in our regressions.
We regressed log10 whole-body mercury concentrations against log10 biopsy mercury concentrations for all (except two pikeminnow outliers) individual fish (n = 208; 13 species) to serve as a predictive ‘‘base model.’’ In addition, we explored two sets of more complex models. In the first set, fish species were grouped by their primary diets into (1) nonpiscivorous (brook trout [Salvelinus fontinalis], brown trout [Salmo trutta], channel catfish [Ictalurus punctatus], cutthroat trout [Oncorhynchus clarki], rainbow trout [O. mykiss], white sucker [Catostomus commersoni]) or (2) piscivorous (largemouth bass [Micropterus salmoides], smallmouth bass [M. dolomieu], northern pike [Esox lucius], northern pikeminnow [Ptychocheilus oregonensis], sauger [Sander canadensis], walleye [S. vitreus], and yellow perch [Perca flavescens]). Separate intercepts and/or log10 biopsy mercury slopes were determined for each dietary group.
In the second set of relationships, separate log10 biopsy mercury slopes and/or intercepts were determined for each of the 10 species having more than 1 fish in our data (n = 204 fish). Both sets of models were compared to the base model using extra-sum-of-squares F tests (Myers 1990).
Results and Discussion
Data Description
Dots in Figure 1 indicate sampling sites, and numbers indicate the number of fish sampled from that site. No fish were collected from streams in Idaho because of collection permit limitations. Similar problems arose in California, Oregon, and Washington after initial sampling. However, sampling sites were reasonably well distributed across the region.
The scatterplot of biopsy mercury versus whole-body mercury for our 210-fish database approximated a linear relationship between biopsy mercury ranging from near detection limits to >2.0 μg/g and whole-body mercury ranging from near detection limits to >1.0 μg/g (Fig. 2). The straight line in the scatter plot is the 1:1 line, which clearly demonstrates that all biopsy mercury concentrations were greater than whole-body mercury concentrations, except for the two anomalous pikeminnow samples (omitted from our regression analysis) in the lower left of Figure 2. An assessment of quality assurance procedures indicated that both biopsy and whole-fish mercury concentrations for those samples were correct as measured. We have no clear explanation for the two outliers. We found nothing unusual about these two fish or other fish from the same site. The result suggested contamination of these two whole-fish samples, which is possible given the numerous sample preparation steps for whole-fish analysis. We are proposing the use of biopsy to predict whole-body mercury concentrations, thus an error or contamination of biopsies would be more problematic than the suspicion of whole-body mercury concentration error as in this case. The two data points in Figure 2 were retained in our database for completeness, but were not part of our data analysis.
Although the relationship between biopsy mercury and whole-fish mercury in Figure 2 appears approximately linear, scatter increases markedly as concentrations increase. Because heteroscedastic residual variance can seriously affect the quality of a normal-theory regression model (Myers 1990), we log-transformed both the biopsy and whole-body mercury concentrations before regression analysis. The log-transformed variables still approximate a linear relation-ship, and the residual variance is effectively stabilized (Fig. 3).
Log10-transformed whole-body mercury versus biopsy mercury concentration (μg/g) for all fish in the sample (13 species, n = 210). Regression line and pointwise 95% CIs on an individual prediction are from model A. in Table 3. CI: Confidence interval.
The histograms in Figure 2 represent the relative frequency of biopsy (top) and whole-body (right) mercury concentrations for fish. The mean, minimum and maximum fish size and their respective biopsy mercury concentrations are summarized by species (Table 1). Both fish sizes and mercury concentrations (filet equivalent to biopsy) in our database were within the ranges of several similar fish-tissue surveys (Schmitt and Brumbaugh 1990; Goldstein, et al., 1995; Brumbaugh et al. 2001), but none of our samples reached the maximum filet mercury concentrations reported for some species (smallmouth bass maximum = 1.05 μg mercury/g; largemouth bass maximum = 4.22 μg mercury/g) by Brumbaugh et al. (2001). There could be several reasons for this, but it is most likely related to the fact that these other studies were targeted toward large rivers and areas of known or suspected high mercury concentrations in fish (Peterson et al. 2002). A probability-based sampling design, such as the one we employed, produces samples more representative of the population at large (potentially fish from any stream appearing on 1:100,000 scale maps) than does a targeted sampling program. Because probability sampling accurately represents the entire population, and extreme values are rare in the population, the extremes are unlikely to be captured by the samples; thus, explaining in part, why mercury tissue concentrations in our survey samples did not reach the high levels observed by Brumbaugh et al. (2001).
Biopsy Stability
Biopsy mercury concentration did not change during a period of 100 days in any of the biopsy storage container types we tested, and there were no significant container dependent differences in mercury concentration among the three container types (Table 2). Based on this information and our own convenience, we selected 20-mL polyethylene scintillation vials for biopsy storage for all fish collected from sites in Figure 1. Biopsy storage times ranged from 59 to 189 days until analysis. The average storage time was 98 days, and the median storage time was 91 days. Sixty-nine percent of the samples were analyzed in <100 days after biopsy.
Predictive Model
The simple base model explained >95% of the variance in log10 whole-body mercury concentration from log10 biopsy mercury alone (model A, Table 3; and Fig 3). No additional variance was explained when separate slopes and intercepts were fitted for piscivorous versus nonpiscivorous groups (F2, 204 = 0.22, p > 0.5). However, when separate intercepts and a common slope were specified for each of the 10 species having >1 observation, there was a slight, but statistically significant, improvement relative to the base model (F9, 194 = 2.38, p = 0.014). Inclusion of separate log10 biopsy mercury slopes for each of 10 species improved neither the base model (F9, 194 = 1.53, p = 0.14) nor the separate-intercepts model (F9, 185 = 1.46, p = 0.17).
As a result, we accepted the separate-intercepts, common-slope model (model B, Table 3) as a potentially useful alternative to the base model. However, we found very little difference in model B intercepts across the species relative to their standard errors (Table 3). Likewise, we saw a great deal of overlap across species-specific confidence intervals (CIs) on predicted values of whole-body mercury (Table 3). Finally, we noted that predictions from the species-specific model (model B) would involve extrapolation if they were used outside the fairly limited range of whole-body and biopsy mercury levels seen for any single species in our data set. The base model, however, has predictive reliability across the full range of mercury concentrations seen across all fish (Fig. 3). For these reasons, we recommend use of the simple base model (model A) for predicting whole-body mercury.
The relationships between biopsy mercury and whole-body mercury concentrations are relatively consistent across a variety of fish species (Table 3; Goldstein et al. 1995). The narrow confidence bands and limited scatter in Figure 3 permit an accurate prediction of whole-body mercury concentrations based on the equation:

Model intercept interpretation is an elementary result of simple linear regression analysis. The −0.2712 intercept from Equation 1 is the predicted value of log10 (whole-body mercury) when biopsy mercury = 1.0 μg/g, i.e., when log10 (biopsy mercury) = 0. In other words, when biopsy mercury = 1.0 μg/g, the model predicts that whole-body mercury will = 10−0.2712 = 0.54 μg/g. Model predicted values of whole-body mercury concentrations and their associated 95% CIs were calculated for model A and for each of the 10 fish species represented by >1 sample. A summary of the whole-body predicted values when plug values equaled 0.5 and 1.0 μg mercury/g are shown in the last 2 columns of Table 3. The 0.5-μg mercury/g level represents the human advisory action level of the World Health Organization, (WHO) whereas the 1.0-μg mercury/g level represents the United States Food and Drug Administration action level for commercially sold fish (Carpenter 1998). Thus, both figures have significance relative to human consumption and health. Although our interest is not the influence of mercury on human health, the health-related benchmarks are useful in denoting potential related effects of mercury on wildlife. For example, note that predicted whole-body mercury concentration from model A (Table 3), and for each fish species, based on even the lowest of the two health-related threshold concentrations (0.5 μg mercury/g), exceeds the critical value for consumption by piscivorous mustelids (0.1 μg mercury/g) by a factor greater than two (Yeardley et al. 1998).
Recommendations and Conclusions
Our biopsies were obtained from previously frozen fish, thus we do not claim to have tested the nonlethal biopsy method per se. However, we have no reason to think that our frozen biopsies would differ from live fish biopsies, which likely would be frozen before analysis anyway. Others (Williams, 1992; Waddell and May, 1995; Pearson 2000; Baker et al 2004) have successfully used nonlethal biopsy procedures on live fish. Pearson (personal communication, June 4, 2003) indicated that some of the fish captured during his study in 2000 exhibited scars from a previous biopsy. Despite the scars, the fish showed no other adverse effects. Baker et al. (2004) found northen pike biopsies nonlethal. Also, there is little reason to suspect a 6 × 8-mm biopsy would be any more lethal than comparably sized fish tags (Everhart et al. 1975; Wydoski and Emery 1983). However, both mortality and frozen versus fresh tissue samples should be evaluated for more species.
To our knowledge, no one has coupled biopsy directly with whole-fish mercury analysis on the same fish across a wide variety of species and fish sizes. Goldstein et al. (1995) focused on a few species of fish with limited sizes, but did not employ biopsies. Our study combined biopsies across 13 fish species of various sizes with direct mercury analysis (the CAAS mercury method). This combination of biopsies (Pearson 2000) with direct mercury analysis (USEPA 1998) offers several advantages over conventional sampling and analysis procedures. Biopsies do not intentionally kill fish. Biopsies are small and thus do not require large freezer storage capacity or high sample-shipping costs. Direct mercury analysis (CAAS method) uses a small sample (0.25 g) with no sample preparation and thus avoids hours of laboratory labor and multiple opportunities for contamination or error.
The regression of whole-body log10 mercury concentrations against log10 biopsy mercury concentrations produced a whole-fish predictive model that is both accurate and robust based on the analysis of 210 various sized fish representing 13 species. Overall, the biopsy-direct mercury analysis procedure is a much simplified and much improved procedure relative to conventional sample preparation and analysis for fish-tissue mercury concentrations for both filet and whole-body analysis.
References
RF Baker PJ Blanchfield MJ Paterson RJ Flett L Wesson (2004) ArticleTitleEvaluation of nonlethal methods for the analysis of mercury in fish tissue Trans Am Fish Soc 133 568–576 Occurrence Handle10.1577/T03-012.1 Occurrence Handle1:CAS:528:DC%2BD2cXlsVCqsLs%3D
Brumbaugh WG, Krabbenhoft DP, Helsel DR, Wiener JG, Echols KR (2001) A national pilot study of mercury contamination of aquatic ecosystems along multiple gradients: Bioaccumulation in fish. USGS/BRD/BSR—2001–0009, pp 25
Carpenter H (1998) Mercury in the Midwest: State public health agency perspective. In: Mercury in the Midwest: Current status and future directions (EPA/905/R-98/003 research report). United States Environmental Protection Agency, Chicago, IL pp 33–38
JV Cizdziel TA Hinners EM Heithmar (2002) ArticleTitleDetermination of total mercury in fish tissues using combustion atomic absorption spectrometry with gold amalgamation Water Air Soil Pollut 135 355–370 Occurrence Handle10.1023/A:1014798012212 Occurrence Handle1:CAS:528:DC%2BD38Xjt1Gls7k%3D
NR Draper H Smith (1998) Applied regression analysis, EditionNumber3 Wiley New York, NY 706
WH Everhart AW Eipper WD Youngs (1975) Principles of fishery science Cornell University Press Ithaca, NY 288
RM Goldstein ME Brigham JC Stauffer (1995) ArticleTitleComparison of mercury concentrations in liver, muscle, whole bodies, and composites of fish from the Red River of the North Can J Aquat Sci 53 244–252 Occurrence Handle10.1139/f95-203
AT Herlihy DP Larsen SG Paulsen NS Urquhart (2000) ArticleTitleDesigning a spatially balanced, randomized site selection process for regional stream surveys Environ Monit Assess 63 95–113 Occurrence Handle10.1023/A:1006482025347 Occurrence Handle1:CAS:528:DC%2BD3cXltFShsL8%3D
RH Myers (1990) Classical and modern regression with applications EditionNumber2 PWS-Kent Boston, MA 488
Pearson E (2000) The analysis of mercury in fish tissue plugs for the purpose of evaluating a potentially non-lethal sampling method. North Dakota Department of Health Report. North Dakota Department of Health, Bismarck, ND, pp 9
DV Peck DK Averill JM Lazorchak DJ Klemm (2003a) Field operations and methods for non-wadeable rivers and streams, draft method United States Environmental Protection Agency Corvallis, OR 198
DV Peck JM Lazorchak DJ Klemm (2003b) Field operations manual for wadeable streams, draft method United States Environmental Protection Agency Corvallis, OR 255
SA Peterson AT Herlihy RM Hughes KL Motter JM Robbins (2002) ArticleTitleLevel and extent of mercury contamination in Oregon, USA, lotic fish Environ Toxicol Chem 21 2157–2164 Occurrence Handle10.1002/etc.5620211019 Occurrence Handle1:CAS:528:DC%2BD38XntlGjtLk%3D
WE Ricker (1973) ArticleTitleLinear regressions in fishery research J Fish Res Board Can 30 409–434 Occurrence Handle10.1139/f73-072
CJ Schmitt WG Brumbaugh (1990) ArticleTitleNational contaminant biomonitoring program: Concentration of arsenic, cadmium, copper, lead, mercury, selenium, and zinc in U.S. freshwater fish, 1976–1984 Arch Environ Contam Toxicol 19 731–747 Occurrence Handle10.1007/BF01183991 Occurrence Handle1:CAS:528:DyaK3cXlvFOgu70%3D
CJ Schmitt SE Finger (1987) ArticleTitleThe effects of sample preparation on measured concentrations of eight elements in edible tissues of fish from streams contaminated by lead mining Arch Environ Contam Toxicol 16 185–207 Occurrence Handle10.1007/BF01055800 Occurrence Handle1:CAS:528:DyaL2sXhtlegtLs%3D
RR Sokel FJ Rohlf (1981) Biometry EditionNumber2 WH Freeman New York, NY 859
P Sprent GR Dolby (1980) ArticleTitleThe geometric mean functional relationship Biometrics 36 547–550 Occurrence Handle10.2307/2530224
JK Taylor (1987) Quality assurance of chemical measurements Lewis Publishers Chelsea, MI 329
Tsubaki T, Irukayama K, (eds) (1977) Minamata disease: Methylmercury poisoning in Minamata and Nigata, Japan Elsevier, Amsterdam, The Netherlands, pp 317
InstitutionalAuthorNameUnited States Environmental Protection Agency (1997) Environmental monitoring and assessment program: Integrated quality assurance project plan for surface waters research activities, section 2 data quality objectives (pp 13-22) United States Environmental Protection Agency Corvallis, OR 139
InstitutionalAuthorNameUnited States Environmental Protection Agency (1998) Mercury in solids and solutions by thermal decomposition, amalgamation, and atomic absorption spectrophotometry, draft method 7473 United States Environmental Protection Agency Washington, DC
InstitutionalAuthorNameUnited States Geological Survey (1989) Digital line graphs from 1:100,000-scale maps; Users guide 2 United States Geological Survey Reston, VA 88
B Waddell T May (1995) ArticleTitleSelenium concentrations in the razorback sucker (Xyrauchen texanus): Substitution of non-lethal muscle plugs for muscle tissue in contaminant assessment Arch Environ Contam Toxicol 28 321–326 Occurrence Handle10.1007/BF00213109 Occurrence Handle1:CAS:528:DyaK2MXltFegt7w%3D
JG Weiner DJ Spry (1996) Toxicological significance of mercury in freshwater fish WN Beyer GH Heinz AW Redmon-Norwood (Eds) Environmental contaminants in wildlife: Interpreting tissue concentrations Lewis Boca Raton, FL 297–339
JH Williamson (1992) Colorado squawfish genetic survey—Tissue sampling protocol 1992 United States Fish and Wildlife Service, Fisheries and Federal Aid, Denver Federal Center Denver, CO 12
R Wydoski L Emery (1983) Tagging and marking LA Nielsen DL Johnson (Eds) Fisheries techniques American Fisheries Society Bethesda, MD 215–237
RB Yeardley SuffixJr JM Lazorchak SG Paulsen (1998) ArticleTitleElemental fish tissue contamination in northeastern U.S. lakes: Evaluation of an approach to regional assessment Environ Toxicol Chem 17 1875–1884 Occurrence Handle10.1002/etc.5620170931 Occurrence Handle1:CAS:528:DyaK1cXlslGrtbs%3D
Acknowledgments
The USEPA funded this research as part of the Environmental Monitoring and Assessment Program supported by Contract No. 68D01005 with Dynamac Corporation. Mention of commercial products in this article does not constitute endorsement by the USEPA. This article was submitted to the USEPA’s peer and administrative review and approved for publication. We thank several people who assisted with the preparation of this paper: M. Hails-Avery, K. Baxter, W. Brumbaugh, R. Hjort, J. Lazorchak, F. McCormick, K. Motter, S. Pierson, C. Schmitt, and anonymous reviewers whose comments helped considerably in the final preparation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Peterson, S.A., Van Sickle, J., Hughes, R.M. et al. A Biopsy Procedure for Determining Filet and Predicting Whole-Fish Mercury Concentration. Arch Environ Contam Toxicol 48, 99–107 (2004). https://doi.org/10.1007/s00244-004-0260-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00244-004-0260-4