Abstract
In this paper, we compare the populations of the Lusitanian toadfish Halobatrachus didactylus along the Portuguese shore, using a putative fragment of the mitochondrial control region and the first intron of the S7 ribosomal protein gene. This demersal species ranges from the tropical West African coast to the Iberian Peninsula and the Mediterranean, having its northern limit in the Tagus River. For the putative control region, a single haplotype occurs in all fish from Tagus and Sado (the location immediately to the south) with a clear increase in diversity to the South, peaking at Algarve (south Portugal). The data seem to point to a very recent Holocenic colonization of Tagus and Sado from the South (possibly Algarve). We suggest that even small oscillations in sea surface temperature (SST) may cause local extinctions and subsequent recolonizations in populations of thermophilic fish that are at the cold limit of their distributions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Many phylogeographic studies of marine organisms with larval dispersal have detected little structure along stretches of coastlines that span hundreds of kilometres or more (e.g. Nephrops norvegicus, Stamatis et al. 2004; Homarus gammarus, Triantafyllidis et al. 2005; Petromyzon marinus, Almada et al. 2008; Sprattus sprattus Debes et al. 2008; Lipophrys pholis, Francisco et al. 2011). This is assumed to result from the presence of planktonic eggs and/or larvae capable of dispersing over distances of kilometres or hundreds of kilometres per generation (however, see Ayre et al. 2009). Most studies assume that the Pleistocene and specially the Last Glacial Maximum (LGM) were critical times when populations were absent from the frozen north and suffered severe bottlenecks in their refugial habitats. In temperate areas like the west coast of Portugal and most of the south-west European shore, however, the conditions seem to have been more complicated. Although sea surface temperatures (SSTs) during the LGM along the coast of Portugal were not significantly different from what they are today (Abreu et al. 2003), several cold episodes towards the end of the Last glaciation considerably lowered mean summer SST (as much as 5 °C).
During the Holocene the oscillations in SSTs continued to be a salient feature of this geographic area, although their amplitudes were smaller. Some cold pulses such as the 8,400 years ago cold event and the Little Ice Age (LIA) caused temperature drops of one or a few degrees (Casper 2010) and may have affected the distributions of fish populations. A temperature anomaly need not to last very long to extirpate a population, it suffices that the population does not breed during a period that surpasses the lifespan of all its members. So, even short pulses that last for a few fish generations can cause a very substantial local effect. Even in present conditions small interannual oscillations can substantially affect the fish fauna, at least for the populations that are at or near the higher latitude limits of the distribution of species (Thomson and Lehner 1976; Stenseth et al. 2002; Henriques et al. 2007).
The Lusitanian toadfish Halobatrachus didactylus (Batrachoididae) (Bloch and Schneider 1801) offers a very interesting model to investigate the structure of populations that are at the extreme north of the species distribution comparing them with others further to the south, thus more sheltered from SSTs oscillations. The Lusitanian toadfish is a thermophilic demersal species that lives in shallow waters down to 50 m depth, often found buried in muddy and soft sand bottoms or concealed in rock crevices (Roux 1986; Bauchot 1987). It inhabits coastal and estuarine areas and its distribution ranges from the tropical Western African coast to the Mediterranean and the Atlantic shore of the Iberian Peninsula (Bauchot 1987) being virtually absent north of the Tagus River (Costa et al. 2003). At the Portuguese western coast, populations occur only in estuaries, while both coastal and estuarine populations can be found in the south (Costa 2004). It is considered a sedentary species (Roux 1986), but some individuals have been found to perform important displacements of up to 17 km (Costa 2004; Campos et al. 2008). The breeding season in the south of Portugal lasts from late spring to early–mid summer (e.g. from May to August in Algarve) (Modesto and Canário 2003) when water temperature reaches 18 °C, which is apparently the minimum required for breeding to take place (Amorim et al. 2006). H. didactylus is a polygynous species, characterized by low fecundity and high survival of embryos and juveniles (Palazón-Fernández et al. 2001). Males defend nests under rocks in shallow water, where they vocalize to attract females to mate with (Vasconcelos et al. 2011). Each female spawns a single batch of eggs annually, but males can guard the eggs of several females (Modesto and Canário 2003; Amorim et al. 2010). The eggs, that are rich in yolk, are guarded by the males until yolk sac absorption and juveniles venture for themselves when they are about 18 mm (Costa 2004). Thus, development is direct and no planktonic stage exists (Costa 2004).
The absence of a planktonic stage in H. didactylus reduces the likelihood that the signature of a bottleneck is quickly wiped out by migrants from other populations, a condition that is to be expected in marine fish with very large population sizes and high dispersal capabilities (e.g. Debes et al. 2008; Francisco et al. 2011).
In the Tagus estuary, summers with a very limited number of days with SSTs above 18 °C are not uncommon leading to a very limited production of young. This observation that unusual cold summers impact reproduction negatively led us to hypothesize that LIA, 8,400 and other cold events of smaller intensity may have caused local extinction of the population.
In this paper, we compare the genetic structure of the Tagus population with a series of other populations south to Algarve, an area with higher SSTs and probably less exposed to the extremes of cold events. The analysis is based on two markers: a putative fragment of the mitochondrial control region and the first intron of the S7 ribosomal protein gene.
Methods
Sampling
Samples of H. didactylus were collected by beach seining from 5 locations: Morocco (Salé), Algarve (Ria Formosa Estuary and Guadiana) and the estuaries of the Rivers Mira, Sado and Tagus (Fig. 1). A total of 117 individuals were obtained, and the number of individuals collected per site/per fragment is listed in Table 1.
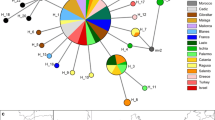
Map with the sampling locations, haplotypes and haplotype network for Halobatrachus didactylus. Left distribution range of H. didactylus. Right sampling locations with a representation of the haplotypes presented and the haplotype network. Coloured circles represent the relative proportion of individuals. In the haplotype network the colours correspond to the sampling locations
DNA extraction, amplification and sequencing
Total genomic DNA was extracted from fin or muscle samples preserved in 96 % ethanol with the REDExtract-N-Amp kit (Sigma-Aldrich) following the manufacturer’s instructions. Voucher specimens are deposited in ISPA (ethanol preserved tissues). PCR amplification of CR and the first intron of the nuclear S7 ribosomal protein gene (S7) were performed with the following pairs of primers: CR—LPro1 (5′-ACTCT CACCC CTAGC TCCCA AAG-3′) and HDL1 (5′-CCTGA AGTAG GAACC AGATG CCAG-3′) (Ostellari et al. 1996) and the first intron of the S7 ribosomal protein gene—S7RPEX1F (5′-TGG CCT CTT CCT TGG CCG TC-3′) and S7RPEX2R (5′-AAC TCG TCT GGC TTT TCG CC-3′) (Chow and Hazama 1998). A search in GenBank yielded no CR sequences for Batrachoididae. In spite of this, the pair of primers used in this study typically amplifies a portion of the CR of other species (e.g. Symphodus melops, Robalo et al. 2012; Salaria pavo, Almada et al. 2009) and succeeded in a considerable proportion of our samples. Because of this and as a note of caution, we refer to this fragment as a putative control region fragment.
PCR amplifications were performed in a 20 μl total reaction volume with 10 μl of REDExtract-N-ampl PCR mix (Sigma-Aldrich), 0.8 μl of each primer (10 μM), 4.4 μl of Sigma water and 4 μl of template DNA. An initial denaturation at 94 °C for 7 min was followed by 35/30 cycles (denaturation at 94 °C for 30/45 s, annealing at 55 °C for 30/45 s and extension at 72 °C for 1 min) and a final extension at 72 °C for 7 min on a BioRad Mycycler thermal cycler (values CR/S7, respectively). The same primers were used for the sequencing reaction, and the PCR products were purified and sequenced by STABVIDA (http://www.stabvida.net/).
DNA analysis
All sequences were edited with Codon Code Aligner (http://www.codoncode.com/index.htm) and aligned using Clustal X (Thompson et al. 1997).
ARLEQUIN software package V.3.5 (Excoffier and Lischer 2010) was used to estimate the genetic diversity within each sample, to access potential population differentiation and to perform neutrality tests. It was also used to perform analysis of molecular variance (AMOVA) (Excoffier et al. 1992) to compute pairwise F STs and corrected pairwise differences between populations. Significance levels of all multiple statistical tests were corrected using the false discovery rate (FDR) approach (Benjamini and Hochberg 2005) implemented in QVALUE package of software R (Dabney et al. 2006). In the case of the S7 intron, the analyses were also run in ARLEQUIN, after allowing the programme to reconstruct the haplotypes present, using the ELB algorithm (Excoffier et al. 2003). The number of migrants was also calculated for each population pair using the same software.
For the control region, the spatial analysis of molecular variance (SAMOVA 1.0) (Dupanloup et al. 2002) was used to identify groups of sampling locations that are geographically and genetically homogeneous and maximally differentiated from each other. This approach relies on a technique of AMOVA (Excoffier et al. 1992). However, in contrast to conventional AMOVA, SAMOVA does not require that the groups’ constitution is defined a priori, allowing instead the groups to emerge from the data. The most likely number of groups was identify by running SAMOVA with two to four groups and choosing the partition scheme with the highest F CT value. For each of the groups of locations identified by SAMOVA, Fu’s Fs (Fu 1997) and Tajima’s D (Tajima 1983) tests were performed to test for population expansion.
A haplotype network was constructed for the putative CR data using Network v4.5 (Bandelt et al. 1999). Past population demography of H. didactylus was inferred using the linear Bayesian skyline plot (BSP) (Drummond et al. 2005) model as implemented in BEAST v.1.6 (Drummond and Rambaut 2007) employing the Bayesian MCMC coalescent method, a HKY + I + G model of substitution, and a strict clock. The Bayesian distribution was generated using results from five independent runs of 150 million MCMC steps obtaining effective samples sizes (ESS) of parameter estimates of over 200, with a burn-in of 10 %. The time to most recent common ancestor (t MRCA) and the median and corresponding credibility intervals of the BSP were depicted using Tracer v.1.4 (Rambaut and Drummond 2007). MtDNA control region mutation rates in fish are widely variable (e.g. 2.2–4.5 %/MY between lineages for East African cichlids—Sato et al. 2003, to 15–20 %/MY for Indo-Pacific sardines—Bowen and Grant 1997). In the absence of a clock calibration for the CR of H. didactylus, we address the uncertainty by tentatively assuming a within lineage mutation rate of 5 %/MY. This value is within the range of values found by (Bowen et al. 2006) after a review of CR molecular clock calibrations for several tropical Atlantic fish species. Nevertheless, these estimations of divergence times must be interpreted with great caution.
Results
The putative control region (CR) data set consisted of a total of 377 characters (92 individuals, Genbank Accession Numbers JQ278604-95). The amplification of the first intron of the S7 gene (S7) yielded a fragment of 713 characters, with 15 polymorphic sites (117 individuals, Genbank Accession Numbers JQ310526-659). The putative CR haplotype network shows only 4 haplotypes (Fig. 1). The most abundant haplotype contains 66 fish from all localities. The locations of Tagus and Sado yielded only this haplotype. The only fish from Morocco included in the CR analysis also displayed this haplotype. The second haplotype (connected to the first by one mutational step) contains 16 fish only from Algarve and the Mira estuary. The same holds for the third haplotype, which contains 9 fish from the same locations. The fourth haplotype is a singleton from Algarve.
For both gene fragments, the genetic diversity indices for each population are summarized in Table 1. When we take together Fig. 1 and Table 1, we conclude that for the putative CR, the location of Algarve showed a strikingly high diversity when compared with all other locations. All haplotypes found in the remaining locations are present in Algarve. The haplotypes of Mira are shared with some fish from Algarve, yielding moderate diversity indices. However, the number of sequences obtained is low, and this finding must be taken with caution. The locations of Tagus and Sado display an impressive null diversity with a single haplotype present in all fish and shared with Algarve and Morocco. We suspect that there may be one or more haplotypes that failed amplification in fish from Algarve, Mira and Morocco. This situation is easily explained if one or more mutations hit the site of primer attachment. Our suspicion is based on the fact that some fish from Algarve, Mira and Morocco repeatedly failed in PCR of the putative CR while yielding good S7 sequences. We believe, however, that the main results of this study were not affected because, if anything, the diversity of Algarve, Mira and Morocco is underestimated. In Tagus and Sado all 49 fish yielded good PCR amplifications of the putative CR and the S7 intron, so that within the limits of our sampling size, no hidden haplotypes could be present as no fish failed amplification. For the S7 intron, no consistent trend of declining diversity from Algarve to Tagus/Sado was detected, which is to be expected if the processes that gave rise to these populations are so recent that a gene with a fourfold number of copies (compared to CR) had not sufficient time to undergo advanced or complete lineage sorting.
The pairwise F STs and corrected average pairwise differences are presented in Tables 2 (putative CR) and 3 (S7). For the putative CR, a very clear pattern emerges that was anticipated by the haplotype network. Tagus and Sado show zero differences, and both are closer to Algarve than to Mira, which in turn shows relatively small differences from Algarve. These results make full sense if the population of Algarve gave rise independently to a population in Mira, on one hand, and another clade that possibly founded the Sado population and later that of the Tagus. For the S7 intron, the smallest F ST and corrected pairwise differences are between Sado and Mira, the pair that also shows the highest number of migrants (28.605). Migration values for the remaining pairs are >1 only for Sado/Algarve = 2.282; Tagus/Algarve = 2.271; Tagus/Morocco = 1.067; Mira/Algarve = 1.238 and Algarve/Morocco = 1.045. Except for the Moroccan fish, Tagus and Sado show the largest differences among them in FST and in average corrected pairwise differences.
SAMOVA (putative CR dataset) did not yield significant FCT for any of the groups analysed (data not shown). In view of this, we rejected the hypothesis that Portuguese populations cluster in well-defined groups. None of the sample localities tested displayed signs of expansion as Tajima’s D and Fu’s Fs tests never yielded significant negative values (results not shown). An AMOVA for the putative CR performed with the 4 locations showed that there is significant interpopulation variation: percentage of variation among populations = 47.17; percentage of variation within populations = 52.83 (overall F ST = 0.471, p < 0.001). A similar result was obtained when we performed an AMOVA for the S7 data (including also individuals from Morocco): percentage of variation among populations = 36.24; percentage of variation within populations = 63.76 (overall F ST = 0.362, p < 0.001).
The BSP (resulting from BEAST) for the putative CR data set of H. didactylus (Fig. 2) reveals that the Lusitanian toadfish, as a whole, experienced a mild growth in the past 40–60 years, reaching a Nef of ~6,000 fish in the present day, the t MRCA being 2,910 years ago (confidence interval: 0.05–32.11 kya, median: 2.91 kya).
Discussion
The results presented above are striking because of the very low number of haplotypes that make up the Portuguese populations and the fact that the populations that are at the species northern limit are extremely impoverished concerning the putative control region. We suggest that H. didactylus, being a warm water fish, may be in a very unstable demographic and genetic situation in Portuguese waters, especially in Tagus. As stated in the introduction, the species does not breed below 18 °C and in cool summers, when temperature drops below this point, male mate attraction calls are suppressed and nests with eggs are scarce (Amorim et al. 2006). Thus, if a cold event, even of moderate intensity, takes place, the populations of Tagus and Sado, and eventually that of Mira, may be extirpated. At Algarve SST is more frequently above 18 °C in May and June than in the Tagus area (NOAA, http://www.nodc.noaa.gov/dsdt/oisst/). Thus, at Algarve the fish are more likely protected from small amplitude lowering of SSTs than at Tagus or Sado. We propose that repeated extinctions in the extreme north of marine species distributions may be more common than usually assumed and likely continue to occur in interglacial times. An interannual survey of 10 years concluded that about 20 % of the species of the inshore fish assemblage at Arrábida (Central Portugal) could change from 1 year to the next, when SST changed from cold to warm years and vice versa (Henriques et al. 2007). When two successive warm years took place, some warm water new comers were even able to breed but disappeared completely from the censuses when a new cold year occurred. Similar results have been widely documented in the literature. For example, Thomson and Lehner (1976) found that for the intertidal fish communities in Baja California, cold water events could, in some years, affect drastically the tropical and subtropical components of the fish fauna. Additionally, Stenseth et al. (2002) reported the same results in a variety of groups of marine animals around Western Europe, after a transition from several mild winters to a cold winter in 1996. These episodes of change in the composition of fish communities are probably underestimated for two reasons: (1) long-term surveys are still insufficient; and (2) most fish have dispersive propagules that allow rapid recolonizations involving large numbers from nearby populations, so that the track of each colonization may be quickly erased.
The absence of a pelagic stage and the sedentary nature of H. didactylus would mean that recolonizations of the north would likely be achieved by a few migrants per generation, helping to make the dynamics of local extinction and recolonizations in the north easier to detect with genetic tools. We propose that the putative CR data are compatible with the following scenario: locations at Algarve (or closer to the entrance of the Mediterranean) could have operated as refugia where toadfish could survive many cold events. Populations more to the north would likely go extinct. When conditions improved, fish from Algarve or more southern locations expanded northwards, colonizing Mira, on one hand, and Sado on the other. From Sado some fish would be able to reach Tagus. All these migration routes will be used by very small numbers of fish, giving rise to strong drift that culminated in the Sado/Tagus clade. How to reconcile this data with those for S7? The significant values of most pairwise F STs for S7 point to a reduced connectivity among populations. Mira and Sado showed the smallest differentiation and the highest number of migrants for S7. For all populations, the diversity indices for S7 were higher than those for the putative CR. This result is to be expected because a marker with 4 times the number of copies (compared to CR) suffers the effects of drift with less intensity and, if the time is short, the process of lineage sorting will tend to occur less markedly. Moreover, if the diversity of the S7 marker was sufficiently high in the source populations, a random process of migration to the north could leave different alleles of S7 in different estuaries. Another interesting possibility is that the males disperse more than females. This would explain the possibility of migration between Sado and Mira, suggested by S7 but apparently contradicted by the putative CR. As males may fight for territories and there are sneakers (small males without territories that try to fertilize eggs in others males nests) described for this species (Modesto and Canário 2003), the possibility of a higher dispersal rate for males seems a possible explanation to the present findings.
How old is the recolonization of Sado and Tagus? We know that in the Holocene, several cold events did take place that could trigger a process like the one hypothesized in this paper. Aside from the recent Little Ice Age (only a few centuries from the present), other cold events were recorded after the end of the Last glaciation (e.g. Casper 2010). With the lack of solid information on mutation rates, it makes no sense to try to ascribe the demographic bottlenecks and probable recolonization of the Tagus and Sado to a specific event. In any case, it seems reasonable to consider that we are dealing with processes that took place after the Last glaciation, probably only a few hundreds or thousands of years from the present. The fact that the populations of Mira, Sado and Tagus acquired few or no mutations of their own and the sharp contrast in diversity between CR and S7 provide evidence compatible with very recent phenomena that allowed no time for population recovery. On a longer timescale even the population of Algarve is not very diverse genetically, suggesting possible marks of older events. However, these older processes must be studied on a broader geographic framework. The Portuguese populations are likely too eroded genetically to preserve an older demographic record.
The present paper addresses events that were probable at the extreme north of the species range and does not intend to replace a full phylogeographic analysis of the species. Indeed, the main caveat of this study is the lack of samples covering a larger part of the geographic distribution of the species, namely tropical and subtropical Africa where samples are very hard to obtain. We hope that, in the future, it would be possible to examine deeper phylogeographic processes affecting this species through international collaboration.
One major conclusion of the present study is the need to combine molecular genetic studies with ecological surveys that allow us to detect and study advances and retreats of species on a rapid basis. In addition, we suggest that more attention must be given to populations that are at the limits of their species distributions, that have reduced or no planktonic stages or have limited dispersal capabilities for any other reason. In general, repeated local extinctions and recolonizations may be a frequent feature of the history of many marine fish populations as it was proposed several years ago (Grant and Bowen 2006).
References
Abreu L, Shackleton NJ, Schönfeld J, Hall M, Chapman M (2003) Millennial-scale oceanic climate variability off the Western Iberian margin during the last two glacial periods. Mar Geol 196:1–20
Almada VC, Pereira AM, Robalo JI, Fonseca JP, Levy A, Maia C, Valente A (2008) Mitochondrial DNA fails to reveal genetic structure in sea-lampreys along European shores. Mol Phylogen Evol 46:391–396
Almada VC, Robalo JI, Levy A, Freyhof J, Bernardi G, Doadrio I (2009) Phylogenetic analysis of Peri-Mediterranean blennies of the genus Salaria: molecular insights on the colonization of freshwaters. Mol Phylogen Evol 52:424–431
Amorim MCP, Vasconcelos RO, Marques JF, Almada F (2006) Seasonal variation of sound production in the Lusitanian toadfish, Halobatrachus didactylus. J Fish Biol 69:1892–1899
Amorim MCP, Simões JM, Fonseca PJ, Almada VC (2010) Patterns of shelter usage and social aggregation by the vocal Lusitanian toadfish. Mar Biol 157:495–503
Ayre DJ, Minchinton TE, Perrin C (2009) Does life history predict past and current connectivity for rocky intertidal invertebrates across a marine biogeographic barrier? Mol Ecol 18:1887–1903
Bandelt H, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Bauchot M-L (1987) Poissons osseux. In: Fischer W, Schneider M, Bauchot M-L.Fiches (Eds) FAO d’Identification des Espèces pour les Besoins de la Pêche. Méditerranée et Mer Noire (Zone de Pêche 37). Révision 1. Vol. II—Vertébrés. FAO, Rome, pp 891–1421
Benjamini Y, Hochberg Y (2005) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300
Bowen BW, Grant WS (1997) Phylogeography of the sardines (Sardinops spp.): assessing biogeographic models and population histories in temperate upwelling zones. Evolution 51:1601–1610
Bowen BW, Muss A, Rocha LA, Grant WS (2006) Shallow mtDNA coalescence in Atlantic pygmy angelfishes (Genus Centropyge) indicates a recent invasion from the Indian Ocean. J Hered 97:1–12
Campos MC, Costa JL, Quintela BR, Costa MJ, Almeida PR (2008) Activity and movement patterns of the Lusitanian toadfish inferred from pressure-sensitive data loggers in the Mira estuary (Portugal). Fish Man Ecol 15:449–458
Casper LK (2010) Global warming cycles: ice ages and glacial retreat. Infobase Publishing, New York
Chow S, Hazama K (1998) Universal PCR primers for S7 ribosomal protein gene introns in fish. Mol Ecol 7:1247–1263
Costa JL (2004) The biology of the Lusitanian toad fish, Halobatrachus didactylus (Bloch & Schneider, 1801), and its role in the structuring and functioning of the biological communities; special reference to the Mira estuary population. University of Lisbon, PhD dissertation
Costa JL, Almeida PR, Costa MJ (2003) A morphometric and meristic investigation of Lusitanian toadfish Halobatrachus didactylus (Bloch & Schneider, 1801): Evidence of population fragmentation on Portuguese coast. Sci Mar 67:219–231
Dabney A, Storey J, Warnes, G: qvalue (2006) Q-value estimation for false discovery rate control. R package version 1
Debes PV, Zachos FE, Hanel R (2008) Mitochondrial phylogeography of the European sprat (Sprattus sprattus L., Clupeidae) reveals isolated climatically vulnerable populations in the Mediterranean Sea and range expansion in the northeast Atlantic. Mol Ecol 17:3873–3888
Drummond AJ, Rambaut A (2007) BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Drummond AJ, Rambaut A, Shapiro B, Pybus OG (2005) Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22:1185–1192
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581
Excoffier L, Lischer H (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10:564–567
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Balding D (2003) Gametic phase estimation over large genomic regions using an adaptive window approach. Human Genomics 1:7–19
Francisco SM, Faria C, Lengkeek W, Vieira MN, Velasco EM, Almada VC (2011) Phylogeography of the shanny Lipophrys pholis (Pisces: Blenniidae) in the NE Atlantic records signs of major expansion event older than the last glaciation. J Exp Mar Biol Ecol 403:14–20
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:437–460
Grant WS, Bowen BW (2006) Living in a tilted world: climate change and geography limit speciation in old world anchovies (Genus Engraulis). Biol J Lin Soc 88:673–690
Henriques M, Gonçalves EJ, Almada VC (2007) Rapid shifts in a marine fish assemblage follow fluctuations in winter sea conditions. Mar Ecol Progress Ser 340:259–270
Modesto T, Canário AVM (2003) Morphometric changes and sex steroid levels during the annual reproductive cycle of the Lusitanian toadfish, Halobatrachus didactylus. Gen Comp Endocri 131:220–231
Ostellari L, Bargelloni L, Penzo E, Patarnello P, Patarnello T (1996) Optimization of single-strand conformation polymorphism and sequence analysis of the mitochondrial control region in Pagellus bogaraveo (Sparidae, Teleostei): rationalized tools in fish population biology. Anim Gen 27:423–427
Palazón-Fernández JI, Arias AM, Sarasquete C (2001) Aspects of the reproductive biology of the toadfish, Halobatrachus didactylus (Schneider, 1801) (Pisces: Batrachoididae). Sci Mar 65:131–138
Rambaut A, Drummond AJ (2007) Tracer v1.4, Available from http://beast.bio.ed.ac.uk/Tracer
Robalo JI, Castilho R, Francisco SM, Almada F, Knutsen H, Jorde PE, Pereira AM, Almada VC (2012) Northern refugia and recent expansion in the North Sea: the case of the wrasse Symphodus melops (Linnaeus, 1758). Ecol Evol 2:153–164
Roux C (1986) Batrachoididae. In: Whitehead PJ, Bauchot M-L, Hureau J-C, Nielsen J, Tortonese E (eds) Fishes of the North-Eastern Atlantic and the Mediterranean, vol III. UNESCO, Paris, pp 1360–1361
Sato A, Takezaki N, Tichy H, Figueroa F, Mayer WE, Klein J (2003) Origin and speciation of haplochromine fishes in East African crater lakes investigated by the analysis of their mtDNA, Mhc genes, and SINEs. Mol Biol Evol 20:1448–1462
Stamatis C, Triantafyllidis A, Moutou KA, Mamuris Z (2004) Mitochondrial DNA variation in northeast Atlantic and Mediterranean populations of Norway lobster, Nephrops norvegicus. Mol Ecol 13:1377–1390
Stenseth NC, Ottersen G, Hurrell JW, Chan K-S, Lima M (2002) Ecological effects of climate fluctuations. Science 297:292–1296
Tajima F (1983) Evolutionary relationship of DNA sequences in finite populations. Genetics 105:437–460
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Thomson DA, Lehner CE (1976) Resilience of a rocky intertidal fish community in a physically unstable environment. J Exp Mar Biol Ecol 22:1–29
Triantafyllidis A, Apostolidis AP, Katsares V, Kelly E, Mercer J, Hughes M, Jørstad KE, Tsolou A, Hynes R, Triantaphyllidis C (2005) Mitochondrial DNA variation in the European lobster (Homarus gammarus) throughout the range. Mar Biol 146:223–235
Vasconcelos RO, Carriço R, Ramos A, Modesto T, Fonseca PJ, Amorim MCP (2011) Vocal behavior predicts reproductive success in a teleost fish. Behav Ecol. doi:10.1093/beheco/arr199
Acknowledgments
We appreciate the skilful technical assistance provided by Sónia Chenu. This study was funded by the European Science Foundation’s MarinERA Project “Marine phylogeographic structuring during climate change: the signature of leading and rear edge of range shifting populations” and by the Eco-Ethology Research Unit (PEst-OE/MAR/UI0331/2011) and CCMAR strategic plans (Fundação para a Ciência e Tecnologia, partially FEDER funded). Collection of specimens complied with the current laws of each country. M Clara Amorim research was supported by a FCT post-doc grant (SFRH/BPD/41489/2007).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. I. Taylor.
Rights and permissions
About this article
Cite this article
Robalo, J.I., Crespo, A.M., Castilho, R. et al. Are local extinctions and recolonizations continuing at the colder limits of marine fish distributions? Halobatrachus didactylus (Bloch & Schneider, 1801), a possible candidate. Mar Biol 160, 2461–2467 (2013). https://doi.org/10.1007/s00227-013-2241-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-013-2241-5