Abstract
Distribution shifts of intertidal species have been thoroughly described for several organisms on the Atlantic coasts of Europe, but comparatively less work has been conducted in the Mediterranean Sea. However, such semi-enclosed basins react faster to global change. The periwinkle Echinolittorina punctata is a littorinid gastropod which has shown a remarkable range expansion during the last few decades. Moreover, its unambiguous taxonomy, accessible habitat and ease of identification qualify it as an ideal descriptor of biotic changes in the Mediterranean Sea. We investigated genetic differentiation among 17 Mediterranean and Atlantic populations covering its current distributional range, using the mitochondrial cox1 marker to test the hypothesis that it experienced a significant range expansion triggered by sea warming after the Last Glacial Maximum (LGM). This would confirm its suitability as descriptor of climatic fluctuations. E. punctata has a weak population structure along its distributional range, with greater haplotype diversity in the western African populations than in the Mediterranean Sea, suggesting a recent population expansion following a bottleneck event. Such range expansion is dated ca. 15,000 years BP, and is, thus, related to the end of LGM. E. punctata planktotrophic development lasts 3–4 weeks, likely triggering the weak population structure and enhanced ability to track environmental change.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Climate change triggers modifications of the geographical ranges of marine organisms (Burrows et al. 2011; Poloczanska et al. 2013). Such changes tend to be faster in intertidal species, because they are living close to their physiological tolerance limits (Birchenough et al. 2015). The observation of distributional shifts in the intertidal is also easier and more reliable because geographical ranges of intertidal species are essentially one-dimensional, as they occupy a narrow strip of coastline between the low and high tide levels, making them optimal indicators of climate and global change in the sea (Mieszkowska 2009). Range extensions of intertidal organisms have been recorded extensively from the European Atlantic coasts (e.g., Mieszkowska et al. 2006 for the United Kingdom; Rubal et al. 2013 for the Iberian peninsula), but there is comparatively less information for the Mediterranean Sea.
The periwinkle Echinolittorina punctata (Gmelin, 1791) (Fig. 1b) is a littorinid gastropod distributed on the northwestern African coasts and in the Mediterranean Sea. Once distributed mainly in the southern sectors of the basin, a 600-kilometer range extension has been recorded in the last 15 years (Albano 2014). Nowadays, populations are present in generally considered cooler areas of the Mediterranean such as southern France (Albano 2015) and Croatia (Milat et al. 2016). Due to its unambiguous taxonomy (Reid 2011), accessible habitat and ease of identification (Albano and Trono 2008; Albano 2010), this species is a candidate as an early-warning descriptor of climate change (Albano et al. 2010).
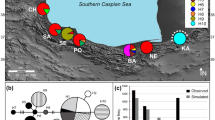
Haplotype network (a) showing the relationship among 26 cox1 haplotypes from populations of Echinolittorina punctata (b) from the Mediterranean Sea and the Atlantic Ocean (c). Locality data in Table 1, haplotype names in Table 2. Each haplotype is indicated by a circle, with circle size proportional to the observed haplotype frequency (the smallest circles correspond to one individual). The length of each line is proportional to the number of substitutions (usually only one mutational step, except H_2 and H_18 with two mutational steps). Image b is reproduced with Editor’s permission from Albano (2010)
The Mediterranean Sea has experienced dramatic climatic fluctuations also in the recent geological past. After the Messinian Salinity Crisis which annihilated the Mediterranean marine fauna, the basin was flooded by Atlantic water and biota in the Pliocene 5.33 Myr ago (Hsü et al. 1977). This fauna was exposed to oceanographic and climatic modifications associated with the change from the tropical–subtropical situation of the Pliocene to the cooler times of the Pleistocene (Sabelli and Taviani 2014). This epoch was characterised by multiple glacial–interglacial cycles that caused dramatic changes in the distribution of organisms (Wilson and Veraguth 2010) with the Last Glacial Maximum (LGM) occurring approximately between 24 and 16.5 kyr BP in the Mediterranean Sea (Cacho et al. 2001).
Thermophilic species sensitive to climatic fluctuations, such as E. punctata, should have extended their ranges into the Mediterranean as a response to the progressive warming following the LGM. Such evidence would confirm a species’ suitability as a descriptor of sea warming. However, the fossil record of intertidal hard substrate species is very poor and unsuitable for testing this hypothesis. Molecular approaches can overcome the lack of fossil data: the genetic structure can provide evidence of a range expansion (Avise 2000) which can be dated with a molecular clock calibration. In particular, rapid range expansion from refugial populations during the LGM might have involved serial bottlenecking with progressive loss of allelic diversity, so that populations in more recently colonized places would contain less genetic diversity. Conversely, it is expected that former refugial areas would contain genetically diversified populations.
Therefore, we test the hypothesis that E. punctata experienced a significant range expansion triggered by sea warming after the Last Glacial Maximum by analyzing the genetic structure of the Mediterranean and nearby Atlantic populations and estimating the timing of the last expansion with a molecular clock calibration.
Materials and methods
Data collection, DNA extraction, and cox1 sequencing
We analyzed 171 specimens from 17 localities across the Mediterranean and Atlantic distribution of E. punctata (Fig. 1c; Table 1). A small piece of tissue was obtained from the mantle of each individual and used for DNA extraction. Total genomic DNA was isolated following the CTAB protocol for molluscs (Winnepenninckx et al. 1993) and quantified using a ND-1,000 spectrophotometer. A fragment of the mitochondrial cox1 gene was amplified for all individuals with the universal primers LCO1490 and HCO2198, designed by Folmer et al. (1994). PCR amplifications were carried out in a 25-μl reaction volume containing 5U/μl of Taq DNA polymerase (Fermentas), 10× load buffer-MgCl2 (Fermentas), 2mM dNTPs (Promega), 2 μM of each primer (Invitrogen), 100× bovine serine albumine (BSA, BioLabs) and approximately 100 ng of template DNA. The PCR amplification was performed on a GeneAmp® PCR System 2700 (Applied Biosystems) based on the following conditions: initial denaturation at 95 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C for 30 s, and extension at 72 °C for 2 min, and a final extension at 72 °C for 10 min. All the amplicons were confirmed by 1.5% TAE 1X agarose gel electrophoresis stained with 10X SYBR Safe concentrate in DMSO (Invitrogen), and purified using the Wizard® SV Gel and PCR Clean-Up System (Promega). The cleaned products were then sent to Macrogen Europe (The Netherlands) for automatic sequencing reactions, which were performed using the PCR primers at 5 μM concentration. To minimize sequencing errors, both strands were sequenced from the cox1 gene for each individual sample.
Genetic and haplotype diversity
For each specimen, forward and reverse cox1 strands were checked in Chromas Lite 2.1.1 (http://www.technelysium.com.au/chromas_lite.html) and combined in BioEdit 7.1.11 (Hall 1999), to obtain consensus sequences to be used in subsequent analyses. The sequences of cox1 were assembled and aligned in MEGA6 (Tamura et al. 2013) using CLUSTALW (Thompson et al. 1994), set at default parameters. cox1 data were translated into amino acid sequences and the substitution saturation test (Xia et al. 2003) was computed in DAMBE 4.2.13 (Xia and Xie 2001) to estimate levels of saturation for the entire cox1 gene and for the first and second versus third codon positions. Genetic diversity was investigated by calculating the number of haplotypes (n), number of polymorphic sites (S), haplotype diversity (h) and nucleotide diversity (π). These indices were estimated for each location as well as for the total sample using DnaSP 5.10 (Librado and Rozas 2009). Identical sequences were collapsed into haplotypes in DnaSP 5.10. The Bayesian information criterion (BIC; Schwarz 1978) and the corrected Akaike information criterion (AICc; Sugiura 1978; Hurvich and Tsai 1989) as implemented in MEGA6 were used to determine the appropriate nucleotide substitution model, which was inferred for the entire cox1 gene as well as for the first, second and third codon positions (models with the lowest BIC scores are considered to best describe the substitution patterns).
Relationships and geographic distribution of haplotypes
To resolve haplotype relationships and to represent their spatial distribution, the minimum spanning network (MSN) method (Excoffier and Smouse 1994) was employed to build a parsimony network, created in the software Network v. 4.5.1.6 (http://www.fluxus-engineering.com) using the median-joining algorithm (Templeton et al. 1992; Bandelt et al. 1999). Haplotype networks better illustrate genetic divergence at the intra-specific level than phylogenetic trees, especially in cases of multiple haplotypes that are derived from a single ancestral sequence (Templeton et al. 1992).
Population structure and spatial analysis
Genetic differentiation between populations was first estimated on the basis of pairwise Φ-statistics (ΦST), computed in Arlequin 3.5 (Excoffier and Lischer 2010) using the Kimura 2-parameter model (K2P) (Kimura 1980). The significance of the statistics was obtained by 10,000 data permutations to generate null distributions of pairwise ΦST values under the hypothesis of no difference between the populations. Significance level (p value) was 0.05; for multiple comparisons, it was adjusted using the Bonferroni correction. Population structure was also examined by the analysis of molecular variance (AMOVA) (Excoffier et al. 1992), which was conducted in Arlequin 3.5 based on the K2P model (AMOVA I). To detect additional hierarchical genetic structure among sampling sites, multilevel AMOVAs using different levels of subdivision were also performed, with molecular variance partitioned into three levels: between groups, among populations within groups and within populations. Therefore, first, populations were classified into six geographic regions, according to their geographic distribution, the main Mediterranean marine currents and the Siculo–Tunisian sill, the Gibraltar sill and Almeria–Oran front (AOF) as barriers to gene flow (AMOVA II, six groups): the Levantine basin (LB: Israel, Turkey, Greece), the Ionian Sea (IS: Salento, Catania, Ragusa), the Tyrrhenian Sea (TS: Palermo, Ischia, Lazio), the northwestern Mediterranean (NW: France, Blanes, Mallorca), the Alboran Sea (AS: Malaga, Gibraltar) and the Atlantic (Atl: Cadiz, Morocco, Senegal). Structure of populations distributed in the eastern (EAST = LB + IS) and western (WEST = TS + AS) Mediterranean basins and in the Atlantic region (Atl) was also explored (AMOVA III, three groups), based on Siculo–Tunisian sill and Gibraltar sill as the only barriers to gene flow. Additional analysis was also used to investigate variation between Mediterranean Sea and Atlantic Ocean, based on the Mediterranean–Atlantic transition (AMOVA IV, two groups). Finally, tests to detect additional geographic structures were performed by grouping the samples into three clusters (AMOVA V, Israel, Senegal and all the other populations), or two clusters (AMOVA VI, Israel and all the other populations; AMOVA VII, Senegal and all the other populations), based on FST results. Indeed, Israel was considered a different group given the significant FST values found with eight other populations. At the same time, the placement of Senegal in a distinct group was justified by significant pairwise ΦST values found with all the other populations and the identifiable cluster formed by all its private haplotypes, which constitute the only geographically restricted clade observed in the haplotype network.
A spatial analysis of molecular variance (SAMOVA) implemented in SAMOVA 1.0 (Dupanloup et al. 2002) was also applied, to test for significant geographic groups of populations that are maximally differentiated from each other based on sequence data. The aim was to determine whether more variation can be explained by other significant groupings of populations than our a priori divisions used in the AMOVA analyses. We tested divisions from two to 15 groups and, because the final configuration was influenced by the initial configuration, 100 simulated annealing steps were used (Dupanloup et al. 2002). For each analysis, we examined the proportion of genetic variance due to differences among groups, ΦCT, and found the K value for which ΦCT was largest and statistically significant, as it is assumed to be the best number of groups and the best population configuration. For population divisions found at each K level, the significance of the ΦCT statistics was assessed by non-parametric randomization tests using 10,000 permutations.
Demographic history
Signatures of demographic changes within E. punctata were first inferred by comparing the observed frequency distribution of pairwise nucleotide differences among individuals (i.e., mismatch distributions; Rogers and Harpending 1992), with distributions expected from a population expansion. Populations at demographic equilibrium or in decline provide a multimodal distribution, while populations that have experienced a sudden demographic expansion display an unimodal distribution (Slatkin and Hudson 1991; Rogers and Harpending 1992). Mismatch distributions for both SAMOVA groups and the total sample were generated using DnaSP 5.10. The fit of the observed distributions to those expected under the sudden expansion model was further assessed by calculating sums of squared deviations (SSD) and the raggedness index r (Harpending 1994), both calculated in Arlequin 3.5 with small SSD and r values typical of an expanding population, and higher values indicating stationary or bottlenecked populations (Harpending 1994). Significance of SSD and r was assessed by 1000 parametric bootstrap replicates.
Demographic changes in E. punctata were also examined by Tajima’s D (Tajima 1989) and Fu’s Fs (Fu 1997) statistics, using Arlequin 3.5, to test whether cox1 fragment evolved under neutrality or departed from neutrality due to factors such as population bottlenecks or expansion events. We also calculated the R2 test using DnaSP 5.10, suitable for small sample sizes (Ramos-Onsins and Rozas 2002). Significance of all tests was assessed by generating 10,000 simulated samples under the hypothesis of selective neutrality and population equilibrium.
For the population groups for which a sudden demographic expansion could not be rejected, the time at which the expansion event took place (t in generations) was dated following the expression t = τ/2μk, where τ is the estimated number of generations since the expansion, μ is the mutation rate per site per generation, and k is the sequence length. A cox1 mutation rate of 2% was considered, following previous studies on the genus Echinolittorina (Williams and Reid 2004; Reid et al. 2006) and works based on cox1 mutation rate in gastropods (Metz et al. 1998; Helleberg and Vacquier 1999; Layton et al. 2014). We assumed a mean generation time of 1 year, in consideration of the species’ reproductive cycle (Palant and Fishelson 1968).
An independent measure of demographic history was estimated using a Bayesian skyline plot (BSP) (Drummond et al. 2005) implemented in BEAST v. 2 (Bouckaert et al. 2014). The Bayesian skyline plot uses MCMC sampling procedures to estimate a posterior distribution of effective population size through time from a sample of gene sequences, given a previously specified nucleotide substitution model. The analysis was made under a strict molecular clock, with time dimension calibrated by fixing mutation rate to 2% per Myr. The dataset was run for 10 million generations and launched from a random starting tree, under an HKY (Hasegawa Kishino Yano) nucleotide substitution model with individual parameters estimated from the data, a constant skyline model with 10 groups, and uniform priors. The chain was sampled every 1000 generations and the first 10,000 generations were discarded as burn-in. The trace plot was inspected to assess convergence and stationarity of the MCMC process in Tracer v 1.6 (Rambaut et al. 2014).
Results
Genetic and haplotype diversity
From an initial 767-bp amplified region, a 636-bp fragment of the mitochondrial cox1 gene was obtained. Analysis of the whole alignment revealed no indels or stop codons, as expected for a coding region. Measure of substitution rate showed insignificant levels of saturation (I SS = 0.0233, ISS.C = 0.7232; P < 0.001), even when the third codon positions were analyzed independently (I SS = 0.6899, I SS.C = 0.9297; P < 0.001). Thirty-one variable sites were found in the alignment, 11 of which were parsimony-informative, yielding an overall h value of 0.57800 ± 0.04300 and a π of 0.00166 ± 0.00023 (Table 1). This diversity resulted in 26 haplotypes (NCBI GenBank A.N. KX866426-KX866451); their geographical distribution is shown in Table 2.
The number of haplotypes in each population ranged between one, in two western Mediterranean localities (Mallorca and Malaga populations) and one nearby Atlantic locality (Cádiz), and six, in the southernmost Atlantic locality (Senegal). Five haplotypes emerged from Ragusa (central Mediterranean basin), while four haplotypes were found in Salento and Ischia (central Mediterranean Sea) and in Morocco. Most haplotypes (22 of 26) were private alleles, i.e., observed in only one population, while the remaining four haplotypes were, instead, shared by more populations: H_1, present at all sampling sites except Senegal, H_3, shared by all the Italian localities, H_7, found in Ragusa and Salento and, finally, H_14, present in Israel and Turkey. Therefore, no private haplotypes were detected in the western Mediterranean region (Mallorca and Malaga), in the more northern Mediterranean and Atlantic localities (Lazio and Cádiz), and in a central site (Catania), while the greatest number of endemic haplotypes was found in Senegal (all the six haplotypes found in this locality), Morocco (three), and Ischia, Ragusa and Gibraltar (two). All the unique haplotypes were singletons, i.e., identified in a single specimen, with the exception of H_2 (Ischia, two individuals), H_8 (Salento, two individuals), H_19 (Senegal, three individuals) and H_21 (Senegal, two individuals). The four shared haplotypes were, instead, the most frequent, with H_1 clearly the most common (64% of the analyzed individuals) and having the highest frequency in the western and eastern Mediterranean localities, and in the localities around the Gibraltar sill (Gibraltar, Cadiz and Morocco). H_3 was the second most frequent haplotype (frequency = 18), followed by H_14 (frequency = 14), while H_7 was found in three individuals.
Haplotype diversity (h ± SD) was very much variable, varying from 0.00000 ± 0.00000 (Malaga, Mallorca and Cadiz) to 0.88900 ± 0.09100 (Senegal), while nucleotide diversity (π ± SD) was low in all the localities, ranging from 0.00000 ± 0.00000 (Malaga, Mallorca and Cadiz) to 0.00463 ± 0.00146 (Senegal) (Table 1). Model of DNA substitution chosen via AICc was the K2P model for the entire cox1 gene as well as for the first, second and third codon positions. The low genetic variation obtained from the cox1 marker showed that the number of individuals was adequate for the present analysis. Increasing the sample size would have produced a marginal increase in dataset information.
Spatial distribution of haplotypes
The minimum spanning network (MSN) (Fig. 1a) did not show any phylogeographical structure among the sampled populations, showing a typical star-like configuration, with the most common haplotype (H_1) at the centre, and less frequent haplotypes, unique or associated with only a few localities, closely related to it. This pattern is often regarded as a signature of a recent population expansion.
All the private haplotypes from Senegal derived from H_1 through a median vector mv1 (with H_19 also connected to a private haplotype from Morocco H_24), forming a distinct haplogroup. Interestingly, haplotype H_19 from Senegal was linked to H_24 from Morocco, by one mutational step. Two derivations emerged from the haplotype network, originated, respectively, from the common haplotype H_3 and the median vector mv1, and interconnected through the central, highly abundant H_1. The first derivation (around H_3) was confined to the central Mediterranean basin and consisted of three haplotypes: H_3, common to all the “Italian” localities (Lazio, Ischia, Palermo, Catania, Ragusa and Salento) and directly derived from H_1, and H_8 (from Salento) and H_11 (from Lazio), instead, recently derived from H_3. The second derivation (around mv1) was endemic to Senegal and it was more divergent from the ancestral H_1, given the additional supposed haplotype (mv1) interconnecting the clade with H_1. It has to be noted that one haplotype from Senegal (H_19) was connetcted to a haplotype from Morocco (H_24). Interestingly, a median vector mv2, derived from H_7 (Ragusa + Salento) and H_4 (Ischia) was drawn between two haplotypes from Ischia (H_2 and H_4). In general, a very short genealogy emerged from the pattern found, with branch lengths of only one mutational step, except haplotypes H_2 and H_18, each linked to mv2 and mv1, respectively, by two mutational steps.
Population structure and spatial analysis
Levels of pairwise ΦST after Bonferroni correction for multiple testing are shown in Table 3. The values were variable, oscillating from −0.00091 to 0.66461, with the lowest value corresponding to the population pair composed by France and Gibraltar, and the highest value found between Senegal and Israel. Interestingly, very low ΦST values were observed not only between neighbouring populations, as expected, but also between distantly spaced locations, such as Greece and Morocco (ΦST = −0.01250). Anyway, most pairwise comparisons were not significant, thus indicating relatively low inter-population divergence. Statistically significant differences were observed only in 23 of the 136 pairwise population comparisons, and particularly in the two populations at the geographic margins of the studied area: Israel, different from Senegal and all the Italian localities, and Senegal, different from all the other sites.
Results of AMOVAs did not identify maximally differentiated groups in our sample (Table 4). The AMOVA analysis performed on the 17 populations (AMOVA I) reflected a lack of genetic structure. The global ΦST value amounted to 0.38028, with 62.0% of the variation being explained by differences within populations, while a lower percentage (38.0%) resulted from differences among populations. A priori division of populations into six geographic regions (LB, IS, TS, NW, AS, Atl), tested with AMOVA II, explained only 8.0% of the total variance, while variation within populations was the major component (61.4%). Separating populations into eastern, western and Atlantic groups (AMOVA III) only reduced the among-groups component (3.8%), as well as a low variation between Mediterranean Sea and Atlantic Ocean (13.7%) was found in AMOVA IV. A priori division of populations into three groups (Senegal, Israel and all the other populations, AMOVA V), explained 56.7% of the total variance, but it was not significant. A low genetic structure was also found when Israel was treated as separate group (AMOVA VI). On the contrary, separating Senegal into single group (AMOVA VII) increased the among-groups component to 71.1%, but this value was not significant.
A search for an alternative partition of local populations without a priori assumptions was performed using SAMOVA (Fig. 2). The sequential tests with K values increasing from two to 15 led to increasing Φ CT values, with the highest Φ CT = 0.0665, observed for four groupings. The combination with five groups produced a slightly lower Φ CT and further increases of k Φ CT values approached a plateau with most groups corresponding to single populations. The differentiation between groups in the division of four was significant and explained 56.2% of total variance, while variance among populations within the groups was only 6.1% (Table 4). Thus, we concluded that only a low level of population structure was evident from our dataset, with the division of populations into four groups best describing this level. In this partition of four groupings, Salento, Ischia and Senegal populations coincided with groups 1, 2 and 3, respectively, while group 4 included all the other populations. Interestingly, Senegal is the only population which did not show the most common haplotype H_1, shared by all the other populations.
Demographic history
The mismatch analyses performed on the total sample and on the four SAMOVA groups are shown in Fig. 2. Total sample, Salento population and the SAMOVA group including 14 populations all produced a skewed unimodal distribution of pairwise differences, consistent with a sudden population expansion model after a bottleneck event. Visually, all the distributions fit an L-shaped curve rather than the typical bell-shaped curve. On the contrary, mismatch distributions for both Ischia and Senegal populations showed a bimodal pattern indicating a demographic equilibrium. Nevertheless, SSD and raggedness r index were not significant for all the analyzed groups showing that all the observed distributions did not differ significantly from expected under a sudden expansion model (Table 5). The Tajima’s D and Fu’s neutrality tests were negative and significant for only two samples, the SAMOVA group formed by 14 populations and the total sample, thus indicating that the hypothesis of sudden population expansion could not be rejected (Table 5). R2 test further evidenced a sudden population expansion for those groups (Table 5). Spatial expansion time parameters τ of 0.8 and 0.7 (95% confidence interval) were found for total sample and SAMOVA 14 populations group, respectively.
The population expansion event occurred approximately 15,000 years BP for the total sample and 13,000 years BP for the SAMOVA group. Consistently, the Bayesian skyline plot (BSP) showed that E. punctata experienced a rapid growth in effective population size following the end of the LGM 16,500 years BP (Fig. 3).
Bayesian skyline plot (BSP) calculated from all cox1 sequences, showing historical demographic trends of Echinolittorina punctata. The solid black line is the mean estimate, while the grey solid lines correspond to the 95% highest posterior density (HPD) limits. The analysis shows a marked population increase following the Last Glacial Maximum (LGM, in grey)
Discussion
Population structure
Our analysis of cox1 sequences of E. punctata in the Mediterranean Sea and the Atlantic Ocean revealed weak population structure along its distributional range. Low levels of differentiation were first indicated by low ΦST values between populations, in most cases close to zero or also negative. AMOVA and SAMOVA analyses also indicated only a very weak structuring of populations into four groups: Salento and Ischia (both Italian localities have been colonized in the last 10–15 years), Senegal (the farthest studied locality), all the other populations.
The Senegal population showed the highest haplotype diversity and the lack of the H_1 haplotype, which is widespread in the Mediterranean and nearby Atlantic (Morocco included). Therefore, it may be the centre of origin of E. punctata and the northern African and Mediterranean populations may have arisen from a bottleneck event. Senegal and Morocco are separated by a 2000-kilometer coastline which is mostly sandy and, therefore, unsuitable for the settlement of periwinkles. This area is also marked by an oceanographic and biogeographic boundary between the temperate north and the tropical south. Upwelling occurs off Mauritania, inducing a difference in water temperature between the Banc d’Arguin and Senegal of 3 °C in winter and 7 °C in summer (Dedah 1993). This barrier is sufficiently strong to have triggered allopatric speciation events (e.g., in nudibranchs, Almada et al. 2016). Such southern populations along the Western African coast are rarely taken into consideration in phylogeographies of Atlanto-Mediterranean organisms.
The main morphological and oceanographic features of the Mediterranean Sea, such as the Gibraltar sill, the Almeria–Oran front and the Siculo–Tunisian sill, are often regarded as barriers to gene flow promoting genetic structuring of marine species (see Patarnello et al. 2007 for a review). The Gibraltar sill favoured the desiccation of the Mediterranean Sea during the Messinian Salinity Crisis; moreover, the dominant surficial current flows into the Mediterranean (Bryden et al. 1994) hindering gene flow across it (e.g., Duran et al. 2004; Lo Brutto et al. 2004; Roman and Palumbi 2004; Palero et al. 2008 for Gibraltar). Eastward, gyres forming a well-defined hydrogeographical boundary of surface waters between Almeria in southeast Spain and Oran in Algeria (the so-called Almeria-Oran oceanographic front (AOF); Tintore et al. 1988) are considered another barrier to gene flow, sometimes most significant than the Gibraltar sill (e.g., Ríos et al. 2002; Calderón et al. 2008; Galarza et al. 2009; Shemesh et al. 2009). During the glacial periods of the Pleistocene, sea level dropped by 120 m, triggering a significant reduction in water flow exchange between the eastern and western basins across the Siculo–Tunisian sill (Shackleton et al. 1984) and, consequently, reducing the gene flow (e.g., Virgilio et al. 2009; Sasson et al. 2012).
Nonetheless, other species show lack of genetic structuring in Atlanto-Mediterranean populations. The pattern described for E. punctata has also been reported for other species of molluscs (Launey et al. 2002), brachiopods (Evangelisti et al. 2010), crustaceans (Stamatis et al. 2004), echinoderms (Zulliger et al. 2009) and fishes (Zardoya et al. 2004).
Echinolittorina punctata spawns pelagic egg capsules. Little is known about the time of development, but the congeneric tropical E. hawaiiensis (Rosewater and Kadolsky 1981) hatches after 3 days and metamorphosis occurs after 21–25 days (Struhsaker and Costlow 1968). A similar duration of planktotrophic development can also be assumed for all Echinolittorina species from their uniform larval shells (Williams and Reid 2004). Species with a pelagic larval stage are expected to show high gene flow among populations as a consequence of their high dispersal potential (i.e., Palumbi 2003). Such gene flow is the most probable cause of the observed low genetic structure.
Range extension following LGM
The demographic analysis provided evidence of a population expansion which can be related to the end of the LGM, dated ca. 16,500 years BP in the Mediterranean Sea (Cacho et al. 2001). At the time, the European ice sheets extended south, creating unfavourable climatic conditions beyond the animal tolerance, which pushed species south, both in the Atlantic and the Mediterranean (Wares and Cunningham 2001; Ilves et al. 2010). During the LGM, annual mean sea surface temperature (SST) was 2 and 7 °C lower than today in the Eastern and Western Mediterranean, respectively (Mikolajewicz 2011). Northerly cold winds of Atlantic origin and the small ice sheet or enhanced glaciation in the Alps triggered an even greater summer cooling, especially in the Western Mediterranean.
Reproduction in E. punctata occurs in summer. Gonad maturation starts in early spring, but most individuals are fertile between June and September (Palant and Fishelson 1968 for Israeli populations). Preliminary data suggest that the recent increase in spring SST along the Italian coastline has triggered its remarkable range extension along the peninsula in the last 15 years (Albano et al. 2010). There is no evidence of the presence of E. punctata in the Mediterranean Sea during the Pleistocene, but such rocky intertidal species have a very limited fossilization potential. Nonetheless, the warming of the Mediterranean Sea following the LGM clearly triggered the population expansion, probably from glacial refugia in northwestern Africa where the temperature drop during LGM was less extreme and where, nowadays, greater haplotype diversity can be observed. Indeed, the most characteristic signature of glacial refugia is high genetic diversity and the presence of private haplotypes/alleles, in contrast to recently colonized areas that have lower diversity and/or only common haplotypes (Avise 2000).
The signature of population expansion lies in the presence of a shared haplotype (H_1) in all the sampled populations in the Mediterranean and nearby Atlantic. The unimodal mismatch distribution obtained for the total sample, together with non-significant SSD and r values, are also indicative of a recent population expansion, with the typical L-shape suggesting a recent genetic bottleneck with the subsequent rapid population expansion creating a “wave” in the distribution that will shift to the right over time (Rogers and Harpending 1992). Negative and significant values of Tajima’s D and Fu’s F statistics, pointing to an excess of low-frequency polymorphisms, also support the hypothesis of population range expansion following a bottleneck (Slatkin and Hudson 1991). All these results fit well with the star-like topology of the haplotype network, which is dominated by a single, most common haplotype (H_1), presumably the ancestral one, and 1-step peripheral ones (Slatkin and Hudson 1991). This is a signature of a species that have recently experienced recent colonization events (bottlenecks and founder effects) (Rogers 1995; Avise 2000).
E. punctata as a descriptor of climate change in the Mediterranean
The rocky intertidal ecosystem is likely to be one of the most sensitive natural systems and shows some of the earliest responses to climate change (Mieszkowska 2009). While such communities have been subject to specific research along the Atlantic European coasts, comparatively less work has been conducted along the Mediterranean shores, maybe also because of the modest extent of the intertidal level in the basin. However, mid-latitude semi-enclosed basins such as the Mediterranean Sea react faster to global change, and knowledge on their response is pivotal to determine the ecological and socio-economic impacts of climate change (Sarà et al. 2014).
The periwinkle E. punctata is a candidate as an early-warning descriptor of Mediterranean Sea warming because of its unambiguous taxonomy, accessible habitat and ease of identification. A major northward range extension has been recorded during the last few decades (Albano 2014), demonstrating its potential to quickly track SST increase. Moreover, we have, here, demonstrated that E. punctata recolonized the Mediterranean Sea after the LGM and currently shows no significant population structure, evidence of a geologically recent colonization event. Such evidence of its distribution history in the recent geological past supports its role as climate change descriptor.
References
Albano PG (2010) Further northward extension of the distribution of Echinolittorina punctata (Mollusca: Gastropoda) along the Italian Tyrrhenian coastline: new stations in Lazio. Cah Biol Mar 51:201–204
Albano PG (2014) Recent changes in the distribution of autochthonous marine molluscs in the Mediterranean Sea. In: Goffredo S, Dubinsky Z (ed) The Mediterranean Sea: its history and present challenges, 1st edn, vol 17. Springer, Dordrecht, pp 307–318
Albano PG (2015) New records of Echinolittorina punctata (Gastropoda: Littorinidae) in the Mediterranean Sea from Italy, France and Greece. Mar Biodivers Rec 8:e3
Albano PG, Trono D (2008) On the occurrence of Echinolittorina punctata (Gmelin, 1791) (Gastropoda: Littorinidae) in Puglia, South-Eastern Italy. Boll Malacol 44:123–126.
Albano PG, Sabelli B, Adani M, Pinardi N (2010) The thermophilous species Echinolittorina punctata as a new descriptor of tropicalization in the Mediterranean Sea-first data. Biol Mar Medit 17(1):90–93
Almada F, Levy A, Robalo JI (2016) Not so sluggish: the success of the Felimare picta complex (Gastropoda, Nudibranchia) crossing Atlantic biogeographic barriers. PeerJ 4:e1561
Avise JC (2000) Phylogeography: the history and formation of species. Harvard University Press, Cambridge, MA
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Ecol 16:37–48
Birchenough SNR, Reiss H, Degraer S, Mieszkowska N, Borja Á, Buhl-Mortensen L, Braeckman U, Craeymeersch J, De Mesel I, Kerckhof F, Kröncke I, Parra S, Rabaut M, Schröder A, Van Colen C, Van Hoey G, Vincx M, Wätjen K (2015) Climate change and marine benthos: a review of existing research and future directions in the North Atlantic. WIREs Clim Chang 6:203–223
Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014) BEAST2: a software platform for Bayesian evolutionary analysis. PLOS Comput Biol 10:e1003537
Bryden HL, Candela J, Kinder TH (1994) Exchange through the Strait of Gibraltar. Prog Oceanogr 33(3):201–248
Burrows MT, Schoeman DS, Buckley LB, Moore P, Poloczanska ES, Brander KM, Brown C, Bruno JF, Duarte CM, Halpern BS, Holding J, Kappel CV, Kiessling W, O’Connor MI, Pandolfi JM, Parmesan C, Schwing FB, Sydeman WJ, Richardson AJ (2011) The pace of shifting climate in marine and terrestrial ecosystems. Science 334:652–655
Cacho I, Grimalt JO, Canals M, Sbaffi L, Shackleton NJ, Schönfeld J, Zahn R (2001) Variability of the western Mediterranean Sea surface temperature during the last 25,000 years and its connection with the Northern Hemisphere climatic changes. Paleoceanography 16(1):40–52
Calderón I, Giribet G, Turon X (2008) Two markers and one history: phylogeography of the edible common sea urchin Paracentrotus lividus in the Lusitanian region. Mar Biol 154:137–151
Dedah SO (1993) Wind, surface water temperature, surface salinity and pollution in the area of the Banc d’Arguin, Mauritania. Hydrobiol 258:9–19
Drummond AJ, Rambaut A, Shapiro B, Pybus OG (2005) Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22:1185–1192
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11(12):2571–2581
Duran S, Palacín C, Becerro MA, Turon X, Giribet G (2004) Genetic diversity and population structure of the commercially harvested sea urchin Paracentrotus lividus (Echinodermata, Echinoidea). Mol Ecol 13:3317–3328
Evangelisti F, Albano PG, Sabelli B (2010) Genetic variation of the brachiopod Gryphus vitreus (Born, 1778) across the Gibraltar sill. Biol Mar Medit 17(1):28–30
Excoffier L, Lischer HEL (2010) Arlequin suite ver. 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol 10(3):564–567
Excoffier L, Smouse PE (1994) Using allele frequencies and geographic subdivision to reconstruct gene trees within a species: molecular variance parsimony. Genetics 136:343–359
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol. Biotech 3:294–299
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Galarza JA, Carreras-Carbonell J, Macpherson E, Pascual M, Roques S, Turner GF, Rico C (2009) The influence of oceanographic fronts and early-life-history traits on connectivity among littoral fish species. Proc Natl Acad Sci 106(5):1473–1478
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Harpending HC (1994) Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol 66:591–600
Helleberg ME, Vacquier VD (1999) Rapid evolution of fertilization selectivity and lysin cDNA sequences in teguline gastropods. Mol Biol Evol 16:839–848
Hsü KJ, Montadert L, Bernoulli D, Cita MB, Erickson A, Garrison RE, Kidd RB, Mèlierés F, Müller C, Wright R (1977) History of the Messinian salinity crisis. Nature 267:399–403
Hurvich CM, Tsai CL (1989) Regression and time series model selection in small samples. Biometrika 76:297–307
Ilves KL, Huang W, Wares JP, Hickerson MJ (2010) Colonization and⁄or mitochondrial selective sweeps across the North Atlantic intertidal assemblage revealed by multi-taxa approximate bayesian computation. Mol Ecol 19:4505–4519
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–120
Launey S, Ledu C, Boudry P, Bonhomme F, Naciri-Graven Y (2002) Geographic structure in the European flat oyster (Ostrea edulis L.) as revealed by micro-satellite polymorphism. J Hered 93:331–338
Layton KKS, Martel AL, Hebert PDN (2014) Patterns of DNA barcode variation in Canadian marine molluscs. PLoS One 9(4):e95003. doi:10.1371/journal.pone.0095003
Librado P, Rozas J (2009) DnaSP v5.10: a software for comprehensive analysis of DNA polymorphism data. Bioinform 25:1451–1451.
Lo Brutto S, Arculeo M, Parrinello N (2004) Congruence in genetic markers used to describe Mediterranean and Atlantic populations of European hake (Merluccius merluccius L. 1758). J Appl Ichthyol 20:81–86
Metz EC, Robles-Sikisaka R, Vacquier VD (1998) Nonsynonymous substitution in abalone sperm fertilization genes exceeds substitution in introns and mitochondrial DNA. Proc Natl Acad Sci USA 95:10676–10681
Mieszkowska N (2009) Intertidal indicators of climate and global change. In: Letcher M (ed) Climate change: observed impacts on planet earth, 1st edn. Elsevier, Oxford, pp 281–296
Mieszkowska N, Kendall MA, Hawkins SJ, Leaper R, Williamson NJ, Hardman-Mountford NJ, Southward AJ (2006) Changes in the range of some common rocky shore species in Britain—a response to climate change? Hydrobiol 555:241–251
Mikolajewicz U (2011) Modeling Mediterranean Ocean climate of the Last Glacial Maximum. Clim Past 7:161–180
Milat T, Dragičević B, Nerlović V, Vrgoč S (2016) First record of Echinolittorina punctata (Gmelin, 1791) (Gastropoda) in the Adriatic Sea. Acta Adriat 57(1):179–182
Palant B, Fishelson L (1968) Littorina punctata (Gmelin) and Littorina neritoides (L.), (Mollusca,Gastropoda) from Israel: ecology and annual cycle of genital system. Israel J Zool 17:145–160
Palero F, Abelló P, Macpherson E, Cristina M, Pascual M (2008) Phylogeography of the European spiny lobster (Palinurus elephas): influence of current oceanographical features and historical processes. Mol Phylogenet Evol 48:708–717
Palumbi SR (2003) Ecological subsidies alter the structure of marine communities. Proc Natl Acad Sci USA 100(21):11927–11928.
Patarnello T, Volckaert FMJ, Castilho R (2007) Pillars of Hercules: is the Atlantic-Mediterranean transition a phylogeographical break? Mol Ecol 16(21):4426–4444
Poloczanska ES, Brown CJ, Sydeman WJ, Kiessling W, Schoeman DS, Moore PJ, Brander K, Bruno JF, Buckley LB, Burrows MT, Duarte CM, Halpern BS, Holding J, Kappel CV, O’Connor MI, Parmesan C, Schwing F, Thompson SA, Richardson AJ (2013) Global imprint of climate change on marine life. Nat Clim Chang 3:919–925
Rambaut A, Drummond AJ, Suchard M (2014). Tracer v1.6. http://beast.bio.ed.ac.uk/Tracer
Ramos-Onsins SE, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Mol Biol Evol 19(12):2092–2100
Reid DG (2011) The genus Echinolittorina Habe, 1956 (Gastropoda: Littorinidae) in the eastern Atlantic Ocean and Mediterranean Sea. Zootaxa 2974:1–65
Reid DG, Lal K, Mackenzie-Dodds J, Kaligis F, Littlewood DTJ, Williams ST (2006) Comparative phylogeography and species boundaries in Echinolittorina snails in the central Indo-West Pacific. J Biogeogr 33:990–1006
Ríos C, Sanz S, Saavedra C, Peña JB (2002) Allozyme variation in populations of scallops, Pecten jacobaeus (L.) and P. maximus (L.) (Bivalvia: Pectinidae), across the Almeria–Oran Front. J Exp Mar Biol Ecol 267:223–244
Rogers AR (1995) Genetic evidence for a pleistocene population explosion. E 49(4):608–615
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9(3):552–569
Roman J, Palumbi SR (2004) A global invader at home: population structure of the green crab, Carcinus maenas, in Europe. Mol Ecol 13:2891–2898
Rosewater J, Kadolsky D (1981) Rectifications in the nomenclature of some Indo-Pacific Littorinidae II. Proc Biol Soc Wash 94:1233–1236
Rubal M, Veiga P, Cacabelos E, Moreira J, Sousa-Pinto I (2013) Increasing sea surface temperature and range shifts of intertidal gastropods along the Iberian Peninsula. J Sea Res 77:1–10
Sabelli B, Taviani M (2014) The making of the Mediterranean molluscan biodiversity. In: Goffredo S, Dubinsky Z (eds) The Mediterranean Sea: its history and present challenges. Springer, Dordrecht, pp 285–306
Sarà G, Milanese M, Prusina I, Sarà A, Angel DL, Glamuzina B, Nitzan T, Freeman S, Rinaldi A, Palmeri V, Montalto V, Lo Martire M, Gianguzza P, Arizza V, Lo Brutto S, De Pirro M, Helmuth B, Murray J, De Cantis S, Williams GA (2014) The impact of climate change on Mediterranean intertidal communities: losses in coastal ecosystem integrity and services. Reg Environ Chang 14(1):5–17
Sasson N, Simon-Blecher N, Achituv Y (2012) New molecular markers for revealing the population structure of Chthamalus stellatus in the Mediterranean and the eastern Atlantic. Mar Ecol Progr Ser 459:99–107
Schwarz G (1978) Estimating the dimension of a model. Ann Stat 6:461–464
Shackleton JC, van Andel TH, Runnels CN (1984) Coastal paleogeography of the central and western Mediterranean during the last 125,000 years and its archaeological implications. J Field Archaeol 11(3):307–314
Shemesh E, Huchon D, Simon-Blecher N, Achituv Y (2009) The distribution and molecular diversity of the eastern Atlantic and Mediterranean chthamalids (Crustacea, Cirripedia). Zool Scr 38(4):365–378.
Slatkin M, Hudson RR (1991) Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129:555–562
Stamatis C, Triantafyllidis A, Moutou KA, Mamuris Z (2004) Mitochondrial DNA variation in northeast Atlantic and Mediterranean populations of Norway lobster, Nephrops norvegicus. Mol Ecol 13:1377–1390
Struhsaker JW, Costlow JD (1968) Larval development of Littorina picta (Prosobranchia, Mesogastropoda), reared in the laboratory. Proc Malacol Soc Lond 38:153–160
Sugiura N (1978) Further analysis of the data by akaike’s information criterion and the finite corrections. Commun Stat Theory. Methods 7:13–26
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Templeton AR, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132(2):619–633
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tintore J, La Violette PE, Blade I, Cruzado A (1988) A study of an intense density front in the eastern Alboran Sea: the Almeria-Oran front. J Phys Oceanogr 18:1384–1397
Virgilio M, Fauvelot C, Costantini F, Abbiati M, Backeljau T (2009) Phylogeography of the common ragworm Hediste diversicolor (Polychaeta: Nereididae) reveals cryptic diversity and multiple colonization events across its distribution. Mol Ecol 18:1980–1994
Wares JP, Cunningham CW (2001) Phylogeography and historical ecology of the north Atlantic intertidal. E 55(12):2455–2469
Williams ST, Reid DG (2004) Speciation and diversity on tropical rocky shores: a global phylogeny of snails of the genus Echinolittorina. E 58:2227–2251
Wilson AB, Veraguth IE (2010) The impact of Pleistocene glaciation across the range of a widespread European coastal species. Mol Ecol 19:4535–4553
Winnepenninckx BT, Backeljau T, Deawachter R (1993) Complete small ribosomal subunit RNA sequence of the chiton Acanthopleura japonica (Lischke, 1873) (Mollusca, Polyplacophora). Nucleic Acids Res 21:1670
Xia X, Xie Z (2001) DAMBE: software package for data analysis in molecular biology and evolution. J Hered 92:371–373
Xia X, Xie Z, Salemi M, Chen L, Wang Y (2003) An index of substitution saturation and its application. Mol Phylogenet Evol 26:1–7
Zardoya R, Castilho R, Grande C, Favre-Krey L, Caetano S, Marcato S, Krey G, Patarnello T (2004) Differential population structuring of two closely related fish species, the mackerel (Scomber scombrus) and the chub mackerel (Scomber japonicus), in the Mediterranean Sea. Mol Ecol 13:1785–1798
Zulliger DE, Tanner S, Ruch M, Ribi G (2009) Genetic structure of the high dispersal Atlanto-Mediterreanean sea star Astropecten aranciacus revealed by mitochondrial DNA sequences and microsatellite loci. Mar Biol 156:597–610
Acknowledgements
FE was funded by the Canziani Fund, Department of Biological, Geological and Environmental Sciences, University of Bologna. F. De Santis, B.S. Galil, S. Gofas, D. Grech, P. Ovalis, B. Öztürk, C. Palumbo, D.G. Reid and D. Trono sent samples from various localities in the Mediterranean Sea and West Africa. M. Passamonti commented on a first draft of the manuscript, M. Marzadori helped with figures, and F. Maltagliati and A. Luchetti provided suggestions on Bayesian analysis. D. Davoult, Executive Editor of Cahiers de Biologie Marine, authorized the reproduction of Fig. 1b, originally published in Albano (2010). Two anonymous reviewers provided useful suggestions to improve the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable national and institutional guidelines for the care and use of animals were followed.
Additional information
Responsible Editor: S. Uthicke.
Reviewed by D. Reid and an undisclosed expert.
Rights and permissions
About this article
Cite this article
Evangelisti, F., Bellucci, A., Sabelli, B. et al. The periwinkle Echinolittorina punctata (Mollusca: Gastropoda) tracked the warming of the Mediterranean Sea following the Last Glacial Maximum. Mar Biol 164, 34 (2017). https://doi.org/10.1007/s00227-017-3071-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00227-017-3071-7