Abstract
The genetic structure of Patagonian toothfish populations in the Atlantic and western Indian Ocean Sectors of the Southern Ocean (SO) were analysed using partial sequences of the mitochondrial 12S rRNA gene and seven microsatellite loci. Both haplotype frequency data (F ST>0.906, P<0.01) and microsatellite genotype frequency data (F ST=0.0141–0.0338, P<0.05) indicated that populations of toothfish from around the Falkland Islands were genetically distinct from those at South Georgia (eastern Atlantic Sector SO), around Bouvet Island (western Atlantic Sector SO) and the Ob Seamount (western Indian Ocean Sector of the SO). Genetic differentiation between these populations is thought to result from hydrographic isolation, as the sites are separated by two, full-depth, ocean-fronts and topographic isolation, as samples are separated by deep water. The South Georgia, Bouvet and Ob Seamount samples were characterised by an identical haplotype. However, microsatellite genotype frequencies showed genetic differentiation between South Georgia samples and those obtained from around Bouvet Island and nearby seamounts (F ST=0.0037, P<0.05). These areas are separated by large geographic distance and water in excess of 3,000 m deep, below the distributional range of toothfish (<2,200 m). No significant genetic differentiation was detected between samples around Bouvet Island and the Ob Seamount although comparisons may have been influenced by low sample size. These localities are linked by topographic features, including both ridges and seamounts, that may act as oceanic “stepping stones” for migration between these populations. As for other species of deep-sea fish, Patagonian toothfish populations are genetically structured at the regional and sub-regional scales.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Benthopelagic fish species, from bathyal depths, often have very wide geographic distributions. Populations occur on continental slopes, the slopes of oceanic islands and on axial and non-axial seamounts that maybe separated by thousands of kilometres of deep ocean. These species are often strong swimmers as adults, are highly fecund and have long-duration pelagic eggs, larvae and/or juvenile stages. As a null hypothesis it is assumed that such life-histories confer a high capacity for dispersal across large distances. Given the lack of obvious physical barriers to dispersal in the oceans, this should lead to high levels of migration between distant populations and genetic homogeneity on regional or even oceanic scales.
In some cases, there is evidence for gene-flow between populations of deep-water benthopelagic fish over large geographic distances. Examples include: slender armourhead (Pseudopentaceros wheeleri Hardy 1983), wreckfish (Polyprion americanus Bloch and Schneider 1801) and alfonsino (Beryx splendens Lowe 1834) (Martin et al. 1992; Sedberry et al. 1996; Hoarau and Borsa 2000). However, in many cases, genetic differentiation has been detected between populations on interoceanic, intraoceanic and even intraregional scales (reviewed in Creasey and Rogers 1999; Rogers 2003; see also Stockley et al. 2005; Aboim et al. 2005).
The Patagonian toothfish, Dissostichus eleginoides, is a benthopelagic species that grows to more than 2 m in length and reaches 95 kg in weight (North 2002). It is an active predator and scavenger that preys on fish, crustaceans (especially prawns), cephalopods and other invertebrates, although the relative importance of these groups in the diet depends on geographic locality and fish size (Pilling et al. 2001; Goldsworthy et al. 2002; Xavier et al. 2002; Arkhipkin et al. 2003). Patagonian toothfish are found at depths of between 200 and 2,500 m around islands and seamounts of the Sub-Antarctic, North of 55°N, and on the shelf and continental slopes of southern Chile and Argentina (Gon and Heemstra 1990).
Patagonian toothfish are thought to spawn at about 1,000–1,500 m depth, and female fish produce between 50,000 and 500,000 large pelagic eggs (Kock et al. 1985; Chikov and Melnikov 1990; Agnew et al. 1999). Eggs and larvae occur in the upper 500 m of the ocean (Evseenko et al. 1995). The duration of the pelagic phase from eggs, through larvae, to juveniles probably lasts for around 1 year (Des Clers et al. 1996). Juvenile fish eventually become associated with the seabed, and age-class 1 between 14 and 25 cm long occur in bottom trawls in waters around 140 m depth (North 2002; British Antarctic Survey unpublished data). Growth of Patagonian toothfish is fairly rapid for the first 10 years and males and females mature at 7 and 12 years, respectively (Horn 2002). Toothfish live for more than 50 years and tend to migrate down-slope, as they grow larger (Horn 2002; Collins et al. 2003). Data on movements of adult Patagonian toothfish are limited and somewhat contradictory. Tagging studies suggest that sub-adult (immature) fish (<85 cm total length) have a tendency to remain within a relatively small area (15 nm; Williams et al. 2002; Tuck et al. 2003; Marlow et al. 2003). However, these studies also indicate that low numbers of sub-adult fish may migrate between locations within regions (see Williams et al. 2002 for Heard/McDonald islands, Kerguelen, Crozet and Marion/Prince Edward Islands). Recent tagging studies around South Georgia, in the Atlantic Sector of the Southern Ocean (SO), have demonstrated that very occasionally toothfish tagged at South Georgia have been recaptured on the Patagonian Shelf (Marlow personal communication Renewable Resources Assessment Group, Imperial College, London). Long-distance, transoceanic dispersal of individual fish has also been reported on one occassion (Møller et al. 2003). It must be noted that because toothfish migrate into increasingly deep waters, as they grow larger (Williams et al. 2002), there is little data on the behaviour of large mature fish.
Several studies on the genetics of Patagonian toothfish have already been completed. These have included the identification of molecular markers for species identification (Smith et al. 2001) but the majority have focused on resolving the genetic population structure of Patagonian toothfish, for the purposes of stock identification (Smith and McVeagh 2000; Appleyard et al. 2002, 2004; Shaw et al. 2004). These studies have demonstrated marked genetic differentiation between populations of Patagonian toothfish located in different geographic regions, namely Falklands, South Georgia, Heard/McDonald Islands and Macquarie Islands. Within regions, a different picture emerges depending on locality. Populations within the area of Prince Edward/Marion Islands, Crozet Islands and Heard/McDonald Islands are genetically homogenous (Appleyard et al. 2004). Comparisons between the Patagonian Shelf and South Georgia/Shag Rocks have revealed marked genetic differentiation (Shaw et al. 2004). These localities are separated by a deep-water channel (more than 2,000 m depth) and by two oceanic fronts, the Polar Front (PF), and the Subantarctic Front (SAF).
Previous studies clearly show that Patagonian toothfish demonstrate marked genetic differentiation on regional and even sub-regional scales. However, a complete picture for the entire distribution of the species is not yet completed on the regional scale. In this study, genetic analysis of populations of Patagonian toothfish from the Patagonian Shelf and Atlantic and western Indian Ocean Sectors of the SO is presented. This includes the first genetically analysed samples of individuals from the southeast Atlantic Sector of the SO around Bouvetρya (Bouvet Island), a locality lying almost exactly between previously analysed localities in South Georgia, and Crozet, Prince Edward/Marion Islands. Bouvet is separated from South Georgia by deep water of abyssal depths. However, Bouvet Island and the islands and seamounts to the east are potentially linked by ridges and seamounts that may act as stepping-stones for dispersal of toothfish either in the adult or larval and juvenile stages. This study therefore provides a test that toothfish may disperse along topographic features such as seamounts and ridges but that deep water presents a barrier to dispersal for this species.
Materials and methods
Samples
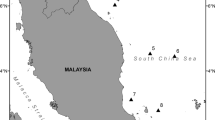
Toothfish samples were collected by long line from commercial fishing vessels over a period of several years but mainly in 1997 and 2003. Samples were collected from several geographic areas including from around the Falkland Islands, Shag Rocks, South Georgia, Bouvet Island, Speiss Seamount, Ob Seamount and Meteor Seamount (Fig. 1). Table 1 presents the location of each sample along with the numbers of toothfish sampled for the present study.
Samples were dissected immediately following capture and a small sample of muscle, liver or gill tissue preserved in 99% ethanol. Samples were transported back to British Antarctic Survey, Cambridge, United Kingdom for genetic analysis.
DNA extraction
In the laboratory, excess ethanol was blotted from the tissue and total genomic DNA was then extracted from small (1–3 mg) sections of tissue using DNeasy kits (Qiagen, Crawley, West Sussex, UK), following the manufacturers instructions for animal tissue. The extracted DNA was resuspended in elution buffer and then checked on a Syngene Gene Genius documentation system (Syngene, Cambridge, UK), with Gene Snap Version 2.60.0.14 documentation software, by comparison with a known standard (1 Kb DNA ladder, Gibco BRL, Rockville, MD, USA) on a 1% agarose gel, following electrophoresis. DNA was also quantified using an Eppendorf Biophotometer (UV spectrophotometer, Eppendorf, Hamburg, Germany) by measuring absorbance at 260 nm. DNA was subsequently stored at -20°C until used for analysis.
Mitochondrial DNA amplification
The genetic variation was assessed between individuals from the different sampling localities for the 16S rDNA and the 12S rDNA mitochondrial partial sequences. Amplification of the 16S rDNA region was carried out using the primers 16AR (5′-CGC CTG TTT ATC AAA AAC AT-3′) and 16SBR (5′-CCG GTC TGA ACT CAG ATC ACG-3′) Palumbi et al. (1991). The reaction components for 16S amplification were as follows: 100 μl reaction; 10 μl 10× PCR buffer (Tris–HCl, KCl pH 7.8, contains 15 mM MgCl2), 20 μl Q-solution, 2 μl dNTPs (10 mM of each dNTP), 1 μl (10 pM) of each primer, 0.5 μl Taq polymerase (2.5u), 64.5 μl dH2O, 1 μl sample DNA (2 ng); all reagents from Qiagen. Reaction conditions were: 94°C for 4 min, 30 cycles of 94°C 30 s, 50°C 1 min, 72°C 1 min, then 72°C for 10 min. All reactions were carried out on an MJ Research Peltier Thermal Cycler 225 DNA Engine Tetrad-Gradient.
Amplification of the partial 12S rDNA region was carried out using the newly developed primers 12STF1 (5′-TCC CTA ACC ACT CTT TAC GC-3′) and 12STR1(5′-CTC CAA CCT CCT TGT TTC G-3′). The reaction components for 12S amplification were as follows: 50 μl reaction; 5 μl 10× PCR buffer, 1 μl dNTPs, 1 μl MgCl2 (25 mM), 0.5 μl each primer, 0.25 μl Taq-polymerase, 41.25 μl dH2O, 0.5 μl sample (1 ng); all reagents Qiagen. Reaction conditions were as for 16S rDNA.
DNA sequencing
Cycle sequencing reactions were carried out using DYEnamic ET terminator reagent premix according to the manufacturer’s instructions (Amersham Biosciences Ltd, Little Chalfont, Buckinghamshire, UK). Samples were purified, following cycle sequencing, using DyeEx dye terminator removal kits (Qiagen) according to manufacturer’s instructions. Products of sequencing reactions were subsequently visualised on a Megabace 500 automated capillary DNA sequencing machine running Sequence Analyzer Version 2.4 software (Amersham Biosciences). Sequence fluorograms were checked by eye, especially where insertion/deletions (indels) were detected by alignment (see below). In all cases, indels were found to result from failure of the software to record nucleotide peaks following multiple “A” or “T” sequences or because of low peaks for “G”.
Microsatellites
Seven microsatellite loci were PCR amplified using the primers developed by Reilly and Ward (1999) and Smith and McVeagh (2000): To2 (forward 5′-CTC TGA AGA TGA ATT GGT GGA TGC-3′ reverse 5′-CAT CAT GTC ACC CTG TCT TTA ACG-3′), To5 (forward 5′-CAC AGA CCA GCA CTA CAA CCC AAG G-3′ reverse 5′-AAG TGT AGT AAT CCA AAT GCA CGC-3′), cmrDe2, cmrDe4, cmrDe9, cmrDe13, cmrDe30. Reverse primers were labelled with a fluorescent dye (To2R—NED, Applied Biosystems, Warrington, Cheshire, UK; To5R—FAM, Transgenomic, Glasgow, UK; De2R—HEX, Transgenomic; De4R—FAM, Transgenomic; De9R—HEX, Transgenomic; De13R—FAM; De30R—FAM, Transgenomic). PCR amplifications were carried out as 10 μl reactions with the following components: 10× PCR buffer 1 μl, Q-solution 2 μl, MgCl2 0.4 μl, dNTPs 0.2 μl, 0.5 μl of each primer, 0.5 μl 1:5 Taq polymerase (0.5u), dH2O 3.9 μl, 1 μl sample DNA (2 ng). Reactions conditions were as follows: 93°C 10 min then 40 cycles of 93°C 30 s, 54°C 1 min, 72°C 2 min, then 72°C 10 min followed by a slow cooling of 60, 50, 40, 30, 20°C for 1 h each. The size of PCR products was estimated using Genotyper Version 1.1 on a Megabace 500 automated capillary DNA sequencing machine.
Data analysis
12S rRNA
Sequence data was aligned using the programme Clustal X (Thompson et al. 1997) using default parameters. Sequences were then trimmed from both ends to allow comparison of good sequence for the maximum number of individuals. Base frequencies were calculated and intrapopulation diversity and diversity across all samples was analysed by estimating gene diversity (h), the probability that two randomly choosen haplotypes are different (Nei 1987), and nucleotide diversity (π), the probability that randomly chosen homologous nucleotides are different (Tajima 1983; Nei 1987).
Overall genetic differentiation between samples from the Falkland Islands, Bouvet Island, Meteor Seamount, Speiss Seamount and Ob Seamount were estimated using pairwise F-statistics (F ST; Wright 1951). Differentiation was also estimated using Exact testing (Raymond and Rousset 1995; Goudet et al. 1996). All sequence analyses were carried out using Arlequin Version 2.0 (Schneider et al. 2000).
Microsatellites
Estimates of observed and expected heterozygosities and Exact tests of conformity to Hardy–Weinberg expectations (Guo and Thompson 1992) were carried out using Arlequin Version 2.0 (Schneider et al. 2000). Tests for linkage disequilibrium between genotypes at each pair of loci was tested using FSTAT 2.9.3.2 (Goudet 1995). This software estimates the significance of association between genotypes at pairs of loci in each sample by using the log-likelihood ratio G-statistic.
Genetic differentiation was analysed using Exact tests of genetic differentiation between each site across all loci using Genepop Version 3.3. Pairwise F STs were estimated using Weir and Cockerham’s (1984) implementation of Wright’s (1978) F-statistics, with significance being estimated using permutation tests in FSTAT. A matrix of F ST values were compared to a matrix of geographic distances in kilometres between the sample areas, again using Genepop. These approaches do not allow the specific testing of hypotheses that relate geographic proximity of populations to genetic population structure. Analysis of molecular variance (AMOVA) was therefore also used to analyse population structure at three levels of hierarchy; (1) within populations, (2) between each population (3) between three groups of populations (Falklands, South Georgia, eastern Atlantic/Indian Ocean Sectors). This analysis was implemented using Arlequin Version 2.0.
Results
Sequences
No variation was found for the 16S rRNA partial sequences (Accession Number: AM180545) so this gene was abandoned for purposes of population genetic analysis. Partial sequences of 249 base pairs (bp) of the 12S rRNA were obtained for 151 individuals across all populations. Overall nucleotide composition was: C=22.58%, T=29.23%, A=22.97% and G=25.22%. A total of three haplotypes (Accession Nos.: AM180546, AM180547, AM180548) were detected in this short region with all mutations being substitutions (two transitions and one transversion). The samples from around the Falklands region showed a gene diversity (h) of 0.1193 (±0.0756) and a nucleotide diversity (π) of 0.000730 (±0.001079). All other populations were fixed for a single haplotype that occurred in one individual in the Falklands samples (see Table 2). For the Falklands samples 32 out of 33 specimens exhibited a different haplotype to all other populations. F STs were highly significant in all pairwise comparisons between the Falklands and all other samples. Pairwise comparisons between populations other than the Falkland Islands showed no differentiation (Table 3).
Microsatellites
A total of 274 individuals were genotyped for seven microsatellite loci. Of these loci, one, To5 was monomorphic in all but one population, Speiss Seamount. Heterozygosity of the other loci was high, ranging from 0.41463, in De13, to 1.0000 in To2 and De2. Numbers of alleles varied from 4 (De30) to 34 (De9). Pairwise genotypic disequilibria were not detected and significant deviations from Hardy–Weinberg expectations were only found in two populations, both for the same locus, De9 (Table 4). It should be noted that this locus also showed the most deviations from Hardy–Weinberg expectations in Appleyard et al. (2002) and significant deviation in a single sample in Shaw et al. (2004). This may reflect difficulties in scoring this locus or occasional occurrence of null alleles although results across the three studies are inconsistent and not thought to be of sufficient magnitude to adversely influence overall conclusions.
Analysis of molecular variance detected significant genetic differentiation between populations but after sequential Bonferroni correction significant differentiation was not detected between groups of populations (Table 5). Exact tests of pairwise genetic differentiation and pairwise F STs (excluding the Bouvet samples because of insufficient sample size) showed significant results for all comparisons between the Falkland Islands and all other populations after Bonferroni correction (Table 6). Comparisons between South Georgia and Speiss seamount also showed significant genetic differentiation although for Exact tests this result was insignificant following Bonferroni correction (Table 6). Comparisons between Meteor, Speiss and Ob seamounts were not significant under any tests prior to or following Bonferroni correction (Table 6). A Mantel test of F ST values versus geographic distance between samples was not significant.
Following these analyses, populations around Bouvet Island (Meteor, Speiss) were pooled and then subsequently pooled with the Ob Seamount sample. F ST analysis and Exact tests for comparisons between the three regional samples (Falklands, South Georgia, West Atlantic/East Indian Ocean Sectors of the SO) were carried out (Table 7). All comparisons between the Falkland Island samples and other populations were significant (F STs P<0.05; Exact tests P<0.01). Comparisons between South Georgia and the pooled samples of Speiss and Meteor Seamounts were significant at the P<0.05 level.
Discussion
This study confirms the findings of Shaw et al. (2004) in demonstrating clear genetic differentiation between populations located on the Patagonian Shelf (Falkland Islands) and South Georgia. This differentiation is apparent in both mitochondrial haplotype data for partial sequences of the 12S rRNA and in analysis of genotype frequencies for microsatellites. As with Shaw et al. (2004), this differentiation is marked in the haplotype data, where the Falkland Islands populations are dominated by individuals with a different haplotype to all other populations in the present study. A single individual out of 33 from the Falkland Islands area showed a “Southern Ocean” haplotype. This may have arisen through a sampling error, arising either on a fishing vessel or in subsequent labelling of collected samples. However, the migration of an individual fish from South Georgia to the Patagonian Shelf is possible given recent data from tagging studies in the Atlantic Sector of the SO (Marlow personal communication).
The question arises as to what mechanisms may prevent dispersal and gene-flow between populations of Patagonian toothfish around the Falkland Islands and South Georgia/Shag Rocks. In terms of larval dispersal it is notable that these two regions are divided by the PF and, further to the north and west, the SAF, the two main current cores of the Antarctic Circumpolar Current (ACC). These are fast flowing bottom-reaching cores, associated with thermohaline fronts and with slower-moving water between. They are constrained to lie within deep passages of the North Scotia Ridge, with the PF lying in the 3,300 m deep Shag Rocks Passage. To the North of these features, around the Falkland Islands and slopes of the Patagonian Shelf, residual current flows are generally towards the north (Glorioso 2000). Therefore it is most unlikely that eggs and larvae produced on the Patagonian Shelf will be advected towards South Georgia.
The oceanography around South Georgia and Shag Rocks is also highly complex. Currents in this region are dominated by the southern sectors of the ACC, most notably the Southern ACC Front (Meredith et al. 2003a), This approaches South Georgia from the southwest, before looping anticyclonically around the island, topography having a very strong influence on direction of flow locally (Thorpe et al. 2005). In addition, an anticyclonic circulation has been observed by deployment of surface drifters and other oceanographic methods on the Northwest Georgia Rise, north of South Georgia (Meredith et al. 2003b). This feature is associated with the formation of a Taylor Column with resultant upwelling and localised increases in productivity. Such features have been associated with larval retention on seamounts and banks, although in this case, this feature in itself would not explain a lack of gene flow between the Falklands and South Georgia (Mullineaux and Mills 1997; Fock and Zidowitz 2004). Instead it is likely that both current–topography interactions and the overall northeastward ACC flow in this region together, mean that it is highly unlikely that larvae in the vicinity of South Georgia are transported towards the Patagonian Shelf. Records of eggs and larvae of Patagonian toothfish are sparse despite extensive sampling in the Atlantic sector of the SO but several eggs and larvae have been captured close to the continental shelf of South Georgia (Evseenko et al. 1995). Furthermore, net-sampling of larvae and juveniles around South Georgia/Shag Rocks suggests that early juvenile fish probably migrate onshore in this region, again retaining the growing fish around the only shallow topography in the area, South Georgia and Shag Rocks (North 2002). This would leave adult dispersal as the only likely means of migration between these populations.
Previous tagging studies have suggested that toothfish migrate in to deeper water as they grow and that small number of sub-adult fish may also undertake migrations. In addition low numbers of toothfish, tagged in South Georgia have been recaptured on the Patagonian Shelf, indicating that at least some fish undertake long-distance migration across oceanic fronts. Microsatellite data from adult fish previously sampled on the North Scotia Ridge, between the Falklands and South Georgia areas, may be intermediate between these two genetically distinct populations (Shaw et al. 2004). This is consistent with adult migration across this zone but levels must be small to prevent genetic homogenisation between the respective shelves in this zone. An alternative possibility is that there is a barrier to reproduction between these populations although there is no evidence to suggest this at the present time. Note that Shaw et al. (2004) also pointed to marked differences between mitochondrial and nuclear microsatellite data. One explanation of this is that male fish are exhibiting different migratory behaviour than females.
For other populations, genetic structure is less resolved. Haplotypes for the partial 12S rRNA sequence for populations at South Georgia, around Bouvet and at the Ob Seamount were identical. Analysis of the separate samples collected from these regions only revealed some differentiation between South Georgia and samples from around Bouvet and the Ob Seamount. Most of these results were insignificant following Bonferroni corrections. Given that no differentiation was detected between the Meteor and Speiss populations, pooling of these samples was carried out. This did show significant genetic differentiation between each of the three regions (Falklands, South Georgia, West Atlantic Sector) using both F ST analysis and Exact testing (note that AMOVA did not suggest significant differentiation attributable to nominal groups of populations). Comparisons between South Georgia and the Ob Seamount by itself were not significant. This contrasts to previous studies that have compared Patagonian toothfish populations in the Indian Ocean Sector of the SO with South Georgia/Shag Rocks (Appleyard et al. 2002). However, as noted by Shaw et al. (2004) the levels of genetic differentiation with microsatellite loci were generally low in most cases. As such, sample size in the Ob Seamount Sample may have influenced the results in the present study. In addition, the times during which samples were collected from these areas (South Georgia versus Bouvet region and Ob Seamount) differed by 5–6 years. Temporal genetic variation in the sampled populations may have influenced comparisons of genotype frequencies. Toothfish are extremely long-lived fish and it is likely that temporal genetic variation in populations will be limited over the timescales represented by this study. No evidence of genetic heterogeneity within populations has been detected in previous studies of toothfish, although these have only spanned 2–3 years (Appleyard et al. 2002, 2004). Temporal genetic variation in fish has been shown to be insignificant or relatively small compared to that related to spatial genetic structure in other species with shorter life-cycles (e.g. Ruzzante et al. 1997; Hansen et al. 2002). Comparisons between the Ob Seamount and pooled Meteor and Speiss Seamount samples also did not show significant genetic differentiation.
That the South Georgia/Shag Rocks populations are genetically distinct from those around Bouvet (junction of the Mid-Atlantic Ridge and South-west Indian Ridge) is unsurprising given previous evidence for regional differentiation in Patagonian toothfish populations (Smith and McVeagh 2000; Appleyard et al. 2002; Shaw et al. 2004). These two areas are separated by a large geographic distance of open-ocean of depths in excess of 3,000 m and no intervening seamounts or islands that allow for step-wise dispersal. Although the evidence for differentiation between the Ob Seamount and South Georgia was not present, previous studies have shown genetic differentiation between Heard and Macdonald Islands, on the Kerguelen Plateau, and South Georgia/Shag Rocks (Appleyard et al. 2002). No consistent significant genetic differentiation has been detected between Marion Prince Edward Islands, Crozet and Kerguelen suggesting that adult migration and/or larval dispersal is sufficient within this region to give a homogenous genetic structure. The present study did not show significant genetic differences between the Speiss/Meteor Seamounts and the Ob Seamount. These sites are potentially linked by oceanic ridge systems and seamounts that may act as oceanic “stepping stones” across this region. Overall this and previous studies support the existence of genetically distinct populations in the Falklands Island area, South Georgia/Shag Rocks and the Bouvet/Crozet/Heard and Macdonald Islands. It should be noted, however, that pooling of the Speiss and Meteor Seamount samples moved comparisons with Ob Seamount samples towards significance. Finer Scale comparisons between the Bouvet area and Marion/Prince Edward Islands, Crozet and Kerguelen would be useful in resolving the question of differentiation between Bouvet and sites to the east.
Patagonian toothfish therefore share the feature of many other species of commercially valuable deep-sea fish in that they demonstrate genetic differentiation at regional and, in the case of the East-Atlantic Sector of the SO, intraregional genetic differentiation. Insignificant correlation between F STs and geographic distance between samples suggest that this differentiation is not simply related to geographic distance between populations and that fronts may play a role in differentiation in the western Atlantic sector of the SO. Fronts have been implicated as barriers to gene flow in other species. For example the subtropical convergence to the south of New Zealand is thought to act as a barrier to gene-flow in populations of orange roughly, Hoplostethus atlanticus (Smith et al. 1996) and pink ling, Genypterus blacodes (Smith and Francis 1982; Smith and Paulin 2003). In other cases, distance has been implicated in the isolation of populations of fish and squid separated by ocean basins. Genetic differentiation has been detected in populations located around the Azores and those on the European continental shelf in black-spot seabream, Pagellus bogaraveo (Stockley et al. 2005) and veined squid, Loligo forbesi (Shaw et al. 1999). The Tasman Sea has been found to be a barrier to gene flow in hoki, Macruronus novaezelandiae (Milton and Shaklee 1987; Baker et al. 1995), pink ling (Smith and Paulin 2003) and black oreo, Allocyttus niger (Ward et al. 1998). Historical patterns of dispersal and colonisation may also strongly influence the population structure of deep-sea fish populations, as indicated in recent studies of the bluemouth, Helicolenus dactylopterus in the North Atlantic (Aboim et al. 2005). In toothfish wide areas of deep water isolate populations inhabiting topographic islands such as ridges, seamounts and the slopes of continental shelves and islands. On intraregional scales, such as the East Atlantic, oceanographic factors are important. The role of history in structuring toothfish populations is unclear from present studies but may also have played a role at intraregional and interregional scales.
References
Aboim MA, Menezes GM, Schlitt T, Rogers AD (2005) Genetic structure and history of populations of the deep-sea fish Helicolenus dactylopterus (Delaroche 1809) inferred from mtDNA sequence analysis. Mol Ecol 14:1343–1345
Agnew DJ, Heaps L, Jones C, Watson A, Berkieta K, Pearce J (1999) Depth distribution and spawning pattern of Dissostichus eleginoides at South Georgia. CCAMLR Sci 6:19–36
Appleyard SA, Ward RD, Williams R (2002) Population structure of the Patagonian toothfish around Heard, McDonald and Macquarie Islands. Antarct Sci 14(4):364–373
Appleyard SA, Williams R, Ward RD (2004) Population genetic structure of Patagonian toothfish in the West Indian Ocean Sector of the Southern Ocean. CCAMLR Sci 11:21–32
Arkhipkin A, Brickle P, Laptikhovsky V (2003) Variation in the diet of the Patagonian toothfish with size, depth and season around the Falkland Islands. J Fish Biol 63(2):428–441
Baker CS, Perry A, Chambers GK, Smith PJ (1995) Population variation in the mitochondrial cytochrome b gene of orange roughy Hoplostethus atlanticus and the hoki Macruronus novaezelandiae. Mar Biol 122:503–509
Chikov VN, Melnikov YS (1990) On the question of the fecundity of the Patagonian toothfish Dissostichus eleginoides in the region of Kerguelen Islands. J Ichthyol 30:122–125
Collins MA, Belchier M, Everson I (2003) Why the fuss about toothfish? Biologist 50(3):116–119
Creasey S, Rogers AD (1999) Population genetics of bathyal and abyssal organisms. Adv Mar Biol 35:1–151
Des Clers S, Nolan CP, Baranowski R, Pompert J (1996) Preliminary stock assessment of Patagonian toothfish longline fishery around the Falkland Islands. J Fish Biol 49(A):145–156
Evseenko SA, Kock K-H, Nevinsky MM (1995) Early life history of the Patagonian toothfish, Dissostichus eleginoides Smitt, 1898 in the Atlantic sector of the Southern Ocean. Antarct Sci 7(3):221–226
Fock HO, Zidowitz H (2004) Episodic recruitment to seamount populations: evidence from Zenopsis conchifer (Lowe, 1852) at the Great Meteor Seamount (subtropical North-east Atlantic). Arch Fish Mar Res 51(1–3):287–293
Glorioso PD (2000) Patagonian Shelf 3D tide and surge model. J Mar Syst 24:141–151
Goldsworthy SD, Lewis M, Williams R, He X, Young JW, van den Hoff J (2002) Diet of Patagonian toothfish (Dissostichus eleginoides) around Macquarie Island, South Pacific Ocean. Mar Freshw Res 53:49–57
Gon O, Heemstra PC (1990) Fishes of the Southern Ocean. J.L.B. Smith Institute of Ichthyology, Grahamstown, pp 462
Goudet J (1995) FSTAT version 1.2: a computer program to calculate F-statistics. J Hered 86:485–486
Goudet J, Raymond M, de Meeüs T, Rousset F (1996) Testing differentiation in diploid populations. Genetics 144:1933–1940
Guo S, Thompson E (1992) Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48:361–372
Hansen MM, Ruzzante DE, Nielsen EE, Bekkevold D, Mensberg KLD (2002) Long-term effective population sizes, temporal stability of genetic composition and potential for local adaptation in anadromous brown trout (Salmo trutta) populations. Mol Ecol 11(12):2523–2535
Hoarau G, Borsa P (2000) Extensive gene flow within sibling species in the deep-sea fish Beryx splendens. C R Acad Sci III Vie 323:315–325
Horn PL (2002) Age and growth of Patagonian toothfish (Dissostichus eleginoides) and Antarctic toothfish (D. mawsoni) in waters from the New Zealand subantarctic to the Ross Sea, Antarctica. Fish Res 56:275–287
Kock K-H, Duhamel G, Hureau J-C (1985) Biology and status of exploited Antarctic stocks: a review. Biomass Sci Ser 6:143
Marlow TR, Agnew DJ, Purves MG, Everson I (2003) Movement and growth of tagged Dissostichus eleginoides around South Georgia and Shag Rocks (Subarea 48.3). CCAMLR Sci 10:101–112
Martin AP, Humphreys R, Palumbi SR (1992) Population genetic structure of the armourhead, Pseudopentaceros wheeleri, in the North Pacific Ocean: application of the polymerase chain reaction to fisheries populations. Can J Fish Aquat Sci 49:2368–2391
Meredith MP, Watkins JL, Murphy EJ, Ward P, Bone DG, Thorpe SE, Grant SD (2003a) Southern ACC front to the northeast of South Georgia: pathways, characteristics and fluxes. J Geophys Res 108(C5); doi 10.1029/2001JC001227, 2003
Meredith MP, Watkins JL, Murphy EJ, Cunningham NJ, Wood AG, Korb R, Whitehouse MJ, Thorpe SE, Vivier F (2003b) An anticyclonic circulation above the Northwest Georgia Rise, Southern Ocean. Geophys Res Letts 30(20):51–55
Milton DA, Shaklee JB (1987) Biochemical genetics and population structure of blue grenadier, Macruronus novaezelandiae (Hector) (Pisces: Merluccidae), from Australian waters. Aust J Mar Freshw Res 38:727–742
Møller PR, Nielsen JG, Fossen I (2003) Fish migration: Patagonian toothfish found off Greenland. Nature 421:599
Mullineaux LS, Mills SW (1997) A test of the larval retention hypothesis in seamount-generated flows. Deep Sea Res I 44(5):745–770
Nei M (1987) Molecular evolutionary genetics. Colombia University Press, New York, pp 512
North AW (2002) Larval and juvenile distribution and growth of Patagonian toothfish around South Georgia. Antarct Sci 14(1):25–31
Palumbi S, Martin A, Romano S, McMillan WO, Stice L, Grabowski G (1991) The simple fool’s guide to PCR version 2.0. Department of Zoology and Kewalo Marine Laboratory, University of Hawaii, Honolulu, 47pp
Pilling GM, Purves MG, Daw TM, Agnew DA, Xavier JC (2001) The stomach contents of Patagonian toothfish around South Georgia (South Atlantic). J Fish Biol 59:1370–1384
Raymond M, Rousset F (1995) An exact test for population differentiation. Evolution 49:1280–1283
Reilly A, Ward RD (1999) Microsatellite loci to determine stock structure of the Patagonian toothfish Dissostichus eleginoides. Mol Ecol 8(10):1753–1754
Rogers AD (2003) Molecular ecology and evolution of slope species. In: Wefer G, Billett D, Hebbeln D, Jørgensen B, Schlüter M, Van Weering T (eds) Ocean margin systems. Springer Verlag, Berlin, Heidelberg, pp 323–337
Ruzzante DE, Taggart CT, Cook D, Goddard SV (1997) Genetic differentiation between inshore and offshore Atlantic cod (Gadus morhua) off Newfoundland: a test and evidence of temporal stability. Can J Fish Aquat Sci 54(11):2700–2708
Schneider S, Roessli D, Excoffier L (2000) ARLEQUIN: a software for population genetics data analysis, version 2.00. Genetics and biometry laboratory, department of anthropology, University of Geneva, Switzerland
Sedberry GR, Carlin JL, Chapman RW, Eleby B (1996) Population structure in the pan-oceanic wreckfish, Polyprion americanus (Teleostei: Polyprionidae) as indicated by mtDNA variation. J Fish Biol 49(Suppl A):318–329
Shaw PW, Arkhipkin AI, Al-Khairulla H (2004) Genetic structuring of Patagonian toothfish populations in the Southwest Atlantic Ocean: the effect of the Antarctic polar front and deep-water troughs as barriers to genetic exchange. Mol Ecol 13(11):3293–3304
Shaw PW, Pierce GJ, Boyle PR (1999) Subtle population structuring within a highly vagile marine invertebrate, the veined squid Loligo forbesi, demonstrated with microsatellite DNA markers. Mol Ecol 8:407–417
Smith P, McVeagh M (2000) Allozyme and microsatellite DNA markers of toothfish population structure in the Southern Ocean. J Fish Biol 57(Suppl A):72–83
Smith PJ, Francis RICC (1982) A glucosephosphate isomerase polymorphism in New Zealand ling Genypterus blacodes. Comp Biochem Physiol 73B:451–455
Smith PJ, Gaffney PM, Purves M (2001) Genetic markers for identification of Patagonian toothfish and Antarctic toothfish. J Fish Biol 58:1190–1194
Smith PJ, McVeagh SM, Ede A (1996) Genetically isolated stocks of orange roughy (Hoplostethus atlanticus), but not of hoki (Macruronus novaezealandiae), in the Tasman Sea and Southwest Pacific Ocean around New Zealand. Mar Biol 125:783–793
Smith PJ, Paulin CD (2003) Genetic and morphological evidence for a single species of pink ling (Genypterus blacodes) in New Zealand waters. NZ J Mar Freshw Res 37:183–194
Stockley B, Menezes G, Pinho M, Rogers AD (2005) Genetic population structure in the black-spot sea bream (Pagellus bogaraveo Brünnich, 1768) from the NE Atlantic. Mar Biol 146(4):793–804
Tajima F (1983) Evolutionary relationship of DNA sequences in finite populations. Genetics 105:437–460
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Thorpe SE, Stevens DP, Heywood KJ (2005) Comparison of two time-variant forced eddy-permitting global ocean circulation models with hydrography of the Scotia Sea. Ocean Model 9:105–132
Tuck GN, de la Mare WK, Hearn WS, Williams R, Smith ADM, He X, Constable A (2003) An exact time of release and recapture stock assessment model with an application to Macquarie Island Patagonian toothfish (Dissostichus eleginoides). Fish Res 63:179–191
Ward RD, Elliot NG, Grewe PM, Last PR, Lowry PS, Innes BH, Yearsley GK (1998) Allozyme and mitochondrial DNA variation in three species of oreos (Teleostei: Oreosomatidae) from Australian waters. NZ J Mar Freshw Res 32:233–245
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Williams R, Tuck GN, Constable AJ, Lamb T (2002) Movement, growth and available abundance to the fishery of Dissostichus eleginoides Smitt, 1898 at Heard Island, derived from tagging experiments. CCAMLR Sci 9:33–48
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Wright S (1978) Evolution and the genetics of populations, vol 4. University of Chicago Press, Chicago and London, pp 590
Xavier JC, Rodhouse PG, Purves MG, Daw TM, Arata J, Pilling GM (2002) Distribution of cephalopods recorded in the diet of the Patagonian toothfish (Dissostichus eleginoides) around South Georgia. Polar Biol 25:323–330
Acknowledgements
We thank Dr Eduardo Balguerías for sampling and provision of toothfish samples from Speiss, Meteor and Ob Seamounts and Bouvet Island. Dr Paul Brickle provided toothfish samples from the Falklands Islands whilst samples from the Shag Rocks/South Georgia regions were provided by scientific observers working on commercial longline vessels and from a survey conducted by the FRV “Dorada”. Simon Morley was funded by the Government of South Georgia and South Sandwich Islands (GSGSSI), through the South Georgia Project. Kester Jarvis received an Undergraduate Research Bursary (Ref: URB/01449/G) from the Nuffield Foundation for a summer internship at British Antarctic Survey, Cambridge. We also acknowledge Dr Michael Meredith for comments on the oceanographic discussion in the paper, Dr Tony North and Dr Martin Collins for comments on toothfish biology, and Prof. Paul Rodhouse and Prof Christopher Rapley for provision of facilities at British Antarctic Survey, Cambridge.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J.P. Thorpe, Port Erin
Rights and permissions
About this article
Cite this article
Rogers, A.D., Morley, S., Fitzcharles, E. et al. Genetic structure of Patagonian toothfish (Dissostichus eleginoides) populations on the Patagonian Shelf and Atlantic and western Indian Ocean Sectors of the Southern Ocean. Mar Biol 149, 915–924 (2006). https://doi.org/10.1007/s00227-006-0256-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-006-0256-x