Abstract
During the last years, molecular studies revealed significant population differentiation and cryptic species within various benthic and pelagic marine Antarctic taxa. This is unexpected due to the lack of obvious barriers to gene flow and strong current systems. Using mitochondrial (COI, 16S rDNA) and nuclear (28S rDNA: D2) gene fragments, we tested whether two circum-Antarctic benthic shrimps with planktotrophic larvae, Chorismus antarcticus and Nematocarcinus lanceopes, show patterns of regional differentiation. For both species, the 16S and the 28S fragment were invariant. However, for COI we found 24 different haplotypes for Chorismus antarcticus and 54 for Nematocarcinus lanceopes. No significant differentiation was observed among populations or regions. Furthermore, we found signatures of a population expansion in the late Pleistocene hinting at an impact of large-scale glaciations in particular on the shallow-water shrimp Chorismus antarcticus, supporting a (re)colonization and demographic expansion of this shrimp species in response to climate oscillation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biogeographic studies on marine Antarctic species have been conducted for over 150 years and represent some of the most fascinating examples in ecology and evolutionary biology. As consequence of the unique tectonic history, the long isolation, and recurrent large-scale glaciations of Antarctica, the Antarctic marine biota are unique in terms of their physiology, ecology, biogeography, and phylogeny (e.g. Clarke and Johnston 2003; Aronson et al. 2007). However, many aspects are still little understood in detail. Prominent examples are differences in species richness in various taxa, which have undergone a significant radiation in continental shelf waters of Antarctica, e.g. notothenioid fishes, ascidians, crinoids, molluscs, pycnogonids, amphipods, and isopods (Brandt 2000; Eastman and McCune 2000; Held 2000; Clarke and Johnston 2003; Raupach and Wägele 2006; Wilson et al. 2007; Near and Cheng 2008; Krabbe et al. 2010; Wilson et al. 2009).
In contrast to these taxa, the number of known decapod shrimp (Caridea) species on the Antarctic continental shelf is less than ten (Macpherson 1988; Gorny et al. 1992; Klages et al. 1995; Gorny 1999). Although low in species number, the Caridea represent an important and most abundant element of the Antarctic shelf and deep-sea benthos (Arntz and Gorny 1991; Gutt et al. 1991; Thatje and Arntz 2004). For example, the shallow-water species Chorismus antarcticus Pfeffer 1887 (Hippolytidae) represents the most abundant shelf inhabiting Antarctic shrimp, distributed around the Antarctic continent (Pfeffer 1887; Kirkwood 1984; Arntz and Gorny 1991; Gutt et al. 1991). The species exhibits a well-defined bathymetric distribution, with abundance values confirming a preference for depths ≤400 m with up to four specimens per m2 (Gutt et al. 1991). In contrast to this species, the deep-sea shrimp Nematocarcinus lanceopes Bate 1888 (Nematocarcinidae) is known from the deeper waters in the Indian Ocean sector of East Antarctica, the Weddell Sea, the Antarctic Peninsula, the Magellan region, the deep waters west of Chile, and off South Africa (Cape and Agulhas basin; Bate 1888; Barnard 1950; Kirkwood 1984; Ledoyer 1989; Guzmán and Quiroga 2005). Large numbers (up to 9 specimens per m2) were recorded between 550 and 1,200 m depth in the Weddell Sea (Arntz and Gorny 1991; Gutt et al. 1991), and it has been found as deep as 4,000 m (Thatje et al. 2005a), indicating a broad bathymetric distribution range from the Antarctic continental slope to the Southern Ocean abyssal plains.
As marine Antarctic species with pelagic larvae (e.g. Caridea) or any kind of drifting stage are often regarded to have a circum-Antarctic distribution (e.g. Knox 1994; Clarke and Johnston 2003; Vermeeren et al. 2004), for both studied species a circumpolar distribution can be expected. This seems particularly likely as no obvious barriers divide the Antarctic continental shelf nowadays, and actually strong current systems, such as the strong Antarctic Circum Polar Current, facilitate the transport of pelagic distribution stages. However, several recent molecular studies demonstrated that species distributions are often much more restricted, and population differentiation is significantly higher than expected according to the physical characteristics of the habitat. For example, various molecular studies of sequence variation of Antarctic Teleostei (Patarnello et al. 2003; Kuhn and Gaffney 2006; Kuhn and Gaffney 2008), Euphausiacea (Zane et al. 1998; Zane and Patarnello 2000; Jarman and Nicol 2002; Jarman et al. 2002) or Cephalopoda (Allcock et al. 1997; Sands et al. 2003) revealed significant genetic differences in geographically separated populations. Hence, in the case of species with pelagic life stages it cannot be assumed a priori that they are broadly distributed and lack genetic differentiation, either due to a lower mobility than expected or characteristic behaviour such as swarming, affecting the spatio-temporal distribution.
In the present study, we assessed and evaluated the genetic structure of the decapod shrimps Chorismus antarcticus and Nematocarcinus lanceopes from various locations around Antarctica in order to test whether disruptive or unifying ecological and evolutionary forces affect both species’ gene pools. Furthermore, we assessed whether populations of the shallow-water species Chorismus antarcticus compared to the deep-sea species Nematocarcinus lanceopes show reduced genetic diversity and stronger signatures of recent expansions following a past population bottleneck or founding event. This can be expected as a feasible consequence of recurring disturbances of the inhabited shelf areas by advancing grounded glaciers during the ice ages in the late Cenozoic, in particular during the Last Glacial period (Thatje et al. 2005b; Janko et al. 2007). We utilized two fragments of fast evolving mitochondrial genes, the cytochrome c oxidase subunit I (COI) gene and the large ribosomal subunit 16S rRNA gene, as well as the D2 expansion fragment of the nuclear 28S rRNA gene, a promising nuclear marker for molecular taxonomy and hybrid detection (Monteiro et al. 2000, Ninet et al. 2003; Schmidt et al. 2006; Sonnenberg et al. 2007), for all analysed specimens.
Materials and methods
Sampling and DNA extraction
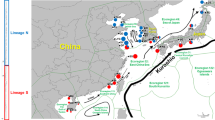
All analysed decapod specimens were collected in the Southern Ocean (Fig. 1) during various expeditions in the years 1996 to 2008. Decapod shrimps were caught using a variety of trawling gear, for example epibenthic sledges, bottom trawls, Rauschert dredges, or Agassiz trawls in depths ranging from 166 to 2,134 m. A detailed sampling record and expedition references are provided in the electronic supplementary Table S1. After collection, samples were stored for at least 24 h at 0° C. Total genomic DNA was extracted from pleon muscle tissue of 178 specimens of Chorismus antarcticus and 187 individuals of Nematocarcinus lanceopes, using a commercial extraction kit (QIAmp© Blood and Tissue Kit, Qiagen GmbH) and following the tissue extraction protocol.
Map of the Southern Ocean showing sample areas. Numbers in parentheses refer to the number of sampled specimens of Chorismus antarcticus (left) and Nematocarcinus lanceopes (right). The black line indicates the Antarctic Polar Front. Circles indicate three geographic groupings of populations (AP Antarctic Peninsula: transparent, EWS Eastern Weddell Sea: light grey; RS Ross Sea: grey) for the analysis of molecular variance (AMOVA). For Nematocarcinus lanceopes, Spiess Seamount and South Sandwich Islands have not been pooled in a geographic group because of their distinct location
DNA amplification and sequencing
Amplification reactions for all gene fragments were carried out on a Progene Thermocycler (Techne Ltd.) or Thermal Cycler GeneAmp© PCR System 2700/2720 (Applied Biosystems) in 20 μl volumes, containing each 4 μl Q-Solution, 2 μl 10× Qiagen PCR buffer, 2 μl dNTPs (2 mmol/μl), 0.1 μl of each primer (both 50 pmol/μl), 1 μl of DNA template, and 0.2 μl Qiagen Taq (5 U/μl) and filled up to 20 μl with sterile H2O. The PCR temperature profile for the mitochondrial COI fragment (~650 bp) using the primers LCO1490 and HCO2198 (Folmer et al. 1994) consisted of an initial denaturation at 94°C (5 min), followed by 38 cycles with 94°C (denaturation, 45 s), 44°C (annealing, 45 s), 72°C (extension, 80 s), and a final extension at 72°C (7 min). The amplification reactions and the temperature profile for the 16S rRNA gene were according to previous studies (Raupach and Wägele 2006). Approximately 200 bp of the 28S rDNA: D2 region was amplified with the newly designed forward primer CD2F (5′-GGACCCGTCTTGAAACAC-3′) and reverse primer CD2R (5′-GCATAGTTCACCATCTTTC-3′), using a PCR protocol of 94°C for 5 min (initial denaturation), 38 cycles with 94°C denaturation for 45 s, 52°C annealing for 45 s, and 72°C extension for 80 s, followed by a final 72°C extension for 7 min.
Negative and positive controls were included in every PCR setup. Three microlitres of amplified product (~0.7 kb) were controlled by electrophoresis on a 1% TAE agarose gel with ethidium bromide using DNA size standards, and the remaining PCR product was purified with a commercial kit (QIAquick© PCR Purification Kit, Qiagen GmbH). Purified PCR products were outsourced for sequencing to a contract sequencing facility (Macrogen, Seoul, Korea) on an ABI3730 XL automatic DNA sequencer, using the same primer set as for PCR. Both sequencing reads were assembled with the program Seqman™ II (DNASTAR, Inc.), while the identity of all new sequences was confirmed with BLAST searches (Altschul et al. 1990). All new sequences were deposited in GenBank (see electronic supplementary Table S2 for accession numbers).
Sequence alignment, haplotype genealogy, and population structure analysis
All sequences of each marker were aligned using Muscle version 3.6 (Edgar 2004) with default settings, generating three individual data sets. Alignments were tested for nucleotide bias using a chi-square test of base composition homogeneity across taxa implemented in PAUP*4.0b10 (Swofford 2002). All aligned COI sequences were translated to amino acid sequences to check for nuclear mitochondrial pseudogenes (numts) using BioEdit 7.0.9.0 (Hall 1999).
For the COI data sets, a statistical parsimony network was constructed with TCS 1.21 (Clement et al. 2000), using default settings (Fig. 2). Population structure analyses for both species were performed using Arlequin 3.11 (Schneider et al. 2000). Haplotype diversity (h) and nucleotide diversity (π; Nei and Miller 1990) were calculated for each population (Table 1). Pairwise F ST (fixation index among populations) were calculated using both haplotype frequencies with and without genetic distances. To estimate differentiation among regions, an analysis of molecular variance (AMOVA) was performed when no geographical differentiation of population within regions was observed (Excoffier et al. 1992; Table 2). To this end, sampled areas were grouped into three geographic regions (“Antarctic Peninsula” vs. “Eastern Weddell Sea” vs. “Ross Sea”) for both species (see Fig. 1). For Nematocarcinus lanceopes, specimens sampled from the South Sandwich Islands and Spiess Seamount (close to Bouvet Island) were not pooled to any of the other regions but remained as a fourth and fifth separate population for the AMOVA due to their distinct and remote geographic location. Covariance components were used to calculate fixation indices (F) among groups of populations (F CT), among populations within groups (F SC), or among populations (F ST). Statistical significances of differences among the geographic regions were assessed by comparing the observed distribution with randomly permuted distributions (10,000 permutations), in which individuals were randomly reallocated to each population.
Statistical parsimony network showing the mutational relationships among the analysed mitochondrial COI haplotypes of the shallow-water shrimp Chorismus antarcticus (above) and the deep-sea species Nematocarcinus lanceopes (below). Each line in the network represents a single mutational change, small black dots indicate missing haplotypes. The colour of the full circles of the network corresponds to the presented colour code of the sampled region (as in the AMOVA). The number of analysed specimens (n) and the depth range are listed [in brackets]. Numbers at the full circles of the networks indicate the haplotype code number (see Table 3 and S1). The diameter of the circles is proportional to the number of haplotypes sampled (see Open circles with numbers). Asterisks indicate amino acid replacement substitutions (colour figure online)
Population history
In order to test whether recent demographic processes have influenced the genetic diversity, we performed four tests to assess whether populations are in mutation-drift equilibrium. First, coalescent modelling of changes in effective population size were analysed using the program Fluctuate vers. 1.4 (Kuhner et al. 1998), which coestimates the parameters θ (the effective population size scaled by the mutation rate, i.e. Neμ) and the exponential rate of population growth or decline relative to the neutral mutation rate “g” for both species. Hence, positive values indicate growth, while negative values give evidence for a population decline. The initial value of θ was estimated using the approach of Watterson (1975). We tested several settings and used 1,000 short Markov chains with 200 generations, and two long Markov chains with 400,000 generations. Sampling increment was 20 for short and long chains. The initial Transition/Transversion ratio was set to 5.
In a second approach, we calculated mismatch distributions of pairwise differences between the specimens using Arlequin 3.11 (Schneider et al. 2000). For each species, the shape of the distribution was compared to a distribution of a simulated data set under a spatial expansion model (Ray et al. 2003; Excoffier 2004) and a sudden population expansion (Rogers and Harpending 1992). The fit between observed and estimated distribution under a sudden expansion model was subjected to two different goodness-of-fit tests (standardized squared differences (SSD) and raggedness index tests; Harpending 1994). The goodness-of-fit tests provide information on how well the simulated model of population expansion fits the observed mismatch distribution, while the parameters Tau (τ), Theta 0, and Theta 1 were inferred directly from the model of demographic expansion in Arlequin 3.11. Furthermore, departures from mutation-drift equilibrium were assessed by calculating Tajima’s D (Tajima 1989) and Fu’s F’s (Fu 1997). Both neutrality tests provide information on possible population expansions or indicate recent selective sweeps. Significance for these two neutrality tests was obtained by simulating 10,000 samples in accordance with the assumptions of selective neutrality and population equilibrium using a coalescent approach.
To approximately date the timing of population expansions (T) for the COI gene according to the formula T = τ/2u following the approach of Rogers (1995) with τ being calculated from the mismatch distribution and u, i.e. for the substitution rate for the 657 bp fragment per year with a bias correction implemented by Excoffier and Schneider (1999), we used several molecular clock estimates. Since no molecular clock for the Antarctic shrimps exists, we used estimates from other decapods that range from 1.4 to 2.6% sequence divergence per MY (Knowlton et al. 1993; Knowlton and Weigt 1998; Schubart et al. 1998). As consequence, we used an average divergence rate of 2% per MY and calculated the lower (more recent) and upper (more ancient) confidence intervals using the divergence rates of 2.6 and 1.4% per MY and the 5 and 95% percentile values for τ, respectively.
Results
Our COI data analyses for 178 individuals of Chorismus antarcticus from 7 populations revealed 24 different haplotypes with a length of 657 bp with no significant differences in base composition (Table 1). For Nematocarcinus lanceopes, a 657-bp fragment was obtained for 187 individuals from 10 different sampling locations. For this deep-sea shrimp species, 54 different haplotypes were found (Table 1). However, there were no intraspecific variations within the analysed mitochondrial 16S rDNA and the nuclear 28S rDNA: D2 sequences (P-distances = 0) for all specimens of both shrimp species. As consequence, population structure analyses were performed only for the COI data sets.
Variability of the COI gene fragment in Chorismus antarcticus
Of 25 polymorphic sites, four (16%) were at the first, three (12%) at the second, and 18 (72%) were at the third codon position. In total, seven amino acid replacement substitutions (first position: 2, second position: 3, and third position: 2) were detected. Uncorrected pairwise genetic distances (p-distances) among haplotypes of Chorismus antarcticus ranged from 0 to 0.0106. Two haplotypes (1 and 2) were present at high frequencies in most analysed populations, representing 48.3 and 35.9% of the entire data set (Fig. 2). Almost all other haplotypes were derived from these haplotypes, while haplotype 1 represented the putative ancestral haplotype, as most other haplotypes derived from this particular haplotype. Of the remaining 22 haplotypes, four were shared among populations, while the other 18 haplotypes were only scored in one specimen (singletons; Table 3). Overall haplotype diversity h was moderate (0.6394 ± 0.0247), while nucleotide diversity π values were low (0.0021 ± 0.0014; Table 1). Most haplotypes differed from each other by only one or two mutations, and there was no evidence for any significant differentiation among populations. Therefore, populations from the three major geographic regions were pooled and analysed in an AMOVA. Results of the AMOVA provided also no evidence for genetic differentiation with >98% of the variation being distributed within populations, supporting the hypothesis of panmixia of the analysed specimens (F CT = 0, F SC = 0.015, F ST = 0; Table 2).
Population history of Chorismus antarcticus based on COI
Sequences of all specimens were pooled as a single group prior to analyses as we found no evidence for population structure. Estimates for the growth rate g were high and positive, with the most likely value for g = 8,212 (SD = 187.9) and θ = 0.0538 (SD = 0.0025), indicating population growth. Replicate runs with alternate random seeds and also different chain settings produced comparable results, ensuring convergence on the correct parameter estimates. Values for g varied in a range of about 3,500–9,000 depending on the number of chains used. Values of both Tajima’s D and Fu’s F’s were negative and significant (Table 4). These negative values indicated an excess of low frequency polymorphisms which is contrary to expected values under a neutral model of sequence evolution.
In line with these findings, distributions of the pairwise differences between sequence pairs showed a bimodal distribution as a result of the two very dominant haplotypes H1 and H2 separated by only two mutation steps (Fig. 2). Both models of expansion cannot be rejected on a 5% confidence level, and also neutrality tests showed significant deviations from mutation-drift equilibrium values (Table 4, Fig. 3). Molecular clock estimates dated the onset of the expansion to 85 KY BP (CI: 0–270 KY BP) for Chorismus antarcticus.
For COI, most replacement substitutions were located at the tips of the network (Fig. 2), indicating an expanding population. Furthermore, the low COI diversity values and the lack of any variation for the mitochondrial 16S rDNA and nuclear 28S rDNA D2 expansion segment indicated that genetic variability was low and not as expected from old viable populations at mutation-drift equilibrium. This genetic population characteristic can be interpreted as a current expansion after a severe past bottleneck but also as a result of a selective sweep.
Variability of the COI gene fragment in Nematocarcinus lanceopes
In total, 43 polymorphic sites were detected: four (9.3%) were first positions, one (2.3%) second position, and 38 (88.4%) third positions, defining 54 distinct haplotypes with no significant differences in base composition (Table 1). Five replacement substitutions (first position: 2, second position: 1, and third position: 2) were detected; all are located at the tips of the network. Pairwise nucleotide differences between sequences ranged from 0 to 0.0152. Three dominating haplotypes, haplotype 1 (19.8% of the entire data set), haplotype 2 (14.4%), and haplotype 3 (18.2%), were detected, with haplotype 1 representing the ancestral haplotype (Fig. 2). Of the remaining 51 haplotypes, 11 were shared among different populations (Table 3). The majority of haplotypes (36 = 66.7%) were singletons (Table 3). Haplotypic diversity h in total was high (0.9025 ± 0.0119), while nucleotide diversity π values were low (0.0057 ± 0.0032; Table 1). Exact test of differentiation among populations revealed significant differences among populations from Balleny Islands and Spiess Seamount for F-statistics (P = 0.0256). Since only two specimens from the Balleny Islands and only five shrimps from Spiess Seamount were sampled, this result can be neglected. For AMOVA, populations from the three major geographic regions were pooled (Fig. 1, Table 2); populations from the South Sandwich Islands and from Spiess Seamount were not pooled but kept as a separate region due to their distinct geographic.
The analysis of molecular variance revealed no significant partitioning of genetic variation within Nematocarcinus lanceopes when stations were grouped by geographic region (F CT = 0.0036, F SC = 0.008, F ST = 0.011; Table 2), giving evidence of a homogeneous population. Furthermore, our data revealed no differences within all analysed mitochondrial 16S rDNA and nuclear 28S rDNA: D2 sequences (P-distances = 0).
Population history of Nematocarcinus lanceopes based on COI
Sequences of all specimens were pooled as a single group. Estimates for the growth rate g were high and positive, with the most likely value for g = 3.783 (SD = 134.1) and θ = 0.2386 (SD = 0.0215), indicating strong and significant population growth. The estimate of the growth rate g was lower and θ higher in Nematocarcinus lanceopes compared to Chorismus antarcticus. As for the Chorismus antarcticus data set, replicate runs with alternate random seeds and different chain settings produced comparable results, ensuring convergence on the correct parameter estimates. Values for g varied in a range of about 1.500–4.000 depending on the number of chains used. Values of both Tajima’s D and Fu’s F’s were negative and significant; only F’s was highly significant (Table 4). Distributions of pairwise differences between sequence pairs showed a unimodal distribution that shifted further to the right when compared to Chorismus antarcticus, hinting at a population expansion that may date back even further in the past. The goodness-of-fit tests cannot reject the model of rapid population expansion (Fig. 3, Table 4). Molecular clock estimates dated the onset of the expansion to have started ~185 KY BP (CI: 20–540 KY). Similar to Chorismus antarcticus, mitochondrial and nuclear patterns strongly deviate from what would be expected from populations at mutation-drift equilibrium.
Discussion
All analysed data sets showed no evidence for cryptic species complexes within Chorismus antarcticus and Nematocarcinus lanceopes. The uncorrected pairwise nucleotide distances were low for both species (Chorismus antarcticus: 0–0.0106; Nematocarcinus lanceopes: 0–0.0152), representing typical intraspecific diversity values of crustaceans (e.g. Lefébure et al. 2006; Bucklin et al. 2007; Costa et al. 2007; Radulovici et al. 2009). Furthermore, there was no evidence for a geographical substructure of genetic variability or any considerable differentiation among any population pair. Although we cannot present an all-encompassing sampling scheme due to limited availability of Antarctic marine specimens, our molecular data provide convincing evidence for a homogeneous genetic structure of the analysed populations, i.e. the absence of significant geographical partitioning of haplotype frequencies. Hence, our results strongly support a circum-Antarctic distribution of Chorismus antarcticus and Nematocarcinus lanceopes.
In most cases, the assumption of circum-Antarctic species distribution is primarily based on morphological evidence only. However, restricted gene flow or even cryptic speciation within species with pelagic distribution stages has been successfully demonstrated for a variety of taxa, e.g. Antarctic Cephalopoda (Brierley et al. 1993; Allcock et al. 1997; Sands et al. 2003), Teleostei (Patarnello et al. 2003; Shaw et al. 2004; Kuhn and Gaffney 2006; 2008), and Euphausiacea (Patarnello et al. 1996; Zane et al. 1998; Jarman and Nicol 2002; Jarman et al. 2002). On the other hand, some molecular studies showed a lack of genetic differentiation within broadly distributed species with pelagic larval stages that inhabit the Southern Ocean. For example, a study analysing mitochondrial and intron DNA markers of the Antarctic toothfish Dissostichus mawsoni revealed low genetic variation (Smith and Gaffney 2005). Two fragments of mitochondrial DNA and additional microsatellites were used to investigate the stock structure of the Patagonian toothfish Dissostichus eleginoides, providing little evidence for population sub-structuring (Appleyard et al. 2002; 2004). Allozymes provided evidence for negligible polymorphisms in the icefish Champsocephalus gunnari (Duhamel et al. 1995), while RFLP analysis of whole mtDNA molecules showed moderate nucleotide diversity but little evidence of population subdivision (Williams et al. 1994). Another study using the partial mitochondrial control region (D-loop) revealed a weak population structure for the abundant Antarctic silverfish Pleuragramma antarcticum (Zane et al. 2006). For invertebrates, evidence for a homogeneous genetic structure of populations sampled south of the Antarctic Polar Front was also shown for the abundant nemertean ribbon worm Parborlasia corrugatus using two mitochondrial gene fragments (Thornhill et al. 2008) and the pycnogonid Colossendeis megalonyx from the Atlantic sector of the Southern Ocean (Krabbe et al. 2010). Finally, a remarkable proof of genetic homogeneity and strong evidence for recent post-glacial recolonization of Southern Ocean habitats has been demonstrated for the widespread Southern Bull Kelp Durvillaea antarctica (Fraser et al. 2009). In summary, these examples across a broad variety of different taxa show that no universal or predictable trend can be seen easily and empirical assessment of the genetic structure is crucial.
Within our study on the genetic structure of two abundant decapod shrimps using mitochondrial and nuclear DNA sequence data, we show that species with low genetic variation (for Nematocarcinus lanceopes only for 16S and 28S rDNA) and pelagic larvae or drifting distribution stages occur in Antarctic waters (Thatje et al. 2005b). Patterns of genetic diversity of both species strongly deviate from the expectations of large and stable populations and show signatures that may result from recent spatial and/or demographic expansion. Haplotype diversity h and nucleotide diversity π was higher for the analysed Nematocarcinus lanceopes specimens (n = 54, h = 0.9025 ± 0.0119, π = 0.0057 ± 0.0032) than for Chorismus antarcticus (n = 24, h = 0.6394 ± 0.0247, π = 0.0021 ± 0.0014; Table 1). In general, genetic variation within a species should be positively correlated with the effective population size. Thus, the observed low mtDNA variation within Chorismus antarcticus and the star-like haplotype network give evidence for a smaller, non-equilibrium population when compared to Nematocarcinus lanceopes and in agreement with a population under expansion following a past bottleneck that eliminated polymorphisms (Nei et al. 1975). This is supported by results of Tajima’s D and the coalescent modelling of population growth rates using Fluctuate, which provide lower estimates for Theta but higher values for the growth rates in Chorismus antarcticus when compared to Nematocarcinus lanceopes. However, absolute values of the growth rates cannot be compared as the algorithm is slightly biased upwards (Kuhner et al. 1998). The results of the mismatch distribution statistics provide somewhat stronger evidence for a spatial expansion than for a pure demographic expansion according to our analyses (not shown). Nevertheless, this should be regarded as first evidence only as a solid statistical framework for these tests is still not available (see Arlequin manual).
For Chorismus antarcticus, the time since population expansion was estimated to be ~85 KY BP (CI: 0–270 KY BP), for Nematocarcinus lanceopes 185 KY BP (CI: 20–540 KY BP). Although molecular clock estimates should be regarded with caution when no strong calibration points exist, our data provide evidence that Chorismus antarcticus may have been experienced a more recent expansion compared to Nematocarcinus lanceopes. The averaged onset estimates calculated with a divergence rate of 2% nucleotide divergence for both species predate the onset of the last deglaciation of the Antarctic shelf. The lower confidence intervals, however, fall within this range. Evidence that values in the lower CI range, i.e. more recent estimates, may reflect better the true demographic scenario, stems from the lack of any variability at the 16S and 28S rRNA genes. Held (2001) demonstrated that mutation rates in the Antarctic could in principle be of similar value compared to non-Antarctic crustaceans. Therefore, the strong deviations of mutation-drift equilibrium conditions in our data support late Pleistocene population expansions following a bottleneck or a founding event.
For Chorismus antarcticus, the more recent estimates for the expansion event suggest a post-glacial expansion after the Last Glacial Maximum of survivors of these species around Antarctica. From the molecular clock data available, it cannot be excluded that the pattern of population expansion predates the Last Glacial Maximum (CI: 0–270 KY BP). The impact of this most recent large-scale glaciation makes this event a good explanation for the data observed. Modelling data suggest that grounded ice masses reached to the continental shelf edge around Antarctica and left no permanent glacial refuges at the Last Glacial Maximum (Huybrechts 2002). On the other hand, both glacial extent and retreat across the continental shelf may have been time transgressive leaving temporary shelters at different locations for benthic organisms (Thatje et al. 2005a). Also, recent geophysical surveys add support to the general small-scale availability of few small-scale shelf refugia (Anderson et al. 2002). Recent molecular genetic data for few benthic, direct-developing invertebrates that lack dispersive distribution stages support the concept of survival on the continental shelf during glacial maxima (Leese 2008; Wilson et al. 2009). Also, due to time transgressive ice extent and retreat patterns in glacial periods, temporarily geographical isolated refugia for benthic organisms may have existed on the continental shelf, allowing species to migrate from one shelter to another (see Thatje et al. 2005b; 2008). However, for shallow-water organisms with planktotrophic larval stages, such as Chorismus antarcticus, it may seem more plausible that they have survived the last ice age in shallow waters of sub-Antarctic islands or Patagonia (but see Thatje et al. 2008) and re-colonized the Antarctic continental shelf during interglacial periods. The strongly reduced or even absent molecular diversity and the much higher population growth rates inferred using the coalescent modelling approach of Fluctuate agree with such a re-colonization and spatial expansion scenario of the Antarctic shelf, possibly from one or very few refugia (Thatje et al. 2008).
In the case of the deep-sea shrimp Nematocarcinus lanceopes it is more likely that earlier events influenced the demography of these species, obviously independent from glacial periods (see Thatje et al. 2008). Nematocarcinus lanceopes basically shows a deep-sea distribution in the Southern Ocean down to about 4,000 m water depth (Thatje et al. 2005a), and only emerges on the Antarctic Continental Slope to about 550 m water depth (Gutt et al. 1991). Based on this distribution pattern, populations of this deep-sea shrimp should have been less affected by the advance of grounded ice sheets across the Antarctic Continental Shelf during glacial periods due to their potential to evade into the deep sea. Hence, they can have maintained a greater diversity when compared to shallow-water species, which is consistent with the findings presented here. These results coincide with other examples of moderate to high genetic variability, which have been observed within various other deep-sea decapod crustaceans (e.g. Shank et al. 1999; Weinberg et al. 2003) or fish (Hoarau and Borsa 2000; Kojima et al. 2001; Danielsdottir et al. 2008). However, as in Chorismus antarcticus, there is also evidence for non-equilibrium as indicated from the neutrality test, the mismatch distribution and the obvious lack of differentiation in the mitochondrial 16S and nuclear 28S rDNA marker. Although in comparison with Chorismus antarcticus, the unimodal distribution of pairwise mismatches in Nematocarcinus lanceopes is shifted to the right being indicative for a bottleneck that predates the Last Glacial Maximum. Thus, our results can provide evidence that in recent history, populations of Nematocarcinus lanceopes were less affected in their genetic diversity, which is consistent with a scenario of recent and recurrent glaciations of the continental shelf that should generally have affected benthic shallow-water shelf species far more than pelagic species (Janko et al. 2007) or primarily deep-sea distributed species. Nevertheless, the basic patterns of mutation-drift disequilibrium are prevailing and obvious. Although we cannot completely rule out the possibility of selective sweeps being responsible for the reduced 16S and 28S rDNA variability, the consistency of both markers and the COI data can explain recent colonization events and expansions of both species around Antarctica quite well.
Additional markers should be tested to understand historical processes and microevolutionary forces leading to the population structure of both analysed species in more detail. Molecular markers with a greater resolution on population level in particular independent SNP sets or microsatellites rather than single locus coding genes alone can reveal more insights into genetic variability and gene flow through migration and dispersal (Ballard and Whitlock 2004; Ballard and Rand 2005) and should be employed in particular for studying population genetics in an Antarctic context (Held and Leese 2007). Additional specimens from other regions of the Southern Ocean (e.g. Davis Sea or Amundsen Sea) are also needed to further verify a circum-Antarctic distribution.
In conclusion, our data provide important first insights into the population structure of two highly abundant species of Antarctic decapod shrimps that constitute significant faunal elements of the Antarctic benthos. The analysed mitochondrial and nuclear data sets reveal genetic homogeneity among populations with the dominance of a few haplotypes and a possible circum-Antarctic distribution of both analysed decapod shrimp species. We interpret the signatures of a population bottleneck dated to the late Pleistocene that in particular for shallow-water species such as Chorismus antarcticus the Last Glacial period with large-scale glaciations of shelf habitats may have had a severe impact and possibly have expelled populations temporarily from the High Antarctic Continental shelf. This effect is less prominent for the deep-sea species Nematocarcinus lanceopes, although the non-equilibrium in the genetic data is indicative also for an impact that may, however, predate the Last Glacial Maximum.
References
Allcock AL, Brierley AS, Thorpe JP, Rodhouse PG (1997) Restricted gene flow and evolutionary divergence between geographically separated populations of the Antarctic octopus Pareledone turqueti. Mar Biol 129:97–102. doi:10.1007/s002270050150
Altschul SF, Gish W, Myers WMEW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Anderson JB, Shipp SS, Lowe AL, Smith Wellner J, Mosola AB (2002) The Antarctic Ice Sheet during the last glacial maximum and its subsequent retreat history: a review. Quat Sci Rev 21:49–70. doi:10.1016/S0277-3791(01)00083-X
Appleyard SA, Williams R, Ward RD (2002) Population structure of the Patagonian toothfish around Heard, McDonald and Macquarie Island. Antarct Sci 14:364–373
Appleyard SA, Williams R, Ward RD (2004) Population genetic structure of Patagonian toothfish in the West Indian Ocean sector of the Southern Ocean. CCAMLR Sci 11:21–32
Arntz WE, Gorny M (1991) Shrimp (Decapoda, Natantia) occurrence and distribution in the eastern Weddell Sea, Antarctica. Polar Biol 11:169–177. doi:10.1007/BF00240205
Aronson RB, Thatje S, Clarke A, Peck LS, Blake DB, Wilga CD, Seibel BA (2007) Climate change and invasibility of the Antarctic benthos. Annu Rev Ecol Evol Syst 38:129–154. doi:10.1146/annurev.ecolsys.38.091206.095525
Ballard JWO, Rand DM (2005) The population biology of mitochondrial DNA and its phylogenetic implications. Annu Rev Ecol Evol Syst 36:621–642. doi:10.1146/annurev.ecolsys.36.091704.175513
Ballard JWO, Whitlock MC (2004) The incomplete natural history of mitochondria. Mol Ecol 13:729–744. doi:10.1046/j.1365-294X.2003.02063.x
Barnard KH (1950) Descriptive catalogue of South African decapod Crustacea. Ann S Afr Mus 38:1–837
Bate CC (1888) Report on the Crustacea Macura collected by HMS Challenger during the years 1873–76. Part 1. Rep Sci Res Voy HMS Challenger 24:1–929
Brandt A (2000) Hypotheses on the Southern Ocean peracarid evolution and radiation (Crustacea, Malacostraca). Ant Sci 12:269–275. doi:10.1017/S095410200000033X
Brierley AS, Rodhouse PG, Thorpe JP, Clarke MR (1993) Genetic evidence of population heterogeneity and cryptic speciation in the ommastrephid squid Martialia hyadesi from the Patagonian Shelf and Antarctic Polar Frontal Zone. Mar Biol 116:593–602. doi:10.1007/BF00355478
Bucklin A, Wiebe PH, Smolenack SB, Copley NJ, Beaudet JG, Bonner KG, Färber-Lorda J, Pierson JJ (2007) DNA barcodes for species identification of euphausiids (Euphausiacea, Crustacea). J Plankton Res 29:483–493. doi:10.1093/plankt/fbm031
Clarke A, Johnston NM (2003) Antarctic marine benthic diversity. Oceanogr Mar Biol Annu Rev 29:47–114
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1660. doi:10.1046/j.1365-294x.2000.01020
Costa F, deWaard JR, Boutillier J, Ratnasingham S, Dooh RT, Hajibabaei M, Hebert PDN (2007) Biological identifications through DNA barcodes: the case of the Crustacea. Can J Fish Aquat Sci 64:272–295. doi:10.1139/F07-008
Danielsdottir K, Gislason D, Kristinsson K, Stefansson O, Johansen T, Pampoulie C (2008) Population structure of deep-sea and oceanic phenotypes of deepwater redfish in the Irminger Sea and Icelandic continental slope: are they cryptic species? Trans Am Fish Soc 137:1723–1740. doi:10.1577/T07-240.1
Duhamel G, Ozouf-Costaz C, Cattaneo-Berrebi G, Berrebi P (1995) Interpopulation relationships in two species of Antarctic fish Notothenia rossii and Champsocephalus gunnari from the Kerguelen Islands: an allozyme study. Antarct Sci 7:351–356. doi:10.1017/S0954102095000496
Eastman JT, McCune AR (2000) Fishes on the Antarctic continental shelf: evolution of a marine species flock? J Fish Biol 57:84–102. doi:10.1111/j.1095-8649.2000.tb02246.x
Edgar RC (2004) Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Excoffier L (2004) Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Mol Ecol 13:853–864. doi:10.1046/j.1365-294X.2003.02004.x
Excoffier L, Schneider S (1999) Why hunter-gatherer populations do not show sign of Pleistocene demographic expansions. Proc Natl Acad Sci USA 96:10597–10602
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome C oxidase subunit I from metazoan invertebrates. Mol Marine Biol Biotechnol 3:294–299
Fraser CI, Nikula R, Spencer HG, Waters JM (2009) Kelp genes reveal effects of subantarctic sea ice during the Last Glacial Maximum. P Natl Acad Sci USA 106:3249–3253. doi:10.1073/pnas.0810635106
Fu Y-X (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–923
Gorny M (1999) On the biogeography and ecology of the Southern Ocean decapod fauna. Sci Mar 63:367–382
Gorny M, Arntz WE, Clarke A, Gore DJ (1992) Reproductive biology of caridean decapods from the Weddell Sea. Polar Biol 12:111–120. doi:10.1007/BF00239971
Gutt J, Gorny M, Arntz WE (1991) Spatial distribution of Antarctic shrimps (Crustacea: Decapoda) by underwater photography. Antarct Sci 3:363–369. doi:10.1017/S0954102091000469
Guzmán G, Quiroga E (2005) New records of shrimps (Decapoda: Caridea and Dendrobranchiata) in deep waters off Chile. Gayana 69:285–290. doi:10.4067/S0717-65382005000200009
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Harpending HC (1994) Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol 66:591–600
Held C (2000) Phylogeny and biogeography of serolid isopods (Crustacea, Isopoda, Serolidae) and the use of ribosomal expansion segments in molecular systematics. Mol Phy Evol 15:165–178. doi:10.1006/mpev.1999.0739
Held C (2001) No evidence for slow-down of molecular substitution rates at subzero temperatures in Antarctic serolid isopods (Crustacea, Isopoda, Serolidae). Polar Biol 24:497–501. doi:10.1007/s003000100245
Held C, Leese F (2007) The utility of fast evolving molecular markers for studying speciation in the Antarctic benthos. Polar Biol 30:513–521. doi:10.1007/s00300-006-0210-x
Hoarau G, Borsa P (2000) Extensive gene flow within sibling species in the deep-sea fish Beryx splendens. Comp Rend Acad Sci 323:315–325. doi:10.1016/S0764-4469(00)901214-0
Huybrechts P (2002) Sea-level changes at the LGM from ice-dynamic reconstructions of the Greenland and Antarctic ice sheets during the glacial cycles. Quat Sci Rev 22:203–231. doi:10.1016/S0277-3791(01)00082-8
Janko K, Lecointre G, DeVries AL, Couloux A, Cruaud C, Marshall C (2007) Did glacial advances during the Pleistocene influence differently the demographic histories of benthic and pelagic Antarctic shelf fishes?—Inferences from intraspecific mitochondrial nuclear DNA sequence diversity. BMC Evol Biol 7:220. doi:10.1186/1471-2148-7-220
Jarman SN, Nicol S (2002) Sources of variance in studies of Krill population genetics. CCAMLR Sci 9:107–116
Jarman SN, Elliott NG, Nicol S, McMinn A (2002) Genetic differentiation in the Antarctic coastal krill Euphausia crystallorophias. Heredity 88:280–287. doi:10.1038/sj.hdy.6800041
Kirkwood JM (1984) A guide to the Decapoda of the Southern Ocean. ANARE Res Notes 11:1–47
Klages M, Gutt J, Starmans A, Bruns T (1995) Stone crabs close to the Antarctic continent: Lithodes murrayi Henderson, 1888 (Crustacea; Decapoda; Anomura) off Peter I Island (68°51’S, 90°51’W). Polar Biol 15:73–75. doi:10.1007/BF00236128
Knowlton N, Weigt LA (1998) New dates and new rates for divergence across the Isthmus of Panama. Proc R Soc Lond B 265:2257–2263. doi:10.1098/rspb.1998.0568
Knowlton N, Weight LA, Solórzano LA, Mills DE, Bermingham E (1993) Divergence in proteins, mitochondrial DNA, and reproductive compatibility across the Isthmus of Panama. Science 260:1629–1632. doi:10.1126/science.8503007
Knox GA (1994) The biology of the Southern Ocean. Cambridge University Press, Cambridge
Kojima S, Segawa R, Hayashi I, Okiyama M (2001) Phylogeography of a deep-sea demersal fish, Bothrocara hollandi, in the Japan Sea. Mar Ecol Prog Ser 217:135–143. doi:10.3354/meps217135
Krabbe K, Leese F, Mayer C, Tollrian R, Held C (2010) Cryptic mitochondrial lineages in the widespread pycnogonid Colossendeis megalonyx Hoek, 1881 from Antarctic and Subantarctic waters. Polar Biol 33:281–292. doi:10.1007/s00300-009-0703-5
Kuhn KL, Gaffney PM (2006) Preliminary assessment of population structure in the mackerel icefish (Champsocephalus gunnari). Polar Biol 29:927–935. doi:10.1007/s00300-006-0134-5
Kuhn KL, Gaffney PM (2008) Population subdivision in the Antarctic toothfish (Dissostichus mawsoni) revealed by mitochondrial and nuclear single nucleotide polymorphisms (SNPs). Antarct Sci 20:327–338. doi:10.1017/S095410200800965
Kuhner MK, Yamato J, Felsenstein J (1998) Maximum likelihood estimation of population growth rates based on the coalescent. Genetics 149:429–434
Ledoyer M (1989) Mysidacea et Caridea (Crustacea) de la campagne MD 42-SIBEX du Marion-Dufresne entre les Iles Kerguelen et la Baie Prydz (Antarctique). Bull Mus Hist Nat (Marseilles) 49:67–70 [in French]
Leese F (2008) The recent evolutionary history of Antarctic and Subantarctic benthic isopods: development and analysis of fast evolving molecular markers. Dissertation, Ruhr-University of Bochum
Lefébure T, Douady CJ, Gouy M, Gibert J (2006) Relationship between morphological taxonomy and molecular divergence within Crustacea: proposal of a molecular threshold to help species delimitation. Mol Phyl Evol 40:435–437. doi:10.1016/j.ympev.2006.03.014
Macpherson E (1988) Three new species of Paralomis (Crustacea, Decapoda, Anomura) from the Pacific and Antarctic oceans. Zool Scr 17:69–75. doi:10.1111/j.1463-6409.1988.tb00087.x
Monteiro FA, Wesson DA, Dotson EM, Schofield CJ, Beard CB (2000) Phylogeny and molecular taxonomy of the Rhodniini derived from mitochondrial and nuclear DNA sequences. Am J Trop Med Hyg 62:460–465
Near TJ, Cheng C-HC (2008) Phylogenetics of notothenioid fishes (Teleostei: Acanthomorpha): inferences from mitochondrial and nuclear gene sequences. Mol Phy Evol 47:832–840. doi:10.1016/j.ympev.2007.11.027
Nei M, Miller SC (1990) A simple method for estimating average number of nucleotide substitutions within and between populations from restriction data. Genetics 125:873–879
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10
Ninet B, Jan I, Botems O, Léchenne B, Jousson O, Panizzon R, Lew D, Monod M (2003) Identification of Dermatophyte species by 28S ribosomal DNA sequencing with a commercial kit. J Clin Microbiol 41:826–830. doi:10.1128/JCM.41.2.826-830.2003
Patarnello T, Bargelloni L, Varotto V, Battaglia B (1996) Krill evolution and the Antarctic Ocean currents: evidence of speciation as inferred by molecular data. Mar Biol 126:603–608
Patarnello T, Marcato S, Zane L, Varotto V, Bargelloni L (2003) Phylogeography of the Chionodraco genus (Perciformes, Channichthydae) in the Southern Ocean. Mol Phyl Evol 28:420–429. doi:10.1016/S1055-7903(03)00124-6
Pfeffer G (1887) Die Krebse von Süd-Georgien nach der Ausbeute der deutschen Station 1882–83. 1. Teil. Mitt Naturhist Mus Hamburg 4:42–150
Radulovici AE, Sainte-Marie B, Dufresne F (2009) DNA barcoding of marine crustaceans from the Estuary and Gulf of St Lawrence: a regional-scale approach. Mol Ecol Res 9:181–187. doi:10.1111/j.1755-0998.2009.02643.x
Raupach MJ, Wägele JW (2006) Distinguishing cryptic species in Antarctic Asellota (Crustacea, Isopoda)—a preliminary study of mitochondrial DNA in Acanthaspidia drygalskii Vanhöffen, 1914. Antarct Sci 18:191–198. doi:10.1017/S0954102006000228
Ray N, Currat M, Excoffier L (2003) Intra-deme molecular diversity in spatially expanding populations. Mol Biol Evol 20:76–86. doi:10.1093/molbev/msg009
Rogers AR (1995) Genetic evidence for a Pleistocene population explosion. Evolution 49:608–615
Rogers AR, Harpending HC (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Sands CJ, Jarman SN, Jackson GD (2003) Genetic differentiation in the squid Moroteuthis ingens inferred from RAPD analysis. Polar Biol 26:166–170. doi:10.1007/s00300-002-0467-7
Schmidt S, Driver F, De Barro P (2006) The phylogenetic characteristics of three different 28S rRNA gene regions in Encarsia (Insecta, Hymenoptera, Aphelinidae). Org Div Evol 6:127–139. doi:10.1016/j.ode.2005.07.002
Schneider S, Roessli D, Excoffier L (2000) Arlequin: a software for population genetics data analysis. vers. 2.000. Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva, Geneva
Schubart CD, Diesel R, Hedges SB (1998) Rapid evolution to terrestrial life in Jamaican crabs. Nature 393:363–365. doi:10.1038/30724
Shank TM, Black MB, Halanych KM, Lutz RA, Vrijenhoek RC (1999) Miocene radiation of deep-sea hydrothermal vent shrimp (Caridea: Bresiliidae): evidence from the mitochondrial cytochrome oxidase subunit I. Mol Phyl Evol 13:244–254. doi:10.1006/mpev.1999.0642
Shaw PW, Arkhipkin AI, Al-Khairulla H (2004) Genetic structuring of Patagonian toothfish populations in the Southwest Atlantic Ocean: the effect of the Antarctic Polar Front and deep-water troughs as barrier to genetic exchange. Mol Ecol 13:3293–3303. doi:10.1111/j.1365-294X.2004.02327.x
Smith PJ, Gaffney PM (2005) Low genetic diversity in the Antarctic toothfish (Dissostichus mawsoni) observed with mitochondrial and intron DNA markers. CCAMLR Sci 12:43–51
Sonnenberg R, Nolte AW, Tautz D (2007) An evaluation of LSU rDNA D1–D2 sequences for their use in species identification. Front Zool 4:6. doi:10.1186/1742-9994-4-6
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sinauer Associates, Sunderland
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Thatje S, Arntz WE (2004) Antarctic reptant decapods: more than a myth? Polar Biol 27:195–201. doi:10.1007/s00300-003-0583-z
Thatje S, Hillenbrand CD, Larter R (2005a) On the origin of Antarctic marine benthic community structure. Trends Ecol Evol 20:534–540. doi:10.106/j.tree.2005.07.010
Thatje S, Bacardit R, Arntz WE (2005b) Larvae of the deep-sea Nematocarcinidae (Crustacea: Decapoda: Caridea) from the Southern Ocean. Polar Biol 28:290–302. doi:10.1007/s00300-004-0687-0
Thatje S, Hillenbrand CD, Mackensen A, Larter R (2008) Life hung by a thread: endurance of Antarctic fauna in glacial periods. Ecology 89:682–692. doi:10.1890/07-0498.1
Thornhill DJ, Mahon AR, Norenburg JL, Halanych KM (2008) Open-ocean barriers to dispersal: a test case with the Antarctic Polar Front and the ribbon worm Parborlasia corrugatus (Nemertea: Lineidae). Mol Ecol 17:5104–5117. doi:10.1111/j.1365-294X.2008.03970.x
Vermeeren H, Vanreusel A, Vanhove S (2004) Species distribution within the free-living marine nematode genus Dichromadora in the Weddell Sea and adjacent areas. Deep Sea Res Part II Top Stud Oceanogr 51:1643–1664. doi:10.1016/j.dsr2.2004.06.028
Watterson GA (1975) On the number of segregating sites in genetical models without m, 6recombination. Theor Popul Biol 7:256–276. doi:10.1016/0040-5809(75)90020-9
Weinberg JR, Dahlgren TG, Trowbridge N, Halanych KM (2003) Genetic differences within and between species of deep-sea crabs (Chaceon) from the North Atlantic Ocean. Biol Bull 204:318–326
Williams R, Smolenski AJ, White RWG (1994) Mitochondrial DNA variation of Champsocephalus gunnari Lönnberg (Pisces: Channichthyidae) stocks on the Kerguelen Plateau, southern Indian Ocean. Antarct Sci 6:347–352
Wilson NG, Hunter RL, Lockhart SJ, Halanych KM (2007) Multiple lineages and absence of panmixia in the “circumpolar” crinoid Promachocrinus kerguelensis from the Atlantic sector of Antarctica. Mar Biol 152:895–904. doi:10.1007/s00227-007-0742-9
Wilson NG, Schrödl M, Halanych KM (2009) Ocean barriers and glaciation: evidence for explosive radiation of mitochondrial lineages in the Antarctic sea slug Doris kerguelenesis (Mollusca, Nudibranchia). Mol Ecol 18:965–984. doi:10.1111/j.1365-294X.2008.04071.x
Zane L, Patarnello T (2000) Krill as a possible model for investigating the effects of ocean currents on the genetic structure of a pelagic invertebrate. Can J Fish Aquat Sci 57:16–23
Zane L, Ostellari L, Maccatrozzo L, Bargelloni L, Battaglia B, Patarnello T (1998) Molecular evidence for genetic subdivision of Antarctic krill (Euphausia superba Dana) populations. Proc R Soc Lond B Biol Sci 265:2387–2391. doi:10.1098/rspb.1998.0588
Zane L, Marcato S, Bargelloni L, Bortolotto E, Papetti C, Simonato M, Varotto V, Patarnello T (2006) Demographic history and population structure of the Antarctic silverfish Pleuragramma antarcticum. Mol Ecol 15:4499–4511. doi:10.1111/j.1365-294X.2006.03105.x
Acknowledgments
We thank D. K. Fütterer for managing the expeditions ANT-XIX/3-4 (ANDEEP I, II), W. E. Arntz for managing ANT XXI/2, E. Fahrbach for managing ANT-XXII/3 (ANDEEP III), and R. Cattaneo-Vietti for managing the 19th expedition of the P.N.R.A. All ANDEEP expeditions were organized by A. Brandt and B. Ebbe. Additional specimens were donated from A. -N. Lörz, C. Held and C. Schubart. We thank K. Linse for providing the map, C. Greve and N. Pernat for laboratory assistance, and two anonymous reviewers for helpful comments. This work is a contribution to MarBEF (GOCE-CT-2003-505446) and is ANDEEP publication # 94. Our study was supported by the Germany Science Foundation (WA 530/28-2 to M.J.R. and RA 1688/2-1 to J.D.).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Riginos.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Raupach, M.J., Thatje, S., Dambach, J. et al. Genetic homogeneity and circum-Antarctic distribution of two benthic shrimp species of the Southern Ocean, Chorismus antarcticus and Nematocarcinus lanceopes . Mar Biol 157, 1783–1797 (2010). https://doi.org/10.1007/s00227-010-1451-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-010-1451-3