Abstract
Phylogenetic, phylogeographic, population genetic and coalescence analyses were combined to examine the recent evolutionary history of the widespread Indo-Pacific parrotfish, Scarus psittacus, over a geographic range spanning three marine biogeographic realms. We sequenced 164 individuals from 12 locations spanning 17,000 km, from 55ºE to 143ºW, using 322 base pairs of mitochondrial control region (D-loop). S. psittacus displayed high haplotype (h = 0.83–0.98), but low nucleotide (<1%) diversity. Most (>83%) genetic variation was within populations. AMOVA revealed significant partitioning and identified five geographic groups. These included one central population and four populations peripheral to the centre. The central population occupied reefs from Western Australia to Tahiti and represented the central Indo-Pacific biogeographic realm. Cocos Keeling was distinct from central and western Indo-Pacific biogeographic realms occupying a position intermediate to these. Peripheral populations (Hawaii, Marquesas) represented the eastern Indo-Pacific biogeographic realm, while Seychelles represented the western Indo-Pacific biogeographic realm. All but the central population expanded (<163 kya). Whilst all populations experienced major sea level and SST changes associated with Pleistocene glaciation cycles, the genetic structure of the central population was relatively homogenous unlike the remaining genetically distinctive populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Indo-Pacific reef fishes have broad longitudinal distributions associated with coral reef systems of the Pacific and Indian Oceans. Their habitat has been visualized as a series of biogeographic realms; very large regions of coastal, benthic, or pelagic ocean with biotas that are coherent at higher taxonomic levels and share evolutionary histories (Spalding et al. 2007). This hierarchical and evolutionary definition of Indo-Pacific biogeographic realms provides an appropriate framework for investigating the phylogeography of Indo-Pacific fishes. Populations of species with broad geographical distributions will be exposed to a wide range of environments and most will have been subject to historical influences of climatic fluctuations in the Indo-Pacific, including sea level changes. This combination, added to the complex geographic configuration of Indo-Pacific reef systems, suggests that populations of Indo-Pacific fishes will have high levels of population structure. Indeed genetic partitioning of populations among and within oceans is common, despite examples of genetic homogeneity observed across thousands of kilometers in reef fishes (Rocha and Bowen 2008).
The most pervasive geographical pattern revealed by phylogeographic analysis of Indo-Pacific populations is differentiation between the Indian and Pacific oceans. This genetic signature, generated by the Indo-Pacific Barrier (IPB) has been identified in a majority of widely distributed Indo-Pacific fishes (Gaither et al. 2010). The variety of taxa investigated ranges from small reef-associated fishes like pomacentrids (Lacson and Clark 1995), acanthurids (Planes and Fauvelot 2002) and scarine labrids (Bay et al. 2004), to larger reef fishes like pelagic foraging acanthurids of the genus Naso (Klanten et al. 2007; Horne et al. 2008). For a minority of species there is no evidence of an Indian/Pacific ocean partition, such as trumpetfish (Bowen et al. 2001), a nasiid surgeon fish (Horne et al. 2008) and a lutjanid (Gaither et al. 2010).
Despite common distribution patterns and histories (all share the impact of Pliocene/Pleistocene glacial cycles), these Indo-Pacific species manifest a wide variety of population structures within and between biogeographic realms. Small reef-associated species including labrids, pomacentrids (Drew et al. 2008; Le Ray et al. 2010) and acanthurids (Planes and Fauvelot 2002) show strong population partitioning with evidence of restricted gene flow within the Pacific Ocean. Two of these species Acanthurus triostegus (Planes and Fauvelot 2002) and Dascyllus trimaculatus (Le Ray et al. 2010) also show partitioning between the Indian and Pacific Oceans. In contrast the holocentrid Myripritis berndti (Craig et al. 2007) and the nasiid acanthurids, Naso vlamingii, N. unicornis (Horne et al. 2008) manifested high levels of gene flow across thousands of kilometers and relatively weak to no population structure within oceans.
A striking example of disparity in population structures of similar species is provided by Gaither et al. (2010), where two co-distributed, phylogenetically related species of the genus Lutjanus, L. kasmira and L. fulvus (Miller and Cribb 2007) revealed contrasting patterns. L. fulvus displayed high levels of population structure at all geographical scales (including the IPB), while L. kasmira lacked genetic structure across the IPB and throughout 12,000 km of its entire central Indo-Pacific range. Clearly the evolutionary histories of Indo-Pacific fishes are complex, since phylogeographic patterns and the underlying processes appear to be highly species-specific. However, given the high diversity of Indo-Pacific fishes, the present picture may simply be the consequence of limited sampling. In order to identify general evolutionary features of Indo-Pacific reef fish, additional analyses of phylogenetically related species with common distributions and habitat associations are required.
The parrotfishes (scarine labrids, sensu Westneat and Alfaro 2005) are suited to such analyses due to broad Indo-Pacific distributions. They are associated with shallow reef environments, although some extend into deeper water and share a common environmental history in which diversification of most species occurred relatively recently in the mid to late Pliocene (Alfaro et al. 2009). They are a diverse group dominated in the Indo-Pacific by two genera, Chlorurus and Scarus, containing 69 of the 90 known species of parrotfish, of which at least 54 have an Indo-West Pacific distribution (Parenti and Randall 2001). They have common developmental traits with small and relatively poorly developed larvae compared to other reef fish larvae (Leis and Carson-Ewart 2000), with an average pelagic duration (PLD) of approximately 28 days (e.g. Chen 2002).
A previous study on a widely distributed parrotfish Chlorurus sordidus (Bay et al. 2004) identified strong genetic partitioning and morphological differentiation between the Indian and Pacific Ocean populations and weaker population structure within the Pacific Ocean. However, the geographic location at which the inter-ocean genetic break occurred was not detected. The focus of the present study is on the parrotfish Scarus psittacus, which shares a number of demographic and ecological characteristics with C. sordidus, including high local densities and rapid population turnover (Choat et al. 1996; Choat and Robertson 2002). Both species also have similar geographic ranges (Parenti and Randall 2001). The research described here is aimed at assessing whether S. psittacus exhibits a similar geographic population structure to C. sordidus, including differentiation between Indian and Pacific Ocean populations.
The sampling and analysis of S. psittacus populations was based on the concept of biogeographic realms (Spalding et al. 2007), including the Eastern, Central and Western Indo-Pacific. This sampling distribution includes areas that are transitional between the Central and Western Indo-Pacific (the location of the Indo-Pacific barrier); the core region of the central Indo-Pacific biogeographic realm, (the geographically complex Coral Triangle, Veron 2000, Allen 2008); and regions that are at the eastern and western range margins of S. psittacus. Other studies (Planes and Fauvelot 2002; Gaither et al. 2010) have identified peripheral regions as important areas of genetic diversity. This study considered four questions: (1) at the broadest spatial scale, is there evidence of S. psittacus population partitions between the three Indo-Pacific biogeographic realms (sensu Spalding et al. 2007)? (2) At the region between the Central and Western realms, what is the pattern and magnitude of population partitions across the IPB? (3) What processes underlie the geographical pattern of population differentiation: is this occurring at the centre and/or at the periphery of the species range? (4) What is the timing of population differentiation: does the structure of S. psittacus populations reflect a relatively ancient pattern driven by pre-Pleistocene events or the impact of relatively recent, Pleistocene events, indicative of a history of recent contractions and expansions?
Methods
Sampling
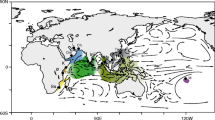
One hundred and sixty-four individuals were collected from 12 localities (Table 1). Geographic coverage was obtained by sampling from the three biogeographic realms in the Indo-Pacific (Table 1; Fig. 1). For three sampling localities Christmas Island, Okinawa and Bali sample sizes were <3. Although these representatives were included in the phylogenetic analyses, the Okinawa and Christmas Island samples were added to the nearby Taiwan and Cocos Keeling populations for the population genetic analysis because they belong to the same ecoregions as those islands (Spalding et al. 2007). The Bali specimens were not included in any of the population genetic analysis, due to the lack of any other representative samples from the centre of the coral triangle in the central Indo-Pacific realm. The Hawaii, Marquesas, Rota, Great Barrier Reef, Western Australia, Cocos Keeling and Seychelles sampling was done by selective spearing between 2001 and 2006. The Tahiti and Taiwan specimens were obtained from markets, as were the specimens from Bali and Okinawa. We are confident that market fish were caught locally at these sites and none were transported from outside the region. Scarids are not transported live and neither do they travel well, as they go off quickly if uncleaned, and the low market value of Scarids means that they are not worth the expense of live shipment. All Taiwan samples were fished in Peng-Hu. Some Tahiti market fish come from the Tuamotos (French Polynesia) but not from beyond. Tissue samples (pectoral fin segments) were immediately placed into 80% ethanol or salt-saturated 20% DMSO.
Scarus psittacus, 164 individuals collected from 12 sites indicated by black dot: Seychelles (SY), Cocos Keeling (CK), Christmas Island (XI), Western Australia (WA), Bali (BL), Taiwan (TW), Okinawa (OK), Rota (RT), Great Barrier Reef (GBR), Hawaii (HI), Tahiti (TH), Marquesas (MQ). Sample sizes are listed in Table 1
Laboratory procedures
Total DNA was extracted from fin tissues of all samples by proteinase K digestion and either standard salt–chloroform extraction procedures (Sambrook et al. 1989) or a 5% Chelex extraction method (Nishiguchi et al. 2002). The D-loop control region of the mitochondrial genome was amplified for all specimens, commonly used for reef fish studies, using Scarus-specific primers Sgh-F (5′ TTA TCC CTG ATC ATC AAG GAA T) and Sghru-R (5′ TAG CTC CCA AAG CTA GAA TC) (Beck et al., unpublished). Polymerase chain reaction (PCR) was performed in a 20-µl reaction volume containing 2.5 mM Tris–Cl pH 8.7, 5 mM KCl, 5 mM (NH4)2SO4, 100 M each dNTP, 2.8 mM MgCl2, 10 M each primer, 2.7 U of Taq Polymerase (Qiagen) and 10 ng template DNA. Temperature profiles for the PCR amplification were set at an initial denaturation at 94°C for 2 min, followed by 5 cycles of denaturation at 94°C for 30 s, annealing at 53°C for 30 s and an extension at 72°C for 1 min 30 s. The remaining 25 cycles were performed as described, but at an annealing temperature of 51°C.
All PCR products were evaluated on a 1.5% agarose gel and purified by standard isopropanol precipitation. The PCR product was sent to Macrogen Inc. (Korea) for sequencing on a 96-capillary ABI 3730XL DNA analyser, in the forward direction only, using the Sgh-F primer. Nucleotide sequences were initially aligned using programs Sequencher v3.12 (Anon 2000) and BioEdit (Hall 1999), while final alignments were checked manually. Sequences were submitted to GenBank, Accession Numbers EU926978 through EU927144 and FJ161085 through FJ161087 (Table 1). The DNA of all specimens was used for phylogenetic, population genetic and coalescent analyses, and was tested for genetic diversity, isolation-by-distance and migration.
Phylogenetic analyses
Phylogenetic analyses were performed using an alignment of all sequences obtained to determine if genetically distinct lineages are identified, which would inform the population genetic analyses. The best substitution model (GTR + I + G) for ML analyses was found using a likelihood approach in Modeltest v3.06 (Posada and Crandall 1998). The Akaike Information Criterion (AIC) identified the most appropriate substitution model for the given data as GTR + I + G with a gamma distribution shape parameter α, of 0.7382, and a proportion of invariable sites, I, of 0.3276. Six substitution types with base frequencies, estimated from the data, were also specified for the sequences. A transition:transversion ratio (T ratio) of 3.4121 was specified. The nucleotide composition was AT-biased (A 28.09%, C 9.68%, G 22.80% and T 39.43%), as is common for fish mitochondrial DNA (Wolstenholme 1992; McMillan and Palumbi 1997). The Shimodaira–Hasegawa test identified six Maximum likelihood (ML) trees that did not differ significantly in their likelihood scores from the best tree obtained (Shimodaira and Hasegawa 2001), the single best ML tree with bootstrap support values is shown in Fig. 2.
The evolutionary relationships between specimens were inferred using distance [neighbour joining (NJ)] and maximum likelihood (ML) methods implemented in PAUP 4.0 (Swofford 2003), with the most appropriate substitution model specified. One thousand bootstrap replicates were performed during the NJ analysis for evaluation of the levels of support for the tree topology. All trees were outgroup rooted using Scarus flavipectoralis and Scarus schlegeli which occur within the same clade as S. psittacus (Kazancioglu et al. 2009). The NJ tree topologies were confirmed by maximum likelihood (ML) and Bayesian analysis, using PAUP 4 (Swofford 2003), GARLI (Zwickl 2006) and MrBayes (Huelsenbeck and Ronquist 2001), respectively. For the Bayesian analysis a Markov Chain Monte Carlo (MCMC) search was run with four chains for one million generations. Trees were sampled every 100 generations and the first 106,000 generations were discarded as “burn-in”. A consensus tree was derived from the best 100 Bayesian trees only and both the likelihood and majority rule values were displayed on the best ML tree from the Bayesian analysis.
Coalescence analyses
A coalescence approach was used to date the most recent expansion of each region in thousands of years before the present. This method assumes a molecular clock and neutrality (Schneider et al. 2000) and estimates ages of coalescence from the terminal nodes of the groups. The pairwise mismatch distribution was plotted for all individuals of S. psittacus to determine if the observed distribution of the average number of pairwise sequence differences corresponded with the expected distribution. The raggedness index, also computed in Arlequin v3.0 (Schneider et al. 2000; Excoffier et al. 2005), uses the method of Harpending (1994) to test if the observed and simulated curves differ significantly or not.
The divergence time (in generations), tau (τ), was estimated using two formulae, τ = 2ut and u = 2μk, where u is the mutation rate for the whole sequence, t is the generation time and k is the length of the sequence (Rogers and Harpending 1992; Harpending 1994). The mutation rate per nucleotide, μ, was estimated as per methodology of Alvarado Bremer et al. (1995), where the parsimony informative sites of the swordfish Xiphias gladius mtDNA control region, gathered from MEGA (Tamura et al. 2007), evolve at a rate of 12.9% substitutions per million year and the rest of the control region, which is conserved, evolves at 1.1% substitutions per million year (Alvarado Bremer et al. 1995; Messmer et al. 2005). Effective population sizes are unknown; therefore mu (μ) is only an estimate. The species-specific mutation rate calculated for S. psittacus D-loop was (12.9 × 0.08) + (1.1 × 0.92) to obtain a rate of 2.04 substitutions per million years, that represents both variable and conserved sites in S. psittacus control region. Finally, the generation time (T) for S. psittacus was calculated as T = (α + ω)/2, where α is the age at first reproduction and ω is the age at last reproduction (Pianka 1978) and assumes that fish are reproductively active throughout this period. Female generation times were estimated at 3.38 years.
Population genetic analyses
Population genetic analyses were performed to measure genetic connectivity within and among Indian and Pacific populations of S. psittacus using Arlequin v3.1 (Schneider et al. 2000; Excoffier et al. 2005). To quantify genetic differentiation between regions and among populations within regions, hierarchical measures of genetic structure were used, as determined by analysis of molecular variance (AMOVA) in Arlequin. This analysis computes the proportion of variation among regions (ϕ ct), the proportion of variation among populations within regions (ϕ sc) and the proportion of variation within populations relative to that overall (ϕ st). In the AMOVA analyses populations were grouped based on the distribution of pairwise F st values. In addition, we grouped populations between oceans to determine if there were clear indications of inter-ocean partitioning.
Diversity was based on genetic distances among haplotypes and frequency differences of haplotypes. Biogeographic regions were partitioned in different ways to determine which accounted for greater genetic variation. AMOVA analyses were run on populations that were grouped based on pairwise F st to assess whether population partitioning could be detected and, if so, which partitioning accounted for the most variation. The three partitioning regimes were as follows: (1) maintaining the Seychelles, Cocos Keeling, Marquesas and Hawaiian populations as four separate groups and the rest of the Western Pacific populations, including Western Australia as a single region as suggested by significant pairwise F st values; (2) grouping populations by geography with Pacific Ocean populations grouped together and populations in the Indian Ocean grouped together; and (3) grouping populations based on known biogeographic breaks with the Pacific populations and the Western Australian population together and the rest of the Indian Ocean populations together.
Population diversity
A minimum spanning tree (MST) was constructed to illustrate spatial and historical patterns of haplotype distributions within and among populations and to explore geographic partitioning of the data using Arlequin (Schneider et al. 2000; Excoffier et al. 2005) as per Rohlf (1973). The unbiased haplotype diversity index (h) was calculated, as per Nei (1987). The nucleotide diversity index (π%) was calculated in Arlequin (Schneider et al. 2000; Excoffier et al. 2005).
Isolation-by-distance and migration
Genetic structure among population pairs (using pairwise F st values) was examined as a function of straight-line geographic distances, as implemented in IBD v1.4 (Bohonak 2002), because geographically separated populations may be partitioned through effects of isolation-by-distance, which produces clinal distributions of traits (cf Slatkin 1993). Migration estimates were calculated in MIGRATE v2.3 (Beerli and Felsenstein 2001) to determine the degree and direction of migration between individual populations. However, these analyses failed to meet assumptions that all populations have the same effective population size or, alternatively, that there is an equal amount of gene flow between each population and are therefore not presented.
Results
There were 322 alignment positions in the mitochondrial data set (including gaps), 23 of which were parsimony informative, for 164 individuals from 12 geographic locations. There were 77 haplotypes in total.
Phylogenetic analyses
The data show that S. psittacus is monophyletic and lacks structure in the deeper nodes, as is evident from the numerous polytomies. There is no clear evidence of a phylogenetic break at the largest geographical scale, between the Indian and Pacific Ocean basins. A total of 13 subclades, of which 7 had more than 50% majority rule support in ML and Bayesian analyses, were evident in the tree structure (Fig. 2). Each of the three of these subclades (4, 6 and 7) was found at only a single peripheral site (Hawaii or the Marquesas), while each of the remaining four (1, 2, 3 and 5) had representatives at 3 or more locations.
Coalescence analyses
The unimodal mismatch distribution suggested recent expansion (Fig. 3) and this was confirmed by non-significant raggedness indices (Table 2) for all but one (central) of the populations. From the coalescence analysis the mean ages of all the populations sampled and the total population ranged from 65 to 115 kya (Table 2). The Cocos Keeling population recorded the greatest mean age (108 kya) with expansion commencing sometime between 31 and 159 kya (Table 2). The Seychelles and Marquesas population expansion time estimates were intermediate based on the mean coalescent estimates, but Marquesas had a much greater possible age range with the largest 95% confidence interval of the expansion parameter, τ. The Hawaiian population expansion was the youngest of the four populations for which coalescent ages could be estimated (Table 2). Because all individual populations have overlapping estimates of expansion time ranges it is not possible to ascertain with confidence the sequence of individual population expansions. What is clear is that the total contemporary S. psittacus population coalescent age estimates are during the last ten percent of the 2 My long Pleistocene, between 94 and 130 kya (mean 115).
Population genetic analyses
Pairwise F st values (Table 3) are significant and substantially higher for the Marquesas population (0.166–0.300) compared to all other population pairwise comparisons (0–0.153). Furthermore, Tahiti shows no significant variation from other West Pacific populations, yet has the second largest pairwise F st (0.292) when paired with the geographically closest neighbour population sampled here, the Marquesas (Table 3). Other populations for which pairwise F st values were significant included Hawaii, Cocos Keeling and the Seychelles. AMOVA analyses (Table 4) showed a significant amount of genetic variation among regions when the Seychelles, Cocos Keeling, Hawaii and Marquesas populations were segregated as individual groups, while the Rota, Tahiti, Taiwan, Great Barrier Reef and Western Australia populations were grouped together in a central region (Table 4A). However, this variation among regions accounted for none of the genetic variation. When populations were grouped by ocean basin there was no significant variation among regions (Table 4B, C), with this grouping accounting for none of the genetic variation. There was a significant amount of genetic variation among populations within regions regardless of the specified a priori grouping pattern (Table 4), although this variation accounted for 14–27% of the total genetic variation. The highest levels of genetic variation detected by AMOVA analyses were for individual populations versus the total population (Table 4), which accounted for 83–90% of the genetic variation. This pattern largely reflects the genetic distinctiveness of insular populations at Hawaii, the Marquesas, the Seychelles and Cocos Keeling (see Table 3).
Population structure and genetic diversity
The minimum spanning tree (Fig. 4) showing the relationships among mtDNA haplotypes within the Pacific and Indian Ocean regions was characterized by a series of star-like topologies with the majority of individuals separated by a single substitution. The haplotypes from both the Indian and Pacific Ocean sites were distributed throughout the tree, in a mixed star topology, based around the most abundant haplotype [32 individuals from all but one (Marquesas) of the nine adequately sampled locations]. A Seychelles grouping (6/20 individuals, within four haplogroups) was separated by 1–3 substitutions from the most abundant haplotype, while a Marquesas grouping (14/20 individuals, within three haplogroups) was separated by 1–2 substitutions. The Cocos Keeling population exhibits the largest number of haplotypes, with only two haplotypes shared and all others represented by a single individual.
Minimum spanning tree of Scarus psittacus, 164 individuals in 12 populations. Circles represent unique haplotypes. Circle sizes are relative to the number of individuals sharing the specific haplotype, also indicated by number of pie pieces. Each haplotype is colour coded, depending on the location from which it was collected, as shown in the key. Lines connecting haplotypes in the minimum spanning tree contain crossbars of varying thickness, depending on the number of substitutions represented, varying from 1 to 10 substitutions per crossbar. The total number of substitutions separating haplotypes is therefore obtained by summing up the inter-connecting cross bars. Key details numbers of individuals sharing haplotypes and number of substitutions for each bar thickness
Haplotype diversities were high in all sampled locations (0.83–0.99), the highest being the Cocos Keeling population (Table 5). Nucleotide diversities were generally low and ranged from 0.52% (Hawaii) to 0.88% (Rota). Values for Marquesas nucleotide and haplotype diversity (0.56% and 0.83) were lower than values for all other populations, except Hawaii’s nucleotide diversity. The central population had low nucleotide and haplotype diversities, despite the collective sampling across a very large area with four times as many individuals as any peripheral population.
Isolation by distance
To test for a correlation between geographic and genetic distances a Mantel test was performed (Mantel 1967) using IBD (Bohonak 2002), which determines the level of correlation between pairwise genetic and geographic distances between all sampled populations of S. psittacus. The greatest geographic distance between populations sampled here was 18,095 km between the Seychelles and Marquesas, and the shortest 1,380 km between Tahiti and the Marquesas. The various positive correlations between these two distance matrices were all non-significant, regardless of whatever transformation of the data were used. Overall, the r 2 values ranged from 0.015, P = 0.89 (for pairwise genetic distances vs. logged geographic distances) to 0.149, P = 0.98 (for logged pairwise genetic distances vs. geographic distances). Further, non-significant results were obtained with the exclusion of either Cocos Keeling (r 2 = 0.016, P = 0.83), which represents a site of overlap between the Pacific and Indian Ocean populations in other taxa (Hobbs et al. 2009); or Marquesas (which was the most distinctive population), r 2 = 0.123, P = 0.94; or both those populations (r 2 = 0.082, P = 0.90). Thus, no evidence for any isolation-by-distance effect in this species was found.
Discussion
An analysis of S. psittacus populations from the full distributional range based on both phylogenetic and phylogeographic methods revealed a complex structure with a number of unexpected features. The first concerned the pattern detected at the broadest spatial scale, that between ocean basins. Based on previous studies of widely distributed Indo-Pacific reef fishes (Bay et al. 2004; Gaither et al. 2010; Planes and Fauvelot 2002), the presence of a well-defined genetic break between the Indian and Pacific Oceans would have been predicted. Indeed the focus of sampling the Western Australia, Cocos Keeling and Seychelles populations (Le Ray et al. 2010) was designed to detect such a break and identify its location. However, the phylogenetic analysis failed to retrieve any coherent pattern of genetic structure at the broadest ocean-level scale. This was re-enforced by the population genetic analyses, which used an AMOVA to specifically investigate partitioning of populations between the Pacific and Indian Ocean systems.
Secondly, genetic analyses revealed distinctive insular population structures at the eastern and western margins of the geographic range of Scarus psittacus, as well as a relatively uniform, non-expanding central region. Pairwise comparisons of F st values showed that this distinctive structure was dominated by the Marquesas population and was also associated with the Hawaii, Seychelles and Cocos Keeling populations. Both Hawaii and the Marquesas represent the eastern margin of the species range and harbour distinctive reef fish faunas with low species diversity and high levels of endemism (Randall 1998). The Seychelles, located within the western Indo-Pacific realm, are within the most western 10% of the entire species range. The Seychelles show reduced species diversity relative to the central Indo-Pacific and are relatively isolated lying 1,800 km to the west of the Maldives in the central Indian Ocean. In contrast, the Cocos Keeling islands in the eastern Indian Ocean lie at the boundary between two biogeographic realms, the Western Indo-Pacific and the Central Indo-Pacific (Spalding et al. 2007; Gaither et al. 2010), and in evolutionary terms represent a junction between Indian and Pacific Ocean reef fish faunas (Hobbs et al. 2009). For the purposes of this analysis the Marquesas, Hawaii and Seychelles are considered peripheral localities as they occur on the outer margins of the species range while Cocos Keeling, following Hobbs et al. (2009), is a transitional locality with respect to Indian and Pacific Ocean populations.
Thirdly, although S. psittacus has an evolutionary history that commenced in the mid-Pliocene (4.5 mya) (Alfaro et al. 2009), the dominant signal of evolutionary change in contemporary populations was confined to the last 163 kya and reflects exclusively Pleistocene events. Coalescence analysis revealed a series of relatively young populations with a signature of recent expansion, with the exception of the central population. The present structure of contemporary populations was defined by bottleneck/colonisation–expansion events less than 0.2 Mya and possibly following the last glacial maximum 18 kya. This signature of recent population expansion is evident from the very few substitutions separating the vast majority of haplotypes, the unimodal mismatch distribution, coalescent analysis and in the starburst pattern of common haplotypes, particularly the most common haplotype that is shared by 20% of the total sample and represents all but one (Marquesas) of nine well-sampled locations.
The analysis of S. psittacus population structure shows that different evolutionary processes are occurring within the complex central region and the relatively isolated and geographically peripheral regions of the species range, Hawaii, Marquesas and the Seychelles. The population structure at these localities indicates the importance of peripheral habitats with respect to the emergence of divergent populations during the Pleistocene. The large area occupied by central populations, is dominated by the most complex arrangement of reef systems found in the world. This area has also been subject to extreme modifications driven by the high amplitude cycles of eustatic sea level changes (the 100 ky world, Sosdian and Rosenthal 2009), which characterized the last 800 ky of the Pleistocene (Sathiamurthy and Voris 2006; Voris 2000). Cyclic changes in sea levels would have resulted in isolated populations throughout the central region during periods of low sea-level stands. During high sea-level stands, previously isolated central-region populations could then have exchanged migrants via the many available stepping-stones in the region, resulting in the apparently stable (non-expanding) population signature, unique to this population.
The absence of a signal of isolation-by-distance, and thus no indication of clinal variation in the distribution of haplotypes, indicates that geography alone does not explain the extent of gene flow between peripheral localities and other areas of the species range. In fact, the strongest partition occurred between the two closest localities sampled, the Marquesas and Tahiti (separated by 1,300 km), which occupy a single biogeographic realm (the eastern Indo-Pacific). Tahiti populations were well connected at evolutionary time scales to those of the central Indo-Pacific biogeographic realm, while the Marquesas population was the most genetically distinct relative to all other populations. The Marquesas population occurs in an area that has depauperate reef fish faunas (Randall 1998), similar to that of the Hawaii population, and is associated with limited coral reef development (Cabioch et al. 2008). The Marquesas is also subject to relatively low water temperatures (Randall 2001) caused by upwelling, which produces episodic phytoplankton blooms (Dickenson 2000; Martinez and Maamaatuaiahutapu 2004), another marked habitat and environmental difference which may contribute to the distinctive genetic structure found with this population. Isolating effects of ocean currents that flow westward, away from rather than towards the Marquesan islands, will also reduce the potential for connectivity between those islands and elsewhere (Gaither et al. 2010; Rocha et al. 2005). In addition, low levels of nucleotide diversity suggest that recent colonisation by founders and population expansions are both important at peripheral locations.
In the Indian Ocean there were a number of distinctive features associated with the Seychelles and Cocos Keeling populations, despite the lack of an explicit genetic break between the Pacific and Indian Oceans. Paired F st comparisons (Table 3) identified distinctive populations at both localities. The Seychelles forms a biological province with the East African coastal system (Spalding et al. 2007) and is at the western periphery of the S. psittacus longitudinal range. The area is isolated from the central and eastern regions of the Indian Ocean and as with most reef localities on the periphery of the Indo-Pacific region it is associated with a regional reduction in the availability of reef habitat (Bellwood and Hughes 2001) and with a depauperate fauna (Mooi and Gill 2001) relative to the central regions of the Indo-Pacific. The Seychelles population differs from those on the eastern periphery of the S. psittacus range in that both the haplotype and nucleotide diversities are higher than those recorded from Hawaii and the Marquesas. Although the Seychelles sample shares the property of a divergent population structure with Hawaii and the Marquesas, the differences in diversity suggest that processes associated with population differentiation differ between the western and eastern margins of the S. psitticus range.
The remaining population identified by the paired comparisons and AMOVA (Tables 3, 4) is Cocos Keeling. This island is geologically recent (Woodroffe et al. 1991), has been subject to major environmental perturbations in recent times (Werner 2008) and is exposed to the westerly flow of the southern equatorial current. It is considered to represent a biogeographic border between the Indian and Pacific Ocean reef fish faunas and is located close to the global centre of coral reef diversity (Indonesia–Philippines) (Hobbs et al. 2009). In geographical terms Cocos Keeling is a transitional locality between two ocean systems and had the highest haplotype diversity of the populations sampled in this study. Horne et al. (2008) suggested that the high genetic diversity found in two acanthurids (Naso spp.) at Cocos Keeling reflected a mixing between Indian and Pacific Ocean derived populations. Unlike the Seychelles, Hawaii and the Marquesas, the fish faunas of Cocos Keeling and Christmas Islands include both eastern and western Indo-Pacific components resulting in a high level of species diversity (Hobbs et al. 2009). The large number of hybridizing species at this locality suggests an evolutionarily dynamic area (Hobbs et al. 2009) and the high haplotype diversity in samples of S. psittacus from Cocos Keeling supports this view. However, the status of S. psittacus populations at this locality requires further investigation, including additional sampling from the adjacent locality of Christmas Island and from the south coasts of Indonesia.
The dominant feature in S. psittacus is the absence of a Pacific/Indian Ocean partition and a recent signature of population expansion. This species has a number of ecological and distributional similarities to the parrotfish Chlorurus sordidus. In both species the Western Australian population did not group with other Indian Ocean populations sampled here (Cocos Keeling, Seychelles). However, S. psittacus lacked any evidence of phylogenetic division between the eastern and western-central Indian Ocean, which was seen in the C. sordidus phylogeography (Bay et al. 2004) and identified as evidence of the action of the Indo-Pacific Barrier (Gaither et al. 2010). The most significant aspect of the comparison between S. psittacus and C. sordidus is that phylogeographic analysis reveals very different recent evolutionary histories, especially with respect to broad-scale patterns of population structure, even though these species have strong similarities in demography, habitat association and distribution. The phylogeographic pattern in S. psittacus is dominated by events that have occurred during the last 5% of its evolutionary age, events that may have obscured an earlier partition between Indian and Pacific Ocean populations as seen in C. sordidus. This mirrors the situation recorded for two Indo-Pacific species of Lutjanus that display divergent patterns of geographic population structure (Gaither et al. 2010). While L. kasmira showed little inter- or intra-oceanic partitioning (only the Marquesas population was genetically distinct), L. fulvus populations in both the Pacific and Indian Oceans showed subdivision at every spatial scale analysed.
The geographic pattern found in S. psittacus suggests a process of peripatric differentiation in which colonising recruits may or may not be experiencing selection when recruiting to isolated and often peripheral locations. This is most relevant to the Marquesas population. Selection may operate if small numbers of founding recruits are exposed to novel biotic and/or abiotic environmental conditions when they invade peripheral habitats. Alternatively, differentiation may result without invoking selection, due to genetic drift in the small founding population. A similar pattern of peripheral differentiation (and genetic homogeneity between Pacific and Indian Ocean populations in some taxa) occurs in Indo-Pacific hermit crabs of the genus Calcinus (Malay and Pauly 2010). A notable feature of their comprehensive analysis of this genus in particular and hermit crabs in general was that broader patterns observed were a composite of different clade-specific patterns implying that multiple processes are involved in generating evolutionary patterns observed in higher taxa. A similar situation may exist in Indo-Pacific reef fishes where additional analyses of phylogenetically related groups of species are a priority.
References
Alfaro ME, Brock CD, Banbury BL, Wainwright PC (2009) Does evolutionary innovation in pharyngeal jaws lead to rapid lineage diversification in labrid fishes? BMC Evol Biol 9:255
Allen GR (2008) Conservation hotspots of biodiversity and endemism for Indo-Pacific coral reef fishes. Aquat. Conserv. Mar. Freshw. Ecosyst. 18:541–556
Alvarado Bremer JR, Baker AJ, Mejuto J (1995) Mitochondrial DNA control region sequences indicate extensive mixing of swordfish (Xiphias gladius) populations in the Atlantic Ocean. Can J Fish Aquat Sci 52:1720–1732
Anon (2000) SEQUENCHER Version 4. 1. 2. Gene Code Corporation, Ann Arbor, MI
Bay LK, Choat JH, van Herwerden L, Robertson DR (2004) High genetic diversities and complex genetic structure in an Indo-Pacific tropical reef fish (Chlorurus sordidus): evidence of an unstable evolutionary past? Mar Biol 144:757–767
Beerli P, Felsenstein J (2001) Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc Natl Acad Sci USA 98:4563–4568
Bellwood DR, Hughes TP (2001) Regional-scale assembly rules and biodiversity of coral reefs. Science 292:1532–1535
Bohonak AJ (2002) IBD (isolation by distance): a program for analyses of isolation by distance. J Hered 93:153–154
Bowen BW, Bass AL, Rocha LA, Grant WS, Robertson DR (2001) Phylogeography of the trumpet fishes (Aulostomus): ring species complex on a global scale. Evolution 55:1029–1039
Cabioch G, Montaggioni L, Frank N, Seard C, Sallé E, Payri C, Pelletier B, Paterne M (2008) Successive reef depositional events along the Marquesas foreslopes (French Polynesia) since 26 ka. Mar Geol 254:18–34
Chen L (2002) Post-settlement diet shift of Chlororus sordidus and Scarus schlegeli (Pisces: Scaridae). Zool. Stud. 41:47–58
Choat JH, Robertson DR (2002) Age-based studies on coral reef fishes. In: Sale PF (ed) Coral reef fishes. Dynamics and diversity in a complex ecosystem. Academic Press, New York
Choat JH, Axe LM, Lou DC (1996) Growth and longevity in fishes of the family Scaridae. Mar Ecol Prog Ser 145:33–41
Craig MT, Eble JA, Bowen BW, Robertson DR (2007) High genetic connectivity across the Indian and Pacific Oceans in the reef fish Myripristis berndti (Holocentridae). Mar Ecol Prog Ser 334:245–254
Dickenson WR (2000) Geomorphology and geodynamics of the Cook-Austral Island-Seamount Chain in the South Pacific Ocean: implications for hotspots and plumes. tectonic Studies of Asia and the Pacific Rim: a tribute to Benjamin M. Page (1911–1997). Geol. Soc Am 3:17–32 International Book Series
Drew J, Allen G, Kaufman L, Barber PH (2008) Endemism and regional colour and genetic differences in five putatively cosmopolitan reef fishes. Conserv Biol 22:965–975
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.1: an integrated software package for population genetics data analysis. Evol Bioinforma Online 1:47–50
Gaither MR, Toonen RJ, Robertson DR, Planes S, Bowen BW (2010) Genetic evaluation of marine biogeographical barriers: perspectives from two widespread Indo-Pacific snappers (Lutjanus kasmira and Lutjanus fulvus). J Biogeogr 37:133–147
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Harpending HC (1994) Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol 66:591–600
Hobbs JP, Frisch AJ, Allen GR, van Herwerden L (2009) Marine hybrid hotspot at Indo-Pacific biogeographic border. Biol. Lett. 23:258–261
Horne JB, van Herwerden L, Choat JH, Robertson DR (2008) High population connectivity across the Indo-Pacific: congruent lack of phylogeographic structure in three reef fish congeners. Mol Phylogenet Evol 49:629–638
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Kazancioglu E, Near TJ, Hanel R, Wainwright PC (2009) Influence of sexual selection and feeding functional morphology on diversification rate of parrotfishes (Scaridae). Proc. R Soc. B 276:3439–3446
Klanten OS, Choat JH, van Herwerden L (2007) Extreme genetic diversity and temporal rather than spatial partitioning in a widely distributed coral reef fish. Mar Biol 150:659–670
Lacson JM, Clark S (1995) Genetic divergence of Maldivian and Micronesian demes of the damselfishes Stegastes nigricans, Chrysiptera biocellata, C. glauca and C. leucopoma (Pomacentridae). Mar Biol 121:585–590
Le Ray M, Beldade R, Holbrook SJ, Schmitt RJ, Planes S (2010) Allopatric divergence and speciation in coral reef fish: the three spot damselfish Dascyllus, Dascyllus trimaculatus, species complex. Evolution. doi:10.1111/j.1558-5646.2009.00917.x
Leis JM, Carson-Ewart BM (2000) The larvae of Indo-Pacific coastal fishes: an identification guide to marine fish larvae. Brill, Leiden
Malay MCD, Pauly G (2010) Peripatric speciation drives diversification and distributional pattern of reef hermit crabs (Decapoda:Diogenidae:Calcinus). Evolution. doi:10.1111/j.1558-5646.2009.00848.x
Mantel N (1967) The detection of disease clustering and generalized regression approach. Cancer Res 27:209–220
Martinez E, Maamaatuaiahutapu K (2004) Island mass effect in the Marquesas Islands: time variation. Geophys Res Lett 31:1–4
McMillan WO, Palumbi SR (1997) Rapid rate of control-region evolution in Pacific Butterfly fishes (Chaetodontidae). J Mol Evol 45:473–484
Messmer V, van Herwerden L, Munday P, Jones GP (2005) Phylogeography of colour polymorphism in the coral reef fish Pseudochromis fuscus, from Papua New Guinea and the Great Barrier Reef. Coral Reefs 24:392–402
Miller TI, Cribb TH (2007) Phylogenetic relationships of some common Indo-Pacific snappers (Perciformes:Lutjanidae) based on mitochondrial DNA sequences, with comments on the taxonomic position of the Caesioninae. Mol Phylogenet Evol 44:450–460
Mooi RD, Gill AC (2001) Historical biogeography of fishes. In: Hart PJB, Reynolds JD (eds) Handbook of fish biology and fisheries. Blackwell Publishing, Oxford, pp 43–68
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nishiguchi MK, Doukakis P, Egan M, Kizirian D, Phillips A, Prendini L, Rosenbaum HC, Torres E, Wyner Y, DeSalle R, Giribet G (2002) Methods and tools in biosciences and medicine: techniques in molecular systematics and evolution DNA isolation procedures. Birkhauser, Basal
Parenti P, Randall JE (2001) An annotated checklist of the species of the labroid fish, families Labridae and Scaridae. Ichthyol. Bull. 68:1–97
Pianka ER (1978) Evolutionary ecology. Harper and Row, New York
Planes S, Fauvelot C (2002) Isolation by distance and vicariance drive genetic structure of a coral reef fish in the Pacific Ocean. Evolution 56:378–399
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Randall JE (1998) Zoogeography of shore fishes of the Indo-Pacific region. Zool. Stud 31:227–268
Randall JE (2001) Four new cardinal fishes (Perciformes:Apogonidae) from the Marquesas Islands. Pac Sci. 55:47–64
Rocha LA, Bowen BW (2008) Speciation in reef fishes. J Fish Biol 72:1101–1121
Rocha LA, Robertson DR, Roman J, Bowen BW (2005) Ecological speciation in tropical reef fishes. Proc. R Soc. Lond B 272:573–579
Rogers AR, Harpending HC (1992) Population-growth makes waves in the distribution of pairwise genetic-differences. Mol Biol Evol 9:552–569
Rohlf FJ (1973) Algorithm 76. Hierarchical clustering using the minimum spanning tree. Comput. J. 16:93–95
Sambrook J, Fritch E, Maniatis T (1989) Molecular cloning, a laboratory manual. Cold Spring Harbour Laboratory Press, Cold Spring Harbour
Sathiamurthy E, Voris HK (2006) Maps of Holocene sea level transgression and submerged lakes on the Sunda Shelf. The Natural History Journal of Chulalongkorn University, Supplement 2, pp 1–44
Schneider S, Roessli D, Excoffier L (2000) Arlequin: a software for population genetics data analysis. Genetics and Biometry Laboratory, Department of Anthropology and Ecology, University of Geneva. 111 p. Available via http://anthro.unige.ch/software/arlequin/software/2.000/manual/Arlequin.pdf
Shimodaira H, Hasegawa M (2001) CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246–1247
Slatkin M (1993) Isolation by distance in equilibrium and non-equilibrium populations. Evolution 471:264–279
Sosdian S, Rosenthal Y (2009) Deep-sea temperature and ice volume changes across the Pliocene–Pleistocene climate transitions. Science 325:306–310
Spalding MD, Fox HE, Allen GR, Davidson N, Ferdana ZA, Finlayson M, Halpern BS, Jorge MA, Lombana AL, Lourie SA, Martin KD, McManus E, Molnar J, Recchia CA, Robertson J (2007) Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. Bioscience 57:573–583
Swofford DL (2003) PAUP*. Phylogenetic analysis using parsimony. Sinauer Associates, Sunderland, MA
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evolution 24:1596–1599
Veron J (2000) Corals of the world. Australian Institute of Marine Science, Townsville, QLD
Voris HK (2000) Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. J Biogeogr 27:1153–1167
Werner R (2008) New insights into the geodynamic history of northern Wharton Basin (South-East Indian Ocean) Eos Trans AGU, 89 (53) Fall Meet. Suppl, Abstract
Westneat MW, Alfaro ME (2005) Phylogenetic relationships and evolutionary history of the reef fish family Labridae Mol. Phylogenet. Evol. 36:370–390
Wolstenholme DR (1992) Animal mitochondrial DNA: structure and evolution. Int Rev Cytol 141:173–216
Woodroffe CD, Veeh HH, Falkland AC, McLean RF, Wallensky E (1991) Last interglacial reef and subsidence of the Cocos (Keeling) Islands, Indian Ocean. Mar Geol 96:137–143
Zwickl DJ (2006) GARLI. Program distributed by the author. Section of Integrative Biology, The University of Texas at Austin. Available at http://www.zo.utexas.edu/faculty/antisense/Garli.html
Acknowledgments
Funding and logistic support were provided by The National Geographic Society, (Cocos Keeling, Marquesas), The Queensland Government/Smithsonian Institution Collaborative Research Program on Reef Fishes, The Smithsonian Institution, The Seychelles Fishing Authority, Cocos Keeling and Christmas Island National Parks, Department of Environment and Heritage Australia, The Taiwan National Museum, The Australian Institute of Marine Science, The Lizard Island Research Station, the University of Guam Marine Laboratory; University of Hawaii Marine Laboratory and the James Cook University internal funding scheme. Tahiti specimens were collected by M. Meekan, W. Murray, R. Thorne, J. Robinson, J. Ackerman, W. Robbins, Li Shu Chen and L. Rocha. C. Birkeland provided logistic and field assistance, collection of specimens and access to Museum material. Collections on the GBR were carried out under GBRMPA Permit number G03/3871.1. S. Klanten, D. Blair and the JCU Molecular Ecology and Evolution Laboratory assisted with laboratory work and analysis. The work was carried out under James Cook University Ethics Approval No. A503.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Reusch.
Rights and permissions
About this article
Cite this article
Winters, K.L., van Herwerden, L., Choat, J.H. et al. Phylogeography of the Indo-Pacific parrotfish Scarus psittacus: isolation generates distinctive peripheral populations in two oceans. Mar Biol 157, 1679–1691 (2010). https://doi.org/10.1007/s00227-010-1442-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-010-1442-4