Abstract
Group I introns are mobile RNA enzymes (ribozymes) that encode conserved primary and secondary structures required for autocatalysis. The group I intron that interrupts the tRNA-Leu gene in cyanobacteria and plastids is remarkable because it is the oldest known intervening sequence and may have been present in the common ancestor of the cyanobacteria (i.e., 2.7–3.5 billion years old). This intron entered the eukaryotic domain through primary plastid endosymbiosis. We reconstructed the phylogeny of the tRNA-Leu intron and tested the in vitro self-splicing ability of a diverse collection of these ribozymes to address the relationship between intron stability and autocatalysis. Our results suggest that the present-day intron distribution in plastids is best explained by strict vertical transmission, with no intron losses in land plants or a subset of the Stramenopiles (xanthophyceae/phaeophyceae) and frequent loss among green algae, as well as in the red algae and their secondary plastid derivatives (except the xanthophyceae/phaeophyceae lineage). Interestingly, all tested land plant introns could not self-splice in vitro and presumably have become dependent on a host factor to facilitate in vivo excision. The host dependence likely evolved once in the common ancestor of land plants. In all other plastid lineages, these ribozymes could either self-splice or complete only the first step of autocatalysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Group I introns are intervening sequences that are able to catalyze their own removal from pre-RNA (i.e., are “self-splicing” [Cech 1990]). This ability, coupled with their capacity for information storage, suggests that some of these elements may be remnants of the RNA world (Pace and Marsh 1985; Gilbert 1986; Moore et al. 1993). However, the majority of existing introns are more likely to be relatively recent additions to eukaryotic genomes (e.g., Goddard and Burt 1999; Bhattacharya et al. 2001, 2002) with one striking exception, the group I intron that interrupts the anticodon loop (U–intron–AA) of the tRNA-Leu gene in many cyanobacteria and the plastids of plants and algae (Kuhsel et al. 1990; Xu et al. 1990; Paquin et al. 1997; Besendahl et al. 2000).
The sister group relationship between the widespread tRNA-Leu intron in cyanobacteria and that in plastids suggests that eukaryotes gained the intron through primary endosymbiosis (Kuhsel et al. 1990; Xu et al. 1990; Besendahl et al. 2000). The putative single plastid endosymbiosis occurred at least one billion years ago (Butterfield 2000; Yoon et al. 2002), and the intron may be significantly older, perhaps present in the common ancestor of all cyanobacteria (i.e., 2.7–3.5 billion years old [Schopf 1993; Paquin et al. 1997; Brocks et al. 1999; Summons et al. 1999]). These characteristics make the tRNA-Leu intron the most ancient intervening sequence known and a model for studying the long-term evolution of autocatalytic RNAs.
Splicing of group I introns is dependent upon a conserved RNA secondary structure consisting of about 10 paired RNA-elements (P1–P10 [Michel and Westhof 1990]). The primary and secondary structure conservation can be an important tool in evolutionary studies because it permits the construction of phylogenies using alignments centered on the intron catalytic core (P, Q, R, and S domains [Cech 1988]). The tRNA-Leu intron lacks structural elements thought to be essential for auto-excision in other group I introns (Price et al. 1985; Barfod and Cech 1988; Beaudry and Joyce 1990; Van der Horst et al. 1991). Despite this characteristic, a cyanobacterial intron (Anabaena) has been shown to self-splice efficiently in vitro (Zaug et al. 1993). Conversely, limited studies in land plants suggest loss of auto-catalysis in these lineages (Xu et al. 1990; Daros and Flores 1996; Besendahl et al. 2000). In this study, we assessed the in vitro self-splicing ability of the tRNA-Leu intron across its entire phylogenetic distribution. Broad patterns in self-splicing ability were determined by mapping this character onto a phylogeny of the tRNA-Leu intron in cyanobacteria and algae. Our study shows that this ancient intron has followed a number of different evolutionary trajectories over its history that form a model for understanding group I intron evolution in general.
Materials and Methods
Strains Used in the Analyses. The algae and plants used in PCR and/or Southern analyses to determine presence/absence of the tRNA-Leu intron are listed in Table 1. In this study, 79 new taxa were tested for intron presence, including 11 streptophytes, 25 chlorophytes, 28 Stramenopiles, 13 rhodophytes, and 2 glaucophytes.
DNA Extraction, PCR, Slot-Blot Analysis, and Sequencing. Total genomic DNA was extracted according to the instructions of the manufacturer, using either the Plant DNeasy Mini Kit (Qiagen) or the Invisorb Plant Spin Kit (Invitrogen). Cells were frozen in liquid nitrogen and mechanically disrupted using a combination of a minipestle and a cell homogenizer with glass beads. A combination of PCR and slot blot techniques was used to determine the presence or absence of the intron. Initial PCRs employed primers matching conserved exon sequences based on those in Kuhsel et al. (1990). The 5′ and 3′ primers are as follows: LeuF, 5′TGGYGRAATYGGTAGACGCWRCGGAC3′ and LeuR, 5′TGGGGRYRGAGRGACTYGAACYCTCACG3′. Two new sets of intron primers were designed, one specific for the chlorophytes and the second for the Stramenopiles: IntF_CLeu, TGMATGCTCWCAAARTCAGG; IntR_CLeu, 5′TGGACTCTATCTTTATCC3′ IntF_HLeu, AGTAAGTTCTCAAATTCAAG; IntR_HLeu, 5′TTGGACTCTYTCTTTACCSA3′. All combinations of primer sets (i.e., IntF_HLeu/IntR_HLeu, LeuF/IntR_HLeu, IntF_HLeu/Leu R, etc.) were used in all cases when no apparent band was found in the initial PCR. The PCR cycling parameters were as follows: 35 cycles of 94°C for 60 s, 50°C for 60 s, and 72°C for 60 s. Amplification was initiated with a 4-min denaturation at 94°C and concluded with a 7-min extension at 72°C. The PCR product encoding the uninterrupted tRNA-Leu gene was expected to be approximately 78 bp in length. For two red algal species (Porphyridium sordidum, Rhodella violacea) examined in this study, the PCR product was verified by sequencing. In addition, Besendahl et al. (2000), using the same PCR primers, verified the presence of an uninterrupted tRNA-Leu gene from the green alga, Cylindrocystis brebissonii, and the red alga, Compsopogon coeruleus. Products longer than ~78 bp were gel-purified on a 1% agarose gel and sequenced either directly or after cloning into the pGEM-T vector (Promega). Both strands of the insert were sequenced using the ALFexpress II (Amersham Pharmacia), L4200 (Licor), or ABI 3100 (Applied Biosystems) automated sequencers.
In the absence of an obvious band indicating either presence or absence of the intron, slot-blot analysis was done according to the manufacturer’s instructions (BioRad). Genomic DNA from each species, ranging from approximately 200 ng to 5 µg, was transferred onto a nylon membrane (Amersham). The membrane was divided into two sections, one containing only chlorophyte species and the other only Stramenopiles. All probes were random-prime labeled with 32P-dCTP. The membranes were first probed with intron PCR products. For the chlorophyte section, we used the intron from Bracteacoccus minor, and for the Stramenopiles section, we used a mixture of Scytosiphon lomentaria and Vaucheria bursata intron DNA. Hybridizations were done at 55°C for 18 h in 0.24 M Na2HPO4 (pH 7.4), 1 mM EDTA (pH 8.0), 1% BSA, and 0.7% SDS. Membranes were washed twice for 15 min at 55°C and once for 30 min at 60°C in 2× SSC, 0.1% SDS prior to autoradiography. The membranes were subsequently stripped of bound probe using a boiling solution of 1 mM EDTA (pH 8.0), 0.1% SDS and were then rehybridized. Probes were made of the chloroplast psbA gene, using primers psbAF and psbAR2 (see Yoon et al. 2002). The species used for the probes were as follows: chlorophyta, mixture of Chlorella vulgaris, Cladophora albida, and Trentepohlia sp.; and Stramenopiles, mixture of Skeletonema costatum, Bumilleriopsis filiformis, Vacuolaria virescens, and Eustigmatos magnus.
Splicing Assays. The tRNA-Leu intron from a selection of cyanobacteria and plastids was amplified using a modified LeuF and the original LeuR primers. A T7-promoter site was included on the 5′-terminus of the forward primer and used to generate RNA transcripts for in vitro splicing assays. The PCR products contained sufficient flanking exon sequence (5′–28 nt, 3′–50 nt) to allow for the required P1 and P10 intron–exon interactions (Cech et al. 1994). The Klenow enzyme (5 U/µg) was used to fill in the 3′-overhang produced by the PCR reaction. Transcriptions were then performed using the Riboprobe in vitro Transcription System (Promega). To promote splicing, 0.5 µl of 250 mM HEPES and 0.75 µl of 500 mM MgCl2 were added and the reaction was further incubated at 50°C for 15 min. Thereafter, 0.5 µl of 100 µM rGTP was added, and the mixture incubated for an additional 15 min. These conditions are based on the method of Zaug et al. (1993). RNA splicing products were precipitated in 100% ethanol, resuspended in loading buffer, and separated on 6% polyaciylamide/7 M urea gels. The gels were blotted onto nylon membranes (Amersham) and probed with PCR products containing both intron and exon sequences. Probe preparation was done as described above.
An additional splicing experiment was done to clearly distinguish between the first step of splicing (which would produce a 5′ exon fragment and an intron plus 3′ exon fragment) and the first step of the alternative hydrolysis pathway (3′ exon fragment and 5′ exon plus intron fragment [Inouye et al. 1986]). The PCR product from the stramenopile, Bodanella lauterborni (used in the splicing assay above), was cloned into the pGEM-T vector (Promega). This construct was digested with the restriction enzymes ApaI and AlwNI, resulting in a fragment that would produce significantly different 5′ (57-nt) and 3′ (918-nt) exon sizes. This construct allowed us to distinguish between the first step of splicing and hydrolysis pathways. Transcription and splicing assays of the B. lauterbornii were done as previously described using the digested construct as a template.
Phylogenetic Analyses. For the tRNA-Leu group I intron data, a set of 80 sequences (both novel [see Fig. 2] and from GenBank) was manually aligned using SeqApp V1.9a169 (Gilbert 2001). Primary and secondary structure similarity was used to juxtapose homologous intron regions (e.g., Michel and Westhof 1990; Bhattacharya et al. 1994, 2001; Damberger and Gutell 1994). The intron alignment is available upon request from D.B. The intron sequences of Bracteacoccus minor, Hormotilopsis tetravacuolaris (Chlorophyceae) and Bryopsis plumosa, Derbesia marina (Ulvophyceae) were excluded from the phylogenetic analyses because of their high divergence rates (for the ulvophytes, see Besendahl et al. [2000]). In addition, the insertions from the apicomplexians (Plasmodium falciparum, Toxoplasma gondii, Neospora caninumare) could not be used because of their high AT content (>85%). The extreme nucleotide bias precluded the reliable alignment of the apicomplexan introns leaving unresolved their origin. The final data set of unambiguously aligned sequences (201 nt) was submitted to a pairwise distance analysis using the Tajima–Nei (TN; 1984) model with equal divergence rates across sites and the transition/transversion ratio = 2. Missing data and gaps were excluded from each pair-wise comparison. This distance matrix was used as input for neighbor-joining (NJ) tree building. Five hundred bootstrap replicates (Felsenstein 1985) were analyzed with the distance method. The NJ-TN tree was then used as input for estimating the parameters of the general time reversible model (Rodríguez et al. 1990), the proportion of invariant sites that are unable to accept substitutions, and the shape parameter of the gamma distribution to accommodate rate variations across sites. The GTR +I + Γ (GTR) model was used in a NJ analysis to calculate bootstrap values (500 replications) for monophyletic groups identified by the TN-NJ tree. We also generated bootstrap support values (500 replications) for the tRNA-Leu intron data using LogDet (LD) distances and the NJ method to address the possible misleading effects of base composition bias in the data set. And, finally, we did Bayesian analysis of the intron data (MrBayes V2.0 [Huelsenbeck and Ronquist 2001]) using the GTR + Γ model with intron sites partitioned into two groups; the first (50 nt) encoded the most highly conserved P, Q, R, and S regions, whereas the second (151 nt) encoded the remaining, more divergent sequence. Site-specific rates were calculated for each intron partition and incorporated into this analysis. Metropolis-coupled Markov chain Monte Carlo (MCMCMC) from a random starting tree was initiated in the Bayesian inference and run for 500,000 generations. Three independent MCMCMC analyses were run and sampled every 100 generations. After discarding the first 200 trees (“burn-in”), consensus trees were made with the remaining 4800 trees in each run and compared to test for the consistency of posterior probabilities at the different nodes. The cyanobacterial tRNA-Leu introns were used to root the phylogenies.
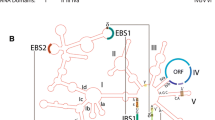
Neighbor-joining–TN tree of tRNA-Leu group I introns from cyanobacteria and plastids. This tree has been rooted on the branch leading to the cyanobacterial introns. The values above the branches on the left of the slash-marks show the results of a NJ–TN bootstrap analysis (500 replications), whereas the values shown on the right of the slash-marks are from a NJ-GTR bootstrap analysis. The bootstrap support values shown below the branches are from a NJ-LD analysis. Only bootstrap values ≥60% are shown. The thicker branches indicate clades that had ≥95% posterior probability in a Bayesian inference. The taxonomic position of the different introns is shown for each clade. The branch lengths are proportional to the number of substitutions per site (see scale in figure). The introns shown in the larger, underlined text were determined in this study.
The distribution of the tRNA-Leu intron was mapped on published host trees of the Chlorophyta and the Stramenopiles. The host phylogenies were adapted from Friedl et al. (1997, 2002) and Buchheim et al. (2001) for the Chlorophyta (nuclear rRNA) and Andersen et al. (1998, 1999), Draisma et al. (2001), Daugbjerg and Guillou (2001), and Yoon et al. (2001) for the Stramenopiles (rbcL, ITS, and rRNA). The parsimony principle was used to choose the loss pattern that minimized the total number of events. To test the vertical evolution of the tRNA-Leu intron, we directly compared intron and existing rbcL “host” sequences from 12 Stramenopiles (see Yoon et al. 2002). Intron and host trees were reconstructed using the maximum likelihood (ML) method with 10 random stepwise additions and optimization done with the TBR branch-swapping algorithm. The site-specific gamma model was used with the intron data, and the GTR + I + Γ model, over all three codon positions, was used with the rbcL sequences. The model parameter values were estimated as described above. Maximum likelihood and NJ-LD bootstrap analyses were done with each data set using 500 replicates. Posterior probabilities were also calculated for both data sets as described above. All phylogenetic analyses, except for Bayesian inference, were done using PAUP*4.0b8 (Swofford 2002).
Results and Discussion
Known Distribution of the tRNA-Leu Intron
Previous analyses show that the tRNA-Leu intron, which is widespread in cyanobacteria and plastids, is present in all studied land plants (>330) and has been vertically inherited within this group (Besendahl et al. 2000). A limited amount of sampling in other plastid containing lineages has revealed a more patchy distribution with an unknown inheritance pattern (Paquin et al. 1997; Besendahl et al. 2000; Costa et al. 2002). A sporadic intron distribution has often been interpreted as evidence for intron movement (e.g., see Bhattacharya et al. 1994, 2002). It is unclear, however, whether the tRNA-Leu intron has experienced strict vertical inheritance with uneven losses occurring in lineages, resulting in the appearance of intron mobility, or whether introns have in fact been frequently laterally transferred. Some authors have suggested that the distribution of the tRNA-Leu intron in cyanobacteria and plastids is best explained by the lateral transfer of the intron into the homologous position in unrelated organisms (Daros and Flores 1996; Rudi and Jakobsen 1997, 1999; Rudi et al. 2002). In this study, we have conducted an extensive survey to determine the phylogenetic distribution of the intron in a diverse group of algae, including the Chlorophyta, Stramenopiles, and Rhodophyta, as well as additional glaucophytes and streptophytes. This study resulted in the discovery of 32 new introns and the determination of 24 new intron sequences. As with other published tRNA-Leu intron sequences, none of these were found to contain any significant open reading frames.
Distribution of the tRNALeu Intron
It is well established that green algae and land plants share a unique common ancestor (e.g., Friedl 1997). There are four classes within the Chlorophyta: the Chlorophyceae, Trebouxiophyceae, Ulvophyceae, and Prasinophyceae. The Streptophyta includes the charophytes, bryophytes, Lycophytina, and all other multicellular land plants. In this study, 10 of 11 streptophytes were found to contain the tRNA-Leu intron (Table 1). The single species lacking the intron was the charophyte Klebsormidium nitens. This is consistent with previous work showing the overwhelming majority of streptophytes to possess this intron, with the few exceptions occurring within the charophytes (i.e., Chara hispida, Cylindrocystis brebissonii [Kuhsel et al. 1990; Besendahl et al. 200]). These results suggest that the tRNA-Leu intron was present at least in the common ancestor of the charophytes and has been retained for the 470 million-year history of land plants (Kenrick and Crane 1997). In contrast, just 10 of the 25 chlorophytes examined contained introns (see Table 1). The extant distribution may be the result of either widespread intron loss or some combination of loss and lateral transfer of introns between species. Inspection of Fig. 1A shows that, barring lateral transfers, a minimum of 13 independent losses is necessary to explain the observed intron distribution in chlorophytes.
Group I intron presence/absence in the Chlorophyta and the Stramenopiles. The host phylogenies are based on the published results of Friedl et al. (1997, 2002) and Buchheim et al. (2001) for the Chlorophyta and on Anderson et al. (1998, 1999), Draisma et al. (2001), Daugbjerg and Guillou (2001), and Yoon et al. (2001) for the Stramenopiles. Presence of the tRNA-Leu intron is denoted with a + and absence with a −. Independent intron losses, under the assumption of intron presence in the ancestor of these algal lineages and the criterion of maximum parsimony, are indicated with vertical bars. The Chlorophyta and Stramenopiles have minimally lost the intron 13 and 4 times, respectively, in these trees.
The plastids of rhodophytes are closely related to the plastids in cryptophytes, Stramenopiles, and haptophytes (the latter taxa are grouped in the Chromista). This relationship is explained by the origin of the plastid in the Chromista common ancestor through a single red algal secondary endosymbiosis (for details, see Yoon et al. 2002). Our work here, together with previous studies, has shown that 28 red algae lack the tRNA-Leu intron (Kuhsel et al. 1990; Reith and Munholland 1993; Ohta 1997; Besendahl et al. 2000; Glockner et al. 2000). Additionally, all cryptophytes (three taxa) and haptophytes (four taxa) studied to date also lack the intron (Douglas and Penny 1999; Besendahl et al. 2000). The only rhodophyte-derived plastid lineage found to contain the tRNA-Leu intron is the Stramenopiles. Here, taxa both with and without the intron have been identified (see Table 1, Fig. 1B). Interestingly, the intron was found exclusively within three closely related lineages, the Phaeophyceae, Xanthophyceae, and Phaeothamniophyceae (see Potter et al. 1997; Anderson et al. 1998). No intron losses appear to have occurred within the multiple Phaeophyceae and Xanthophyceae that we have sampled.
Phylogeny of the tRNA-Leu Intron
Given the large number of taxa (80), small number of characters (201 nt), and ancient origin of the first algal plastids (≥1 billion years ago [Butterfield 2000; Yoon et al. 2002]), the tRNA-Leu intron phylogeny shown in Fig. 2 is remarkably accurate in reconstructing predicted plastid relationships (e.g., Bhattacharya and Medlin 1995; Helmchen et al. 1995; Delwiche and Palmer 1997; Martin et al. 1998; Turner et al. 1999; Yoon et al. 2002). The three primary plastid lineages that have been identified in multiple studies are the green algal/land plant plastids (chloroplasts), the cyanelles of the glaucophytes, and the red algal plastids. All three of these lineages are likely to have arisen from a single common cyanobacterial endosymbiosis (Bhattacharya and Medlin 1995; Palmer and Delwiche 1996; Delwiche and Palmer 1997). Consistent with this idea, the tRNA-Leu intron sequences, representing all three lineages of primary plastids (or derived from these lineages [i.e., Stramenopiles]), form a monophyletic group that is sister to the cyanobacteria (Fig. 2).
The vertical evolution of the tRNA-Leu intron is best supported in the Streptophyta, where the phylogeny is generally congruent with what is currently known about the branching order of land plants (e.g., Nickrent et al. 2000). The Euphyllophytina, Lycophytina, and charophytes are monophyletic with the latter forming the sister group to land plants (Nickrent et al. 2000; Pryer et al. 2001; Karol et al. 2002). Our results are also consistent with numerous microevolutionary studies in land plants (e.g., Wallander and Albert 2000; Asmussen and Chase 2001; Soliva et al. 2001) and with the analyses of Besendahl et al. (2000) that failed to find evidence of lateral transfer of the tRNA-Leu intron. Minimally, these data provide evidence for the stable inheritance of the tRNA-Leu intron in the Streptophyta. In contrast, the evolutionary history of this intron in the chlorophyte green algae remains open. Our sparse intron phylogenetic data do not allow us to confidently address the vertical transmission of the intron. If lateral transfers have occurred in the recent history of the green algae, then one would expect, however, to see a specific and strong phylogenetic association in the intron tree between the source (e.g., cyanobacterial or algal) of the intron and its green algal recipient. Inspection of Fig. 2 provides no obvious evidence of such a transfer.
The Stramenopiles intron data set is more robust, and comparison of the intron and host trees shows congruency. This supports the strict vertical inheritance of the intron at least since the origin of the phaeophytes, xanthophytes, and phaeothamniophytes (Fig. 3). However, the absence of the intron in all other red or red-derived plastid lineages is puzzling. This distribution could be explained in two ways. Under the first scenario, the intron was lost in the rhodophytes, after the chromist secondary endosymbiosis, with subsequent losses in the cryptophytye and haptophyte common ancestors. This view asserts that the intron was retained only within the phaeophytes, xanthophytes, and phaeothamniophytes in the Stramenopiles. Alternatively, the tRNA-Leu intron was lost in the red algal common ancestor before secondary endosymbiosis or never existed in this lineage and was recently laterally transferred into the common ancestor of the Phaeophyceae, Xanthophyceae, and Phaeothamniophyceae. Although the second scenario is more parsimonious, the early branching position of the Stramenopiles introns in Fig. 2 suggests otherwise. In this tree, the Stramenopiles introns form an independent lineage that is sister to introns in the green and glaucophyte algae. This is consistent with an ancient phylogenetic split between the Stramenopiles tRNA-Leu intron and this sequence in all other plastids, rather than a recent lateral transfer into the common ancestor of the Phaeophyceae, Xanthophyceae, and Phaeothamniophyceae. The finding of the tRNA-Leu intron in other chromists or in the red algae will provide critical evidence to unambiguously distinguish between the two scenarios discussed here.
Comparison of ML trees of tRNA-Leu group I introns and rbcL coding regions from selected Stramenopiles. The intron tree has been rooted on the branch leading to the chlorophyteintrons (e.g., Chlorella vulgaris), whereas the rbcL tree has been rooted on the branch leading to Phaeothamnion confervicola. The values above the branches show the results of a ML bootstrap analysis (500 replications [see text for details]), whereas the values shown below the branches are from a NJ-LD analysis. Only bootstrap values ≥60% are shown.The thicker branches indicate clades that had ≥95% posterior probability in a Bayesian inference for each data set. The light gray box contains the Xanthophyceae and the dark gray box the Phaeophyceae. The phaeothamniophyte (Phaeothamnion confervicola) is not boxed. The branch lengths are proportional to the number of substitutions per site (see scales in figure). The intronsshown in the larger text were determined in this study.
Self-Splicing Ability of the tRNA-Leu Intron
Based on limited in vitro assays, it was previously hypothesized that the tRNA-Leu intron in chloroplasts had lost the ability to self-splice and, thus, must be dependent upon the host to facilitate excision (Xu et al. 1990; Besendahl et al. 2000). To test the validity of this hypothesis, we did self-splicing assays using tRNA-Leu introns from the cyanobacteria, basal streptophytes and land plants, chlorophytes, Stramenopiles, and glaucophytes (see Fig. 4). As previously predicted, all cyanobacterial tRNA-Leu introns could self-splice efficiently in vitro (e.g., Zaug et al. 1993), strongly suggesting that the intron was auto-catalytic at the time of plastid primary endosymbiosis. More surprisingly, the introns from the glaucophytes, Stramenopiles, and chlorophytes also showed some autocatalytic capacity. The intron from the charophyte, Chlorokybus atmophyticus, was able to fully self-splice in vitro, whereas many of the remaining introns from this group showed partial catalytic activity. The presence of extra bands in these assays (see Fig. 4B), when not clearly due to complete self-splicing, could result from either completion of the first-step of the splicing reaction (in which cleavage occurs at the 5′ splice site, leaving the free 5′ exon plus the intact intron and 3′ exon) or the alternative hydrolysis pathway (in which cleavage occurs at the 3′ splice site, leaving the free 3′ exon plus the intact 5′ exon and intron [Inoue et al. 1986; Cech 1990]). Based on the results from splicing experiments with the Stramenopile, Bodanella lauterborni, we suggest that the first step of splicing (and not hydrolysis) occurs in these taxa. This reflects the finding of a second band of size 1127 nt with this taxon that matches the expectation when the first step of splicing is completed (i.e., intron [209 nt] + 3′ exon [918 nt]). Hydrolysis of the B. lauterbornii intron would have resulted in a second band of size 266 nt. The land plant introns that were tested could neither fully splice nor complete the first step. Whereas the assay conditions were designed to favor splicing, it is still possible that under different reagent or temperature conditions, splicing may have occurred in some of these taxa (Uhlenbeck 1995; Pan et al. 1997). However, in support of our results, previous analyses of plastid tRNA-Leu introns have also shown that the intron from Marchantia polymorpha (Xu et al. 1990), Annona cherimola (Daros and Flores 1996), Psilotum triquetrum, and Cosmarium botrytis (Besendahl et al. 2000) cannot self-splice in vitro.
The distribution of self-splicing ability mapped on the phylogeny of tRNA-Leu group I introns. A Schematic representation of the phylogeny of the tRNA-Leu group I intron (see Fig. 2 and Besendahl et al. [2000]). The taxa that were tested for in vitro self-splicing ability are shown, as is their ability to complete the splicing reaction (+), to complete only the first step (no mark), and to have no apparent splicing ability (−). B Examples of typical results in our experiments for tRNA-Leu introns that were self-splicing in vitro (i.e., the cyanobacteria Arthrospira PCC 8005 and Nodularia sphaerocarpa SAG 50.79) and those that could only complete the first step of splicing (the glaucophyte Cyanoptyche gloeocystis SAG 4.97 and the Stramenopiles Bodanella lauterborni SAG 123.79). The pre-RNA is shown, as are the free intron and the ligated exons. The expected size of the ligated exons in our intron constructs was 78 nt.
The observed pattern of splicing capacity, although not exhaustively tested under different biochemical conditions, is nonetheless broadly consistent with the idea that self-splicing ability was either modified or lost after primary endosymbiosis. However, given the ability of some of the earlier branching plastid introns to self-splice or complete the first step, we speculate that the loss of auto-catalysis, and presumed dependence on a protein-mediated splicing pathway (for review, see Lambowitz et al. 1999), was a gradual process. Consistent with the idea of host-mediated splicing in land plants, chloroplast ribonucleoproteins (cpRNPs) have been demonstrated to associate with unspliced tRNA-Leu transcripts in Nicotiana tabacum (Nakamura et al. 1999). The cpRNP-tRNA complexes confer stability and ribonuclease resistance to the RNAs and may act as a scaffold for the specific catalytic machinery involved in splicing of the intron-containing tRNAs (Nakamura et al. 1999).
Evolution of the tRNA-Leu Intron
Our study suggests that the tRNA-Leu intron has experienced different lineage-dependent fates in its greater than 1 billion-year history in plastids. At the time of primary endosymbiosis, this intron was likely able to self-splice and has retained this ability in the cyanobacteria. The land plants are the only intron lineage that has completely lost self-splicing ability. Notably, there has been strict vertical ancestry of the intron with no detected losses in these taxa. These two characteristics may indicate a reciprocal dependence between the intron and its host. A second evolutionary path taken by the tRNA-Leu intron is found within the Charophyta and Chlorophyta. The evolutionary history of the intron within these lineages is putatively marked by multiple losses combined with the ability of the ribozyme to complete the first step of splicing. Finally, within the red plastid lineage there has been a combination of loss and stable inheritance. We postulate that the intron was ancestrally present in the Stramenopiles and has been lost in all taxa except for the Phaeophyceae, Xanthophyceae, and Phaeothamniophyceae. Interestingly, the intron appears to have been fixed in these Stramenopiles but, unlike in the streptophytes, can complete the first step of splicing. Taken together, these data underline our working hypothesis that no single inevitable fate accounts for the evolutionary history of group I introns. Our study is, however, only the first step in understanding the complexities of intron-host cell coevolution over millions of years and additional phylogenetic and splicing studies are needed to test the hypotheses generated by this research.
References
RA Anderson D Potter RR Bidigare M Latosa K Rowan CJ O’Kelly (1998) ArticleTitleCharacterization and phylogenetic position of the enigmatic golden alga Phaeothamnion confervicola: Ultrastructure, pigment composition, and partial SSU rDNA Sequence. J Phycol 34 286–298 Occurrence Handle10.1046/j.1529-8817.1998.340286.x
RA Anderson Y Van de Peer D Potter JP Sexton M Kawachi T LaJeunesse (1999) ArticleTitlePhylogenetic analysis of the SSU rRNA from members of the Chrysophyceae. Protist 150 71–84 Occurrence Handle10724520
CB Asmussen MW Chase (2001) ArticleTitleCoding and noncoding plastid DNA in palm systematics. Am J Bot 88 1103–1117 Occurrence Handle1:CAS:528:DC%2BD3MXltV2qu7s%3D Occurrence Handle11410476
ET Barfod TR Cech (1988) ArticleTitleDeletion of nonconserved helices near the 3′ end of the rRNA intron of Tetrahymena thermophila alters self-splicing but not core catalytic activity. Genes Dev 2 652–653 Occurrence Handle1:CAS:528:DyaL1cXks1yjtLs%3D Occurrence Handle3417146
AA Beaudry GF Joyce (1990) ArticleTitleMinimum secondary structure requirements for catalytic activity of a self-splicing group I intron. Biochemistry 29 6534–6539 Occurrence Handle1:CAS:528:DyaK3cXktlOjsbc%3D Occurrence Handle2207095
A Besendahl Y-L Qiu J Lee JD Palmer D Bhattacharya (2000) ArticleTitleThe cyanobacterial origin and vertical transmission of the plastid tRNALeu group-I intron. Curr Genet 37 12–23
D Bhattacharya L Medlin (1995) ArticleTitleThe phylogeny of plastids: A review based on comparisons of small subunit ribosomal RNA coding regions. J Phycol 31 489–498 Occurrence Handle1:CAS:528:DyaK2MXos1Gls7Y%3D
D Bhattacharya B Surek M Ruesing S Damberger M Melkonian (1994) ArticleTitleGroup I introns are inherited through common ancestry in the nuclear-encoded rRNA of Zygnematales (Charophyceae). Proc Natl Acad Sci USA 91 9916–9920 Occurrence Handle1:CAS:528:DyaK2MXht1Cmsbk%3D Occurrence Handle7937917
D Bhattacharya JJ Cannone RR Gutell (2001) ArticleTitleGroup I intron lateral transfer between red and brown algal ribosomal RNA. Curr Genet 40 82–90 Occurrence Handle10.1007/s002940100227 Occurrence Handle1:CAS:528:DC%2BD3MXlvVaqsbY%3D Occurrence Handle11570520
D Bhattacharya T Friedl G Helms (2002) ArticleTitleVertical evolution and intragenic spread of lichen-fungal group I introns. J Mol Evol 55 74–84 Occurrence Handle10.1007/s00239-001-2305-x Occurrence Handle1:CAS:528:DC%2BD38XlvVCqsbc%3D Occurrence Handle12165844
N Butterfield (2000) ArticleTitle Bangiomorpha pubescens n. gen., n. sp.: Implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiol 26 386–404
JJ Brocks GA Logan R Buick RE Summons (1999) ArticleTitleArchean molecular fossils and the early rise of eukaryotes. Science 285 1033–1036 Occurrence Handle10.1126/science.285.5430.1033 Occurrence Handle1:CAS:528:DyaK1MXlt1Gns7k%3D Occurrence Handle10446042
MA Buchheim EA Michalopulos JA Buchheim (2001) ArticleTitlePhylogeny of the Chlorophyceae with special reference to the Sphaeropleales: A study of 18S and 26S rDNA data. J Phycol 37 819–835 Occurrence Handle10.1046/j.1529-8817.2001.00162.x Occurrence Handle1:CAS:528:DC%2BD3MXosFCgsr4%3D
TR Cech (1988) ArticleTitleConserved sequences and structures of group I introns: Building an active site for RNA catalysis—A review. Gene 73 259–271 Occurrence Handle1:CAS:528:DyaL1MXhsVyku7o%3D Occurrence Handle3072259
TR Cech (1990) ArticleTitleSelf-splicing of group I introns. Annu Rev Biochem 59 543–568 Occurrence Handle1:CAS:528:DyaK3cXltFSjsbY%3D Occurrence Handle2197983
TR Cech SH Damberger RR Gutell (1994) ArticleTitleRepresentation of the secondary and tertiary structure of group I introns. Nature Struct Biol 1 273–280 Occurrence Handle1:CAS:528:DyaK2MXitVaks7g%3D
JL Costa P Paulsrud P Lindblad (2002) ArticleTitleThe cyanobacterial tRNA(Leu) (UAA) intron: Evolutionary patterns in a genetic marker. Mol Biol Evol 19 850–857 Occurrence Handle1:CAS:528:DC%2BD38Xks1Ojtb8%3D Occurrence Handle12032241
SH Damberger RR Gutell (1994) ArticleTitleA comparative database of group I intron structures. Nucleic Acids Res 22 3508–3510 Occurrence Handle1:CAS:528:DyaK2MXhtF2hsbs%3D Occurrence Handle7937050
JA Daros R Flores (1996) ArticleTitleA group I plant intron accumulates as circular RNA forms with extensive 5′ deletions in vivo. RNA 2 928–936 Occurrence Handle1:CAS:528:DyaK28XlslGjsLk%3D Occurrence Handle8809019
N Daugbjerg L Guillou (2001) ArticleTitlePhylogenetic analyses of Bolidophyceae (Heterokontophyta) using rbcL gene sequences support their sister group relationship to diatoms. Phycologia 40 153–161
CF Delwiche JD Palmer (1997) Plastid origin and secondary symbiosis. D Bhattacharya (Eds) Origins of algae and their plastids. Springer Wien 53–86
SE Douglas SL Penny (1999) ArticleTitleThe plastid genome of the cryptophyte alga, Guillardia theta: Complete sequence and conserved synteny groups confirm its common ancestry with red algae. J Mol Evol 48 236–244 Occurrence Handle1:CAS:528:DyaK1MXotVyqsQ%3D%3D Occurrence Handle9929392
SGA Draisma WF Prud’homme van Reine WT Stam JL Olsen (2001) ArticleTitleA reassesment of phylogenetic relationships within the phaeophyceae based on rubisco large subunit and ribosomal DNA sequences. J Phycol 37 586–603 Occurrence Handle10.1046/j.1529-8817.2001.037004586.x
J Felsenstein (1985) ArticleTitleConfidence intervals on phylogenies: An approach using the bootstrap. Evolution 39 783–791
T Friedl (1997) The evolution of the green algae. D Bhattacharya (Eds) Origins of algae and their plastids. Springer Wien 87–102
T Friedl CJ O’Kelly (2002) ArticleTitlePhylogenetic relationships of green algae assigned to the genus Planophila (Chlorophyta): Evidence from 18S rDNA sequence data and ultrastructure. Eur J Phycol 37 373–384 Occurrence Handle10.1017/S0967026202003712
Gilbert D (2001) SeqApp: A Macintosh biosequence editor, analyzer, and network handyman. Available at ftp://iubio.bio. indiana.edu/iubionew///molbio/dna/display/SeqApp/
W Gilbert (1986) ArticleTitleThe RNA world. Nature 319 618
G Glockner A Rosenthal K Valentin (2000) ArticleTitleThe structure and gene repertoire of an ancient red algal plastid genome. J Mol Evol 51 382–390 Occurrence Handle1:CAS:528:DC%2BD3cXotlWnsbk%3D Occurrence Handle11040290
MR Goddard A Burt (1999) ArticleTitleRecurrent invasion and extinction of a selfish gene. Proc Natl Acad Sci USA 96 13880–13885 Occurrence Handle10.1073/pnas.96.24.13880 Occurrence Handle1:CAS:528:DyaK1MXns1OqtLY%3D Occurrence Handle10570167
T Helmchen D Bhattacharya M Melkonian (1995) ArticleTitleAnalyses of ribosomal RNA sequences from glaucocystophyte cyanelles provide new insights into the evolutionary relationships of plastids. J Mol Evol 41 203–210 Occurrence Handle1:CAS:528:DyaK2MXntVSlt7s%3D Occurrence Handle7666450
JP Huelsenbeck F Ronquist (2001) ArticleTitleMrBayes: Bayesian inference of phylogeny. Bioinformatics 17 754–755 Occurrence Handle10.1093/bioinformatics/17.8.754 Occurrence Handle1:STN:280:DC%2BD3MvotV2isw%3D%3D Occurrence Handle11524383
T Inoue FX Sullivan TR Cech (1986) ArticleTitleNew reactions of the ribosomal RNA precursor of Tetrahymena and the mechanism of self-splicing. J Mol Biol 189 143–165 Occurrence Handle1:CAS:528:DyaL28XktlOltrk%3D Occurrence Handle2431151
KG Karol RM McCourt MT Cimino CF Delwiche (2002) ArticleTitleThe closest living relatives of land plants. Science 294 2351–2353 Occurrence Handle10.1126/science.1065156
P Kenrick PR Crane (1997) ArticleTitleThe origin and early evolution of plants on land. Nature 389 33–39 Occurrence Handle10.1038/37918 Occurrence Handle1:CAS:528:DyaK2sXlvVClt7w%3D
MG Kuhsel R Strickland JD Palmer (1990) ArticleTitleAn ancient group-I intron shared by eubacteria and chloroplasts. Science 250 1570–1573 Occurrence Handle1:CAS:528:DyaK3MXhtlOgurY%3D Occurrence Handle2125748
AM Lambowitz MG Caprara S Zimmerly PS Perlman (1999) Group I and group II ribozymes as RNPs: Clues to the past and guides to the future. RF Gesteland TR Cech J Atkins (Eds) The RNA world, 2nd ed. Cold Spring Harbor Laboratory Press Cold Spring Harbor, NY 451–485
W Martin B Stoebe V Goremykin S Hansmann M Hasegawa K Kowallik (1998) ArticleTitleGene transfer to the nucleus and the evolution of chloroplasts. Nature 393 162–165 Occurrence Handle1:CAS:528:DyaK1cXjt1ahsL0%3D Occurrence Handle11560168
F Michel E Westhof (1990) ArticleTitleModelling the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol 216 585–610 Occurrence Handle1:CAS:528:DyaK3MXps1agtw%3D%3D Occurrence Handle2258934
MJ Moore CC Query PA Sharp (1993) Splicing of precursors to mRNA by the spliceosome. RF Gesteland JF Atkins (Eds) The RNA world. Cold Spring Harbor Laboratory Press Cold Spring Harbor, NY 303–357
T Nakamura M Ohta M Sugiura M Sugita (1999) ArticleTitleChloroplast ribonucleoproteins are associated with both mRNAs and intron-containing precursor tRNAs. FEBS Lett 460 437–441 Occurrence Handle10.1016/S0014-5793(99)01390-3 Occurrence Handle1:CAS:528:DyaK1MXntVChtLw%3D Occurrence Handle10556512
DL Nickrent CL Parkinson JD Palmer RJ Duff (2000) ArticleTitleMultigene phylogeny of land plants with special reference to bryophytes and the earliest land plants. Mol Biol Evol 17 1885–1895 Occurrence Handle1:CAS:528:DC%2BD3cXptVWrsbg%3D Occurrence Handle11110905
N Ohta (1997) ArticleTitleAnalysis of a plastid gene cluster reveals a close relationship between Cyanidioschyzon and Cyanidium. J Plant Res 110 235–245 Occurrence Handle1:CAS:528:DyaK2sXls1WisLc%3D
NR Pace TK Marsh (1985) ArticleTitleRNA catalysis and the origin of life. Origins of Life 16 97–116 Occurrence Handle1:CAS:528:DyaL28XlvVGqtrs%3D Occurrence Handle2423941
JD Palmer CF Delwiche (1996) ArticleTitleSecond-hand chloroplasts and the case of the disappearing nucleus. Proc Natl Acad Sci USA 93 7432–7435 Occurrence Handle10.1073/pnas.93.15.7432 Occurrence Handle1:CAS:528:DyaK28XksFSkuro%3D Occurrence Handle8755491
J Pan D Thirumalai SA Woodson (1997) ArticleTitleFolding of RNA involves parallel pathways. J Mol Biol 273 7–13 Occurrence Handle9367740
B Paquin SD Kathe SA Nierzwicki-Bauer DA Shub (1997) ArticleTitleOrigin and evolution of group-I introns in cyanobacterial tRNA genes. J Bacteriol 179 6798–6806 Occurrence Handle1:CAS:528:DyaK2sXnt1arsrc%3D Occurrence Handle9352932
D Potter GW Saunders RA Andersen (1997) ArticleTitlePhylogenetic relationships of the Raphidophyceae and Xanthophyceae as inferred from nucleotide sequences of the 18S ribosomal RNA gene. Am J Bot 84 966–972 Occurrence Handle1:CAS:528:DyaK2sXlsVKksro%3D
JV Price GL Kieft JR Kent EL Sievers TR Cech (1985) ArticleTitleSequence requirements for self-splicing of the Tetrahymena thermophila pre-ribosomal RNA. Nucleic Acids Res 13 1871–1889 Occurrence Handle1:CAS:528:DyaL2MXhvVSjsLw%3D Occurrence Handle4000946
KM Pryer H Schneider AR Smith R Cranfill PG Wolf JS Hunt SD Sipes (2001) ArticleTitleHorsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature 409 618–622 Occurrence Handle10.1038/35054555 Occurrence Handle1:STN:280:DC%2BD3M7islSntw%3D%3D Occurrence Handle11214320
M Reith J Munholland (1993) ArticleTitleA high-resolution gene map of the chloroplast genome of the red alga Porphyra purpurea. Plant Cell 5 465–475 Occurrence Handle10.1105/tpc.5.4.465 Occurrence Handle1:CAS:528:DyaK3sXkvVyju7w%3D Occurrence Handle12271072
F Rodriguez JL Oliver A Marin JR Medina (1990) ArticleTitleThe general stochastic model of nucleotide substitution. J Theor Biol 142 485–501 Occurrence Handle2338834
K Rudi KS Jakobsen (1997) ArticleTitleCyanobacterial tRNA(Leu)(UAA) group I introns have polyphyletic origin. FEMS Microbiol Lett 156 293–298 Occurrence Handle10.1016/S0378-1097(97)00446-1 Occurrence Handle1:CAS:528:DyaK2sXmvFWjtL4%3D Occurrence Handle9513279
K Rudi KS Jakobsen (1999) ArticleTitleComplex evolutionary patterns of tRNA Leu(UAA) group I introns in the cyanobacterial radiation. J Bacteriol 181 3445–3451 Occurrence Handle1:CAS:528:DyaK1MXjs1yrurc%3D Occurrence Handle10348857
K Rudi T Fossheim KS Jakobsen (2002) ArticleTitleNested evolution of a tRNA(Leu)(UAA) group I intron by both horizontal intron transfer and recombination of the entire tRNA locus. J Bacteriol 184 666–671 Occurrence Handle1:CAS:528:DC%2BD38XntVGhsQ%3D%3D Occurrence Handle11790735
JW Schopf (1993) ArticleTitleMicrofossils of the early Archaen Apex chert: New evidence of the antiquity of life. Science 260 640–646 Occurrence Handle1:STN:280:DC%2BD3MnlvVequg%3D%3D Occurrence Handle11539831
M Soliva A Kocyan A Widmer (2001) ArticleTitleMolecular phylogenetics of the sexually deceptive orchid genus Ophrys (Orchidaceae) based on nuclear and chloroplast DNA sequences. Mol Phylogenet Evol 20 78–88 Occurrence Handle10.1006/mpev.2001.0953 Occurrence Handle1:CAS:528:DC%2BD3MXksF2mu70%3D Occurrence Handle11421649
RE Summons LL Jahnke JM Hope GA Logan (1999) ArticleTitle2-Methylhopanoids as biomarkers for cyanobacterial oxygenic photosynthesis. Nature 400 554–557 Occurrence Handle10.1038/23005 Occurrence Handle1:CAS:528:DyaK1MXltFKku7o%3D Occurrence Handle10448856
DL Swofford (2002) PAUP*: Phylogenetic analysis using parsimony (*and other methods) 4.0b8. Sinauer Associates Sunderland, MA
F Tajima M Nei (1984) ArticleTitleEstimation of evolutionary distance between nucleotide sequences. Mol Biol Evol 1 269–285 Occurrence Handle1:CAS:528:DyaL2MXhvF2kt7c%3D Occurrence Handle6599968
S Turner KM Pryer VP Miao JD Palmer (1999) ArticleTitleInvestigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol 46 327–338 Occurrence Handle1:CAS:528:DyaK1MXlslSlu7Y%3D Occurrence Handle10461381
OC Uhlenbeck (1995) ArticleTitleKeeping RNA happy. RNA 1 4–6 Occurrence Handle1:CAS:528:DyaK2MXkvFemtbc%3D Occurrence Handle7489487
G Van der Horst A Christian T Inoue (1991) ArticleTitleReconstitution of a group I intron self-splicing reaction with an activator RNA. Proc Natl Acad Sci USA 66 184–188
E Wallander VA Albert (2000) ArticleTitlePhylogeny and classification of Oleaceae based on rps16 and trnL-F sequence data. Am J Bot 87 1827–1841 Occurrence Handle1:CAS:528:DC%2BD3MXktFCrtQ%3D%3D Occurrence Handle11118421
M-Q Xu SD Kathe H Goodrich-Blair SA Nierzwicki-Bauer DA Shub (1990) ArticleTitleBacterial origin of a chloroplast intron: Conserved self-splicing group-I introns in cyanobacteria. Science 250 1566–1570 Occurrence Handle1:CAS:528:DyaK3MXhsl2nsrc%3D Occurrence Handle2125747
HS Yoon JD Hackett G Pinto D Bhattacharya (2002) ArticleTitleThe single, ancient origin of chromist plastids. Proc Natl Acad Sci USA 99 15507–15512 Occurrence Handle10.1073/pnas.242379899 Occurrence Handle1:CAS:528:DC%2BD3sXjvVOg Occurrence Handle12438651
HS Yoon JY Lee SM Boo D Bhattacharya (2001) ArticleTitlePhylogeny of Alariaceae, Laminariaceae, and Lessoniaceae (Phaeophyceae) based on plastid-encoded RuBisCo spacer and nuclear-encoded ITS sequence comparisons. Mol Phylogenet Evol 21 231–243 Occurrence Handle10.1006/mpev.2001.1009 Occurrence Handle1:CAS:528:DC%2BD3MXotVynurs%3D Occurrence Handle11697918
AJ Zaug MM McEvoy TR Cech (1993) ArticleTitleSelf-splicing of the group I intron from Anabaena pre-tRNA. Requirement for base-pairing of the exons in the anticodon stem. Biochemistry 32 7946–7953 Occurrence Handle1:CAS:528:DyaK3sXkvFSjtrY%3D Occurrence Handle8347600
Acknowledgements
This work was supported by a grant awarded to D.B. from the National Science Foundation (MCB 01-10252) and a grant from the Deutsche Forschungsgemeinschaft to T.F. (Fr 905/7-1,2). D.F. received financial support from the GlaxoSmithKline Corporation and a Sigma Xi Grant-in-Aid award. D.S. was supported by a Stanley Fellowship from the University of Iowa. We also thank Hwan Su Yoon (Iowa) and Peik Haugen (Iowa) for helpful discussions and technical advice.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Simon, D., Fewer, D., Friedl, T. et al. Phylogeny and Self-Splicing Ability of the Plastid tRNA-Leu Group I Intron . J Mol Evol 57, 710–720 (2003). https://doi.org/10.1007/s00239-003-2533-3
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00239-003-2533-3