Abstract
The upside-down jellyfish Cassiopea is a globally distributed, semi-sessile, planktonically dispersed scyphomedusa. Cassiopea occurs in shallow, tropical inshore marine waters on sandy mudflats and is generally associated with mangrove-dominated habitats. Controversy over the taxonomy of upside-down jellyfishes precedes their introduction to the Hawaiian Islands during the Second World War, and persists today. Here we address the global phylogeography and molecular systematics of the three currently recognized species: Cassiopea andromeda, C. frondosa, and C. xamachana. Mitochondrial cytochrome c oxidase I (COI) sequences from Australia, Bermuda, Fiji, the Florida Keys, the Hawaiian Islands, Indonesia, Palau, Panama, Papua New Guinea, and the Red Sea were analyzed. Highly divergent COI haplotypes within the putative species C. andromeda (23.4% Kimura 2-parameter molecular divergence), and shared haplotypes among populations of two separate putative species, C. andromeda and C. xamachana from different ocean basins, suggest multiple anthropogenic introductions and systematic confusion. Two deeply divergent O’ahu haplotypes (20.3%) from morphologically similar, geographically separate invasive populations indicate long-term (14–40 million years ago) reproductive isolation of phylogenetically distinct source populations and cryptic species. Data support at least two independent introductions to the Hawaiian Islands, one from the Indo-Pacific, another from the western Atlantic/Red Sea. Molecular phylogenetic results support six species: (1) C. frondosa, western Atlantic (2) C. andromeda, Red Sea/western Atlantic/Hawaiian Islands (3) C. ornata, Indonesia/Palau/Fiji (4) Cassiopea sp. 1, eastern Australia (5) Cassiopea sp. 2, Papua New Guinea and (6) Cassiopea sp. 3, Papua New Guinea/Hawaiian Islands.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The study of diversification and species boundaries in marine invertebrates presents unique challenges. In spite of considerable data on species distributions, fundamental ecological and evolutionary processes generating observed biogeographic patterns often remain unclear (Palumbi 1997). For example, while factors limiting dispersal and leading to reproductive isolation may be apparent in phylogeographic studies in terrestrial settings (e.g. Holland and Hadfield 2002) such barriers have often proven difficult to detect in marine ecosystems (Palumbi 1997; Knowlton 2000). Yet, the traditional view that morphologically cohesive marine invertebrate species can have natural, global distributions has recently come under scrutiny, as many such species have been shown to comprise cryptic species complexes or to exhibit deep phylogeographic structuring evident at the molecular level (e.g. Gómez and Snell 1996; Collin 2000; Knowlton 2000; Dawson and Jacobs 2001; Gómez et al. 2002). By “cryptic” we refer to species that are difficult or impossible to distinguish on morphological characters alone (Mayr and Ashlock 1993). Further confounding elucidation of isolating mechanisms in marine settings is the ongoing global redistribution of marine taxa by inadvertent anthropogenic translocation, chiefly hull-fouling and ballast water transfer by commercial shipping operations (e.g. Carlton 1989; Holland 2000). In cases where genetic concordance is demonstrated over ocean basin or global scales, questions now arise regarding the possibility of recent anthropogenic transport, in addition to natural distribution mechanisms such as long-distance dispersal via larval entrainment, rafting, or adult migration.
The systematic treatment of Cassiopea spp. in the Pacific has been characterized by confusion and disagreement. Although six nominal species of upside-down jellyfishes have been described from the Pacific, all are currently placed in a single species, C. andromeda (Gohar and Eisaway 1960; Hummelinck 1968). As was recently suggested by Hofmann and Hadfield (2002), however, the systematics of Cassiopea warrants reappraisal.
According to the earliest documented surveys of Hawaiian island scyphozoans, Cassiopea were not present 100 years ago (Mayer 1906). Two, presumably exotic, species of upside-down jellyfish were subsequently reported from the island of O’ahu: C. medusa Light 1914 in Pearl Harbor (leeward O’ahu) between 1941 and 1945 (Doty 1961), and C. mertensi Brandt 1838 (Bishop Museum catalog no. BPBM-D 353) from Kane’ohe Bay (windward O’ahu) in 1964 (Uchida 1970). Cassiopea medusa is believed to have appeared in Honolulu Harbor and the nearby Ala Wai Canal in 1950 (Doty 1961). By the time C. medusa was documented, however, the two putative species had already been synonymized to C. andromeda (Gohar and Eisaway 1960; Hummelinck 1968). It has been assumed that Cassiopea was inadvertently introduced to the Hawaiian Islands from the South Pacific, Indo-Pacific, or Southeast Asia during the Second World War by U.S. Navy vessels.
Due to their potential negative impacts, it is essential that introduced species can be identified reliably. Unfortunately, scyphozoan systematics has traditionally employed morphological characters, which tend to exhibit high plasticity (Gohar and Eisaway 1960) and may be of limited taxonomic and phylogenetic utility (Dawson 2003). In such cases, molecular data may be invaluable. In an effort to address the long-standing systematic uncertainties surrounding Cassiopea in the Hawaiian Islands, we combined morphological and molecular approaches to test the hypothesis that throughout its range on O’ahu, populations of C. andromeda represent a single, genetically concordant species (Hummelinck 1968). It became rapidly apparent, however, that a traditional morphological approach was not systematically informative, despite molecular evidence for unexpected haplotype diversity. We therefore shifted our emphasis to molecular analyses to assess the broader issues of global phylogeography, molecular systematics and potential geographic sources for invasive O’ahu lineages.
Materials and methods
Sampling
Cassiopea andromeda specimens were collected from five O’ahu localities (four windward: one from a Kahuku aquaculture pond, three from within or near Kane’ohe Bay; one leeward: from the Hilton lagoon, Honolulu BPBM-D1074), one location each from Moloka’i (BPBM-D1073), Palau, Fiji, Indonesia, Australia, and the Red Sea, and two locations from Papua New Guinea (PNG) (Emona and Observation Point). Cassiopea xamachana and C. frondosa were obtained from the Florida Keys, C. xamachana was collected from two locations in Bermuda (Walsingham Pond and Richardson Bay), and C. frondosa was collected in the San Blas Islands, Panama. Outgroups included scyphozoan taxa Catostylus mosaicus, Aurelia labiata and Cyanea sp., scleractinian corals Montastraea annularis and M. cavernosa and the hydroid Maeotis marginata (Table 1). Live medusae were held overnight in the absence of food prior to tissue sampling, in order to reduce chances of contamination of extracted DNA by food sources. Gonad tissue was then dissected out and stored in 80–99% ethanol.
Seventy medusae were maintained at Kewalo Marine Laboratory, Honolulu, for 8 weeks. Morphological examinations were conducted for Hawaiian specimens from the following locations: one population from the Hilton Hawaiian Village lagoon, leeward O’ahu; four populations from windward O’ahu; and one population from Kainaone fish pond on the island of Moloka’i (Table 2; Figs. 1, 2). Traditional morphological observations were made including bell diameter, color, and number of rhopalia present.
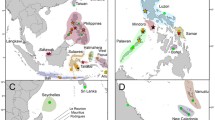
World map showing approximate positions of sampling localities for the three morphologically-defined species of upside-down jellyfishes, Cassiopea andromeda, C. frondosa, and C. xamachana. Numbers in parentheses indicate localities where more than one population was sampled. For more detailed locality names see Table 1
DNA extraction, PCR amplification and sequencing
Genomic DNAs were extracted according to the manufacturer’s protocol using QIAGEN DNeasy nucleic acid extraction kits (QIAGEN, Valencia, Calif.). DNAs were eluted in deionized autoclaved water, and stored at −20°C.
Polymerase chain reaction (PCR) was performed using a PTC-100 thermocycler (MJ Research, Reno, Nev.). Universal COI primers (Folmer et al. 1994) amplified the target fragment consistently under the following PCR conditions: 2 min at 92°C, 30 cycles of 94°C degrees for 30 s, 48°C for 30 s and 72°C for 45 s, with a final 72°C extension for 7 min.
PCR-amplified DNA fragments were purified with QIAquick spin columns (QIAGEN), according to the manufacturer’s protocol, then checked via agarose gel electrophoresis. Forward and/or reverse strands were cycle-sequenced using the PCR primers. ABI Prism DYE Terminator Cycle Sequencing Reaction Kits in a Perkin-Elmer 9700 thermal cycler were used to generate single stranded products, and sequences were determined using an ABI 377 automated sequencer (PE Biosystems, Foster City, Calif.).
Phylogeny reconstruction
A fundamental test of the robustness of a phylogenetic reconstruction is to use various analysis options and to compare the results. In cases where different methods give similar or identical topologies, confidence is increased that the results are representative of the evolutionary history of the sequences comprising the data set (Carranza et al. 2002). Differences in results indicate that more than one interpretation is possible. Therefore a variety of approaches was used for phylogeny reconstruction, including maximum-parsimony (MP), maximum-likelihood (ML), and minimum-evolution (ME) with various models of molecular evolution (e.g. Jukes-Cantor, uncorrected “P”, Tajima-Nei, Kimura 2-parameter, Tamura-Nei, Kimura 3-parameter). Statistical support was assessed with 1,000 bootstrap replicates for MP and ME methods, and for ML 100 replicates were performed (Felsenstein 1985). Trees were rooted with multiple cnidarian outgroups, and resultant topologies were compared for the three methods.
Results
Of 615 aligned nucleotide positions over all taxa, 311 were variable and 283 were parsimony-informative. Within the genus Cassiopea, 226 nucleotide positions were variable and 205 were parsimony-informative. Empirical base frequencies for all cnidarian specimens (n=42) included were: A=27.7%; C=17.1%; G=19.7%; and T=35.5%. Nucleotide composition was biased towards A–T with 63.2%. Base frequencies for Cassiopea spp. (n=32) were also A–T (63.4%) biased: A=28.5%; C=17.1%; G=19.5%; and T=34.9%.
Of 105 pairwise Kimura 2-parameter molecular divergence comparisons shown (Table 3) 17 (16%) were ≤3.9%, 3 (2.9%) were between 10.2% and 10.9%, while 82 (78%) were between 17.6% and 23.4%. Haplotype divergence was often high in among-population comparisons, and genetic distances did not always correlate with geographic distance (Fig. 1, Table 3). The maximum COI divergence observed, 23.4%, was between Cassiopea andromeda Kakaban, Indonesia and C. andromeda Observation Point, PNG. Relatively high divergences were also observed among leeward and windward O’ahu sequences (20.3%), among sequences from the two sampling localities in PNG (Emona and Observation Point) (20.2%), and between two specimens from a single locality, Observation Point PNG (6.9%). Genetic divergence was unexpectedly low in comparisons among certain geographically distant sampling localities, C. andromeda windward O’ahu to C. andromeda Emona PNG (1.3%), C. andromeda leeward O’ahu to C. xamachana Bermuda and the Florida Keys (mean divergence 2.7%), and C. andromeda leeward O’ahu to C. andromeda Red Sea (3.0%). COI haplotypes for C. andromeda Moloka’i were closely related to western Atlantic sequences (mean divergence 0.4%), and to those from leeward O’ahu (3.0%). The C. andromeda Red Sea haplotype was nearly identical to western Atlantic C. xamachana collected at the two Bermuda localities and the Florida Keys (mean divergence 0.3%). Sequences from western Atlantic C. frondosa were genetically distinct from other Cassiopea COI sequences, ranging from 18.6%–23.3% divergent from all other haplotypes, and were similar to one another (≤2.0%).
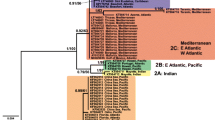
Phylogenetic analyses using three approaches, MP, ML and ME yielded nearly identical ingroup topologies, providing confidence that the relationships inferred among Cassiopea COI sequences represent meaningful patterns and accurate evolutionary relationships (Carranza et al. 2002; Fig. 3). Monophyly of the Cassiopea species complex with respect to cnidarian outgroups was strongly supported (92–98%) in all analyses (MP, ML, ME; Fig. 3). Bootstrap analysis yielded uniformly strong support for clades representing six distinct lineages (100%), and in all but two instances relationships among ingroup clades were well-resolved.
Minimum evolution phylogram based on a 615 basepair fragment of the cytochrome c oxidase I gene of upside-down jellyfish, Cassiopea spp., and cnidarian outgroups. Bootstrap values for major nodes were estimated with 1,000 parsimony, and minimum evolution, replicates, and 100 likelihood replicates, shown on the tree. Operational taxonomic units (OTUs) include traditional scientific names (Gohar and Eisaway 1960; Hummelinck 1968) and geographic information. Abbreviations in OTUs can be found in Table 3, approximate geographic sample localities are shown in Fig. 1, and the six proposed species designations are shown to the right of each clade. Maximum molecular divergence values shown for each ingroup clade were determined using the Kimura 2-parameter substitution model
Discussion and conclusions
Molecular systematics: reconciling morphology and molecules
Taxonomists traditionally have used diagnostic morphological characters to identify species, assuming phenotypic discontinuities reflect underlying genetic distinctions. Evolutionary biologists have generally recognized genetically discrete groups of individuals as species based on the assumption that genetic differentiation reflects reproductive isolation. However, the growing number of exceptions to these generalities, their attendant species concepts, and implied processes of speciation have sparked a revolution in the field of speciation biology (Mallet 2001). A new and more Darwinian perspective, that integrates a greater range of data types in holistic definitions of species (e.g. Templeton 1989; Crandall et al. 2000; Knowlton 2000) and that considers populations, varieties, and species as stages along an evolutionary continuum, is in the ascendant (Mallet 2001). In this new approach, molecular data in particular have an increasingly important role (Hebert et al. 2003; Tautz et al. 2003).
Controversy persists as to how much molecular variation is needed to define species (e.g. Collin 2000). A survey of the literature suggests that 10–20% sequence divergence in COI may be a suitable benchmark (Dawson and Jacobs 2001), which is consistent with the 18.4%–20.9% pairwise sequence divergences (16.0%–17.6% uncorrected) observed between the traditionally recognized moon jellyfish species Aurelia aurita, A. labiata, and A. limbata (reanalysis of data in Dawson and Jacobs 2001). Therefore, we consider molecular divergences at the upper limits of values revealed in this study (10.9–23.4%), which exceed many published maximum values for intraspecific divergences (Table 4), as sufficient for species recognition. Consequently, we propose that COI sequence data support recognition of six Cassiopea species: C. frondosa plus five genetically distinct but morphologically cryptic species (Fig. 3).
Cassiopea frondosa is the only species in the genus that has been reliably and consistently separated from the others based on morphology; mtDNA data confirmed that it is a distinct systematic entity. Cassiopea frondosa sequences from Panama and the Florida Keys formed a well-supported clade with relatively low sequence divergence (2.0%) and were a minimum of 18.6% divergent from all other sequences. Cassiopea frondosa sequences were 20.4% mean pairwise sequence divergence from C. xamachana, which also occurs in the western Atlantic. That C. frondosa and C. xamachana both occur in the Florida Keys, yet can be distinguished morphologically and genetically, provides additional evidence that such molecular divergence reflects reproductively isolated species. High levels of nucleotide divergence were also detected among windward versus leeward populations of the putative single species C. andromeda from O’ahu (20.3%), and between two C. andromeda populations from PNG 20.2% (16.3–17.2% uncorrected) (Table 3). Cassiopea medusae from Fiji, Indonesia and Palau comprised a genetically distinct clade, from which the Port Douglas Australia haplotype was separated by 10.9% (9.7% uncorrected). Molecular divergence values for within-locality comparisons were uniformly low, 1% or less, with the exception of Observation Point, PNG 6.9% pairwise sequence divergence (3.9% uncorrected).
Morphological examination of 70 specimens from multiple sampling sites on O’ahu revealed substantial polymorphism but no informative pattern of differentiation (Table 2). Interestingly, all four windward O’ahu populations sampled were found to be comprised entirely of male individuals, and COI sequences from these populations were identical (0.0% pairwise sequence divergence). In contrast, the leeward O’ahu medusae collected from the Hilton lagoon were either simultaneous hermaphrodites or gonochoristic (Hofmann and Hadfield 2002). The mean within-population sequence variation from the Hilton lagoon sequences was 0.5% (Table 3).
Cryptic invasive species and potential geographic sources
Notwithstanding their original identifications as C. medusa and C. mertensi, invasive populations of Cassiopea on O’ahu have been assumed to comprise a single species, C. andromeda (Gohar and Eisaway 1960; Hummelinck 1968; Hofmann and Hadfield 2002), due to their morphological similarity (Table 2). The presence of two distinct genetic lineages of Cassiopea on O’ahu, with a mean pairwise molecular divergence of 20.3%, therefore provides an example where taxonomic uncertainty and the presence of morphologically similar populations of cryptic marine species has obscured the underlying genetic signal of at least two separate introductions.
Genetic data suggest that populations from windward O’ahu originated in the Indo-Pacific (possibly in or near PNG), and that the leeward O’ahu population sampled originated in either the Red Sea or the western Atlantic Ocean. These routes are consistent with other introductions of marine invertebrates into Hawaiian waters. The two most common biogeographic sources of at least 287 exotic taxa established in Hawai’i are the Indo-Pacific (61 species, 21.3%) and the western Atlantic (30 species, 10.5%) (Eldredge and Carlton 2002). This suggests anthropogenic introductions of Cassiopea are associated with the same mechanisms as other marine invertebrate invasions, namely hull-fouling, ballast-water release, intentional or unintentional direct release (Holland 2000) and similar ecological constraints (e.g. Sax 2001). Like other introduced scyphozoans, Cassiopea are capable of colonizing large nearshore areas and producing problematic blooms (Mills 2001). Coastal eutrophication, habitat disturbance and mangrove filling have been identified as leading causes in large-scale increases of C. xamachana in the Bojorquez Lagoon, Mexico (Arai 2001).
The populations of Cassiopea in Hawai’i representing the two divergent O’ahu haplotypes (20.3%) appear to be geographically separated on windward and leeward shores of the island, which may reflect ecological differences at their sources. Two deeply divergent lineages were also identified from PNG, from Emona and Observation Point (20.2%), again indicating that what has been classed as a single systematic entity on morphological grounds actually has a more complex evolutionary history involving substantial periods of reproductive isolation. At this time, it is impossible to determine whether one of these species has been introduced, also a possibility during the mid-1900s. The currently limited data describing two distinct haplotypes in two different environments (Emona is a muddy reef and mangrove area, while Observation Point is a dark sand slope with adjacent fringing reef and seagrass) near the Indo-West Pacific center of marine biodiversity also would be consistent with multiple evolutionary scenarios including micro-allopatric, parapatric, peripatric, and sympatric speciation, or allopatric speciation followed by range extension. Geographic overlap of divergent genotypes associated with fine-scale ecological separation has also been reported in the jellyfishes Aurelia and Mastigias (Dawson and Jacobs 2001; Dawson 2003). Thus, local adaptation may have contributed to ecological speciation.
Historical biogeography and evolution of Cassiopea
Kramp (1970) proposed that Cassiopea is an evolutionarily ancient genus that originated in the Indo-Pacific and dispersed south of Africa into the Atlantic. The Atlantic range expansion ended in the late Tertiary when the connection was terminated and eastern Atlantic representatives became extinct (Kramp 1970). The phylogeny presented is generally consistent with an older Pacific and more recent Atlantic history of Cassiopea, although greater complexity is implied despite the limited resolution of some internal nodes (Fig. 3). If we apply the rule of thumb of 0.5–1.4% sequence divergence per million years for COI (Knowlton and Weigt 1998), molecular differences among proposed Cassiopea species (23.4–10.9%) suggest divergences on the order of 46–8 million years ago, indicating speciation as early as the mid-Eocene to early Miocene, similar to estimates for cryptic Aurelia species (Dawson and Jacobs 2001). However, estimates of divergence times for scyphozoans are uncertain for several reasons, including: (1) lack of fossil evidence for a local molecular clock calibration; (2) the few substitution rates that are known for cnidarians are lower than those for most other taxa; and (3) divergence estimates may be underestimated since saturation can cause homoplasy in protein coding gene data. In addition, as yet undiscovered cryptic species in unsampled localities may alter phylogenetic relationships and historical biogeographic interpretations.
It is clear that in this evolutionarily ancient group, as in the globally distributed scyphozoan genus Aurelia (Dawson and Jacobs 2001), molecular evolution has outpaced morphological divergence. This phenomenon is in marked contrast to species complexes that exhibit little molecular divergence in spite of dramatic phenotypic polymorphism, such as among African Rift Valley cichlids (Bowers et al. 1994; Meyer et al. 1996) and island radiations of birds (Tarr and Fleischer 1995) and plants (Baldwin and Robichaux 1995). The selective regimes in which morphology has evolved at such an accelerated pace have garnered considerable attention. Examples of morphological similarity persisting despite tens of millions of years of reproductive isolation are equally intriguing, being reminiscent of living fossils, although the underlying selective mechanisms at work may be somewhat less clear. Phenotypic similarity across multiple lineages may result from recent speciation, stabilizing selection, an absence of disruptive selection, or evolutionary convergence. Molecular data reveal that in the case of Cassiopea genetically separable lineages split long ago, refuting similarity due to recent common ancestry, and convergence is unlikely since the Cassiopea species complex is monophyletic (Fig. 3).
Reconstructions of speciation scenarios often rely on multiple assumptions, many of which must be deduced from present day ecology, distribution and divergence patterns (Lessios 1998). In the case of the genus Cassiopea, its ecological niche may hint at the causes behind a pattern of speciation that is characterized by reproductive isolation and long-term maintenance of similar phenotypes. Cassiopea occupies a benthic niche in which relatively clear shallow water is required for exposure of photosymbionts in the oral surface during daylight hours permitting photosynthetic activity. Mangrove-sheltered habitat is favorable for Cassiopea spp. reproduction, as degrading mangrove leaves provide primary settlement sites and possibly metamorphic cues for sexual and asexual propagules (Fleck and Fitt 1999; Hofmann and Hadfield 2002). Mangrove habitat tends to be patchily distributed globally in tropical latitudes, depending on geological and hydrographic conditions. Cassiopea is not a strong natural disperser, and its global distribution may therefore be the result of sweepstakes dispersal in which long-distance gene flow is extremely rare. Divergent Cassiopea haplotypes may provide examples of allopatric speciation driven by the forces of rare dispersal, long periods of ecological stability, geographic isolation, and stabilizing selection. However, highly divergent lineages sampled from different habitats in the same vicinity (e.g. PNG and western Atlantic) also provide evidence for ecological divergence or post-speciation dispersal.
Systematic recommendations
Results of this study indicate a need for global taxonomic revision of the genus Cassiopea. Since this investigation employed a single mtDNA marker and limited geographic sampling, it would be inappropriate to update the systematics of the genus based solely on these data. However, the preliminary systematic implications warrant future reappraisal and clarification using one or more nuclear genetic markers and detailed examination of structures such as subgenital ostia and gastroventricular canals (Thiel 1975).
All three currently recognized species of Cassiopea were sampled: C. andromeda, C. frondosa, and C. xamachana. Phylogenetic results, however, support the existence of at least six species. (1) Genetic results confirm the distinction of western Atlantic C. frondosa (Pallas 1774) sampled from the Florida Keys and Atlantic coast of Panama. (2) The Red Sea/leeward-O’ahu/Moloka’i/western-Atlantic clade most probably represents C. andromeda (Forskal 1775), which originally was described from the Red Sea. (3) For the clade including the Indonesia/Palau/Fiji haplotypes we propose C. ornata, because the type locality for this species includes Palau (Haeckel 1880), which is more precise than the alternative “Neu-Guinea” (Haeckel 1880), and because C. ornata has priority over C. ndrosia described from Fiji (Agassiz and Mayer 1899). We did not sample close to the type localities of the remaining three species, so it is unclear which available names might apply. At the present time, therefore, we propose that these phylogenetic species should be referred to as follows: (4) Cassiopea sp.1 representing the Port Douglas Australia haplotype (5) Cassiopea sp. 2 for the Observation Point PNG clade and (6) Cassiopea sp. 3 representing the windward O’ahu and Emona PNG clade.
References
Agassiz A, Mayer AG (1899) Acalephs from the Fiji Islands. Bull Mus Comp Zool 32:157–189
Arai MN (2001) Pelagic coelenterates and eutrophication: a review. Hydrobiologia 451:68–87
Baldwin B, Robichaux RH (1995) Historical biogeography and ecology of the Hawaiian silversword alliance (Asteraceae): new molecular phylogenetic perspectives. In: Wagner WL, Funk VA (eds) Hawaiian biogeography: evolution on a hot spot archipelago. Smithsonian Institution Press, Washington, pp 259–287
Bowers N, Stauffer JR, Kocher TD (1994) Intra- and interspecific mitochondrial DNA sequence variation within two species of rock-dwelling cichlids (Teleostei: Cichlidae) from Lake Malawi, Africa. Mol Phylogenet Evol 3:75–82
Buckley TR, Simon C, Chambers GK (2001) Phylogeography of the New Zealand cicada Maoricicada campbelli based on mitochondrial DNA sequences: ancient clades associated with Cenozoic environmental change. Evolution 55:1395–1407
Bucklin A, Bentley AM, Franzen SP (1998) Distribution and relative abundance of the copepods, Pseudocalanus moultoni and P. newmani, on Georges Bank based on molecular identification of sibling species. Mar Biol 132:97–106
Caccone A, Sbordoni V (2001) Molecular biogeography of cave life: a study using mitochondrial DNA from bathysciine beetles. Evolution 55:122–130
Carlton JT (1989) Man’s role in changing the face of the ocean: biological invasions and implications for conservation of nearshore environments. Conserv Biol 265–273
Carranza S, Arnold EN, Mateo JA, Geniez P (2002) Relationships and evolution of the North African geckos, Geckonia and Tarentola (Reptilia: Gekkonidae), based on mitochondrial and nuclear DNA sequences. Mol Phylogenet Evol 23:244–256
Castilla JC, Collins AG, Meyer CP, Guiñez R, Lindberg DR (2002) Recent introduction of the dominant tunicate, Pyura praenuptialis (Urochordata, Pyuridae) to Antofagasta, Chile. Mol Ecol 11:1579–1584
Collin R (2000) Phylogeny of the Crepidula plana (Gastropoda: Calyptraeidea) cryptic species complex in North America. Can J Zool 78:1500–1514
Crandall KA, Bininda-Emonds ORP, Mace GM, Wayne RK (2000) Considering evolutionary processes in conservation biology. Trends Ecol Evol 15:290–295
Cywinska A, Hebert PDN (2002) Origins and clonal diversity in the hypervariable asexual ostracod Cypridopsis vidua. J Evol Biol 15:134–145
Dawson MN (2003) Macro-morphological variation among cryptic species of the moon jellyfish, Aurelia (Cnidaria: Scyphozoa). Mar Biol 143:369–379. Erratum. Mar Biol 144:203
Dawson MN, Jacobs DK (2001) Molecular evidence for cryptic species of Aurelia aurita (Cnidaria, Scyphozoa). Biol Bull 200:92–96
Doty MS (1961) Acanthophora, a possible invader of the marine flora of Hawai’i. Pac Sci 15:547–552
Eldredge LG, Carlton JT (2002) Hawaiian marine bioinvasions: a preliminary report. Pac Sci 56:211–212
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fleck J, Fitt WK (1999) Degrading leaves of Rhizophora mangle Linné provide a natural cue for settlement and metamorphosis of the upside down jellyfish Cassiopea xamachana. J Exp Mar Biol Ecol 234:83–94
Flowers JM, Schroeter SC, Burton RS (2002) The recruitment sweepstakes has many winners: genetic evidence from purple sea urchins. Evolution 56:1445–1453
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech 3:294–299
Funk DJ (1999) Molecular systematics of cytochrome oxidase I and 16S from Neochlamisus leaf beetles and the importance of sampling. Mol Biol Evol 16:67–82
Gohar HAF, Eisawy AM (1960) The biology of Cassiopea andromeda (from the Red Sea) (With a note on the species problem). Publ Mar Biol Stat Ghardaqa 11:3–39
Gómez A, Snell TW (1996) Sibling species in the Brachionus plicatilis species complex. J Evol Biol 9:953–964
Gómez A, Serra M, Carvalho GR, Lunt DH (2002) Speciation in ancient cryptic species complexes: evidence from the molecular phylogeny of Brachionus plicatilis. Evolution 56:1431–1444
Haeckel EHPA (1880) Das system der medusen: erster theil einer monographie der medusen. Fischer, Jena
Hart MW, Byrne M, Smith MJ (1997) Molecular phylogenetic analysis of life-history evolution in asterinid starfish. Evolution 51:1846–1859
Hebert PDN, Cywinska A, Ball SL, de Waard JR (2003) Biological identifications through DNA barcodes. Proc R Soc Lond B 270:313–322
Herke SW, Foltz DW (2002) Phylogeography of two squid (Loligo pealei and L. plei) in the Gulf of Mexico and northwestern Atlantic Ocean. Mar Biol 140:103–115
Hofmann DK, Hadfield MG (2002) Hermaphroditism, gonochorism, and asexual reproduction in Cassiopea sp.—an immigrant in the islands of Hawai‘i. Invertebr Reprod Dev 41:215–221
Holland BS (2000) Genetics of marine bioinvasions. Hydrobiologia 420:63–71
Holland BS, Hadfield MG (2002) Islands within an island: phylogeography and conservation genetics of the endangered Hawaiian tree snail Achatinella mustelina. Mol Ecol 11:365–376
Hummelinck PW (1968) Caribbean scyphomedusae of the genus Cassiopea. studies of fauna of Curaçao and other Caribbean Islands 23:1131–1143
Jordal BH, Normark BB, Farell BD, Kirkendall LR (2002) Extraordinary haplotype diversity in haplodiploid inbreeders: phylogenetics and evolution of the bark beetle genus Coccotrypes. Mol Phylogenet Evol 23:171–188
Knowlton N (2000) Molecular genetic analyses of species boundaries in the sea. Hydrobiologia 420:73–90
Knowlton N, Weigt LA (1998) New dates and new rates for divergence across the Isthmus of Panama. Proc R Soc Lond B 265:2257–2263
Kramp PL (1970) Zoogeographical studies on Rhizostomae (Scyphozoa). Vidensk Medd Dan Naturhist Foren Khobenhavn 133:7–30
Lessios HA (1998) The first stage of speciation as seen in organisms separated by the Isthmus of Panama. In: Howard DJ, Berlocher SH (eds) Endless forms: species and speciation. Oxford University Press, New York, pp 186–201
Mallet J (2001) The speciation revolution. J Evol Biol 14:887–888
Mayer AG (1906) Medusae of the Hawaiian Islands collected by the steamer Albatross in 1902. Bull US Fish Comm 23:1131–1143
Mayr E, Ashlock PD (1993) Principles of systematic zoology, 2nd edn. McGraw-Hill, New York, N.Y.
Meyer A, Knowles LL, Verheyen E (1996) Widespread geographical distribution of mitochondrial haplotypes in rock-dwelling cichlid fishes from Lake Tanganyika. Mol Ecol 5:341–350
Meyran JC, Monnerot M, Taberlet P (1997) Taxonomic status and phylogenetic relationships of some species of the genus Gammarus (Crustacea, Amphipoda) deduced from mitochondrial DNA sequences. Mol Phylogenet Evol 8:1–10
Mills CE (2001) Jellyfish blooms: are populations increasing globally in response to changing ocean conditions? Hydrobiologia 451:55–68
O’Foighil D, Gaffney PM, Wilbur AE, Hilbish TJ (1998) Mitochondrial cytochrome oxidase I gene sequences support an Asian origin for the Portuguese oyster Crassostrea angulata. Mar Biol 131:497–503
Palumbi SR (1997) Molecular biogeography of the Pacific. Coral Reefs 16:47–52
Peek AS, Gaut BS, Feldman RA, Barry JP, Kochevar RE, Lutz RA, Vrijenhoek RC (2000) Neutral and nonneutral mitochondrial genetic variation in deep-sea clams from the family Vesicomyidae. J Mol Evol 50:141–153
Rundell RJ, Holland BS, Cowie RC (2004) Molecular Phylogeography of the endemic Hawaiian Succineidae (Gastropoda: Pulmonata). Mol Phylogenet Evol 31:246–255
Sax DF (2001) Latitudinal gradients and geographic ranges of exotic species: implications for biogeography. J Biogeogr 28:139–150
Tarr CL, Fleischer RC (1995) Evolutionary relationships of the Hawaiian honeycreepers (Aves: Drepanidae). In: Wagner WL, Funk VA (eds) Hawaiian biogeography: evolution on a hot spot archipelago. Smithsonian Institution Press, Washington, pp 147–159
Tautz D, Arctander P, Minelli A, Thomas RH, Vogler AP (2003) A plea for DNA taxonomy. Trends Ecol Evol 18:70–74
Templeton AR (1989) The meaning of species and speciation: a genetic perspective. In: Otte D, Endler JA (eds) Speciation and its consequences. Sinauer, Sunderland, Mass., pp 3–27
Thiel VME (1975) Bemerkungen zur Systematik der Gattung Cassiopea (Cepheida, Scyphomedusae). Mitt Hamb Zool Mus Inst 72:25–46
Trewick SA, Wallis GP, Morgan-Richards M (2000) Phylogeographical pattern correlates with Pliocene mountain building in the alpine scree weta (Orthoptera, Anostostomatidae). Mol Ecol 9:657–666
Uchida T (1970) Occurrence of a rhizostome medusa, Cassiopea mertensii Brandt from the Hawaiian Islands. Annot Zool Jpn 43:102–104
Williams ST (2000) Species boundaries in the starfish genus Linckia. Mar Biol 136:137–148
Acknowledgements
We are grateful to Professor Mike Hadfield for providing support and research facilities at the Kewalo Marine Laboratory, University of Hawai’i, and to Professor John Benzie for facilities at the Centre for Marine and Coastal Studies, University of New South Wales (UNSW). We thank William Puleloa, Tom Iwai, Lori Colin, Bert Hoeksema, Harilaos Lessios, Don de Maria, Laura Martin, David Miller, Kylie Pitt, Alan Nelson, Kirk Murakami, and Dan Lindstrom for providing medusae specimens. Thanks to Kualoa Ranch and Mid-Pacific Golf Course for providing access. François Seneca assisted in laboratory DNA extractions. Thanks to Dr. Jonathan Gardner, Victoria University, New Zealand for valuable comments on an earlier draft of this manuscript. We also thank Aquarium Solutions International, Ruhr-Universität Bochum, and the Vice-Chancellor’s Post-doctoral Fellowship scheme (UNSW) for financial support. The experiments performed during the course of this study are in full compliance with the current laws of the United States.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P.W. Sammarco, Chauvin
Rights and permissions
About this article
Cite this article
Holland, B.S., Dawson, M.N., Crow, G.L. et al. Global phylogeography of Cassiopea (Scyphozoa: Rhizostomeae): molecular evidence for cryptic species and multiple invasions of the Hawaiian Islands. Marine Biology 145, 1119–1128 (2004). https://doi.org/10.1007/s00227-004-1409-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-004-1409-4