Abstract
In this study, we developed a multiplex PCR system mediated by “universal primers” (UP-M-PCR) method that could not only effectively reduce the concentration of species-specific primers but also increase the detection flux. This method was used to detect components of dog, chicken, cattle, pig, horse, donkey, fox, and rabbit in foodstuffs simultaneously by amplifying gene fragments with different sizes. The amplified fragments of dog, chicken, cattle, pig, horse, donkey, fox, and rabbit have sizes of 181, 229, 287, 412, 451, 510, 570, and 678 bp, respectively. The sensitivity of the assay could reach 0.05 ng/μL, which is adequate for food inspection. The accuracy of the test results of 103 commercial meat products from market demonstrated the effectiveness and applicability of the established assay. Accordingly, the specificity, sensitivity, and efficiency of the cost-effective assay developed on the conventional PCR platform make it a great promotion and application value in food inspection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Meat is an important source of protein and trace elements for humans. At present, the total meat consumption in the world exceeds 300 million tons annually [1]. It accounts for about one quarter of the world’s meat consumption in China. According to statistics, the production of Chinese meat products exceeded 1.6 million tons in 2017 [2] and the market is still growing. The increase in demand has led to the doping and adulteration of various meat products, which has become one of the main problems of food safety in China. Illegal traders partially or even entirely substitute beef or donkey with cheap chicken, pork, and even fox meat [3]. This not only undermines the interests of consumers, but also reduces food nutritional value and causes food safety issues [4], which may directly affect consumers’ health, especially consumers who are allergic to certain foods. In addition, the adulteration of pork in halal food involves religious issues [5] and is not conducive to social harmony and stability. Therefore, the establishment of an accurate, sensitive, and rapid identification method for adulterated meat has important practical significance for ensuring meat safety and protecting consumers’ rights and interests [6].
Currently, the main methods for meat identification include immunology, nucleic acid, and metabolite-based detection. Immunology-based [7] methods use antibodies to distinguish proteins from various meat sources. However, because the processed food proteins are susceptible to damage and decomposition, the test results are prone to yield false negative result and thus this method has poor reproducibility. Metabolite-based detection methods, such as the near infrared spectroscopy analysis [8], reversed-phase high-performance liquid chromatography (HPLC) [9], combination of tryptic peptide-specific labeling methods and HPLC–MS/MS technology [10] and electronic nose detection technology [11] detect characteristic peaks or response curves for different species to identify specific meat sources [12]. Although these analytical methods are efficient and accurate, these methods require expensive equipment and have high testing cost. In addition, biochips [13] loop-mediated isothermal amplification (LAMP) [14] and DNA barcode technology [15] are not widely available for detection of meat in the short term.
Polymerase chain reaction (PCR) is a classical molecular biology technique. PCR has now proven to be a core technique for detecting small amounts of DNA and can be used to determine the origin of meat species for deep-processed products [16, 17]. In China, according to an investigation in the Suzhou market in 2013, eight beef samples were found adulterated with pork [18]. Similarly, a high ratio 27.0% of beef was found to be adulterated with pork and chicken in a survey performed in the Beijing market in 2014 [19]. Multiplex PCR detecting multiple nucleic acids in a single PCR tube has attracted wide attention in food safety because of its high efficiency, sensitivity, low cost, and simplicity. However, the development and application of multiplex PCR in food detection may be restricted by the interference of primers and the low detection flux (the number of species that can be distinguished) in multiplex PCR detection system. To improve the throughput of the multiplex PCR, universal primers were used to reduce the concentration of species-specific primers and thus diminish the cross-reactivity between primers. The working principle of universal primers is shown in Fig. 1.
This study established a systematic and high-throughput multiplex PCR system mediated by “universal primers” based on the variation of different animal mitochondrial genes. This assay can be used to rapidly, specifically, sensitively, and cost-effectively detect the presence of dog, chicken, cow, pig, horse, donkey, fox, and rabbit-derived components in meat products.
Materials and methods
Raw and commercial samples collection
Raw dog, chicken, cattle, pig, horse, donkey, fox, and rabbit samples were purchased from the farmers’ markets in Suzhou, China. Approximately 500–800 mg each fresh tissue samples were collected at room temperature and subsequently stored at − 20 °C.
To assess the application feasibility of the newly established method, a total of 103 meat products were purchased from different retail markets in Suzhou or online shops.
DNA extraction
Mitochondrial DNA was extracted from meat tissues using an animal mitochondrial DNA extraction kit (PCR level, Beijing Biolebo) according to the manufacturer’s instructions. The purity and concentration of the extracted DNA were assessed on a UV–Vis spectrophotometer (Nanodrop 2000, Thermo Fisher). The extracted DNA samples were diluted to a final concentration of 20 ng/μL and stored at − 20 °C.
Polymerase chain reaction
DNA samples extracted from meats were further subjected to PCR for the analysis of 16S rRNA gene using a pair of control primers. PCRs were carried out on a A-300 PCR Thermal Cycler (LongGene®) in a total of 25 μL mixture containing 12.5 μL Taq MasterMix, 1 μL of DNA extraction (20 ng for meat or 50 ng for meat product), 1 μL (10 μM) each forward and reverse primers, and 9.5 μL ddH2O. PCR amplification with internal reference primers was carried out according to the following cycling conditions: denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s for 35 cycles followed by a final extension at 72 °C for 10 min. PCR amplification products were analyzed by 2% agarose gel electrophoresis. To evaluate the sensitivity of the established multiplex PCR, the extracted DNA (dog, chicken, cattle, pig, horse, donkey, fox, and rabbit) with the initial concentration of 20 ng/μL was diluted with deionized water to 2, 0.5, 0.25, 0.1, 0.05 ng/μL.
Design and validation of PCR primers
Primers were designed using the Software Primer 5 according to the following criteria. All primers were designed on a conserved DNA sequence of a mitochondrial gene. All reverse primers (Dog-UP-R, Chicken-UP-R, Cattle-UP-R, Pig-UP-R, Horse-UP-R, Donkey-UP-R, Fox-UP-R, Rabbit-UP-R) contained a species-specific reverse primer (Dog-R, Chicken-R, Cattle-R, Pig-R, Horse-R, Donkey-R, Fox-R, Rabbit-R) at the 3′-end and the universal primer at the 5′-end, respectively. Moreover, a pair of primers for the 16S rRNA gene was used not only to serve as a positive control but also to evaluate the quality of the extracted DNA. The designed primers were further assessed for the specificity and the cross-species binding with other animal or plant species using the online BLAST alignment tool in the NCBI database. The specificity and sensitivity of all designed primers were repeatedly verified by PCR experiments, and the primers suitable for the UP-M-PCR method were finally determined. All finally confirmed primers (Table 1) were synthesized by Suzhou GenWiz Biotechnology Co., Ltd.
Simplex PCR
To verify the specificity of the primers, the optimized PCR amplification reaction in a total of 25 μL mixture containing 12.5 μL Taq MasterMix, 1 μL of DNA extraction mixture (concentration of each meat was 20 ng/μL), 1 μL (10 μM) each UP, forward and reverse primers, 8.5 μL ddH2O, was performed with the following cycling conditions: denaturation at 95 °C for 30 s, annealing at 70 °C for 10 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s for 10 cycles, then denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s for 25 cycles followed by a final extension at 72 °C for 10 min. PCR products were analyzed by 2% agarose gel electrophoresis.
Multiplex PCR mediated by universal primer (UP-M-PCR)
In the UP-M-PCR reaction, 2 μL of the mixed primers was used. The optimal concentrations of the mixed primers were: UP was 0.083 μM, Dog-F was 0.067 μM,Donkey-F was 0.033 μM, Fox-F was 0.099 μM. Chicken-F, Cattle-F, Pig-F, Horse-F, Rabbit-F were 0.05 μM, Dog-UP-R, Chicken-UP-R, Cattle-UP-R, Pig-UP-R, Horse-UP-R, Donkey-UP-R, Fox-UP-R, Rabbit-UP-R was 0.067, 0.00132, 0.0165, 0.00132, 0.033, 0.033, 0.067, 0.033 μM, respectively. Other reagents were the same as the simplex PCR and the reaction volume was adjusted to 25 μL by adding ddH2O. The PCR conditions and reaction parameters were the same as described above. Negative control PCR without template DNA was set up simultaneously.
Results
DNA extraction
In this study, all the mitochondrial DNA samples were isolated from raw or processed commercial meat samples. Spectrophotometer measurements showed that DNA concentrations varied between 20 and 200 ng/μL and the purity (A260/A280 = 1.72 ~ 2.10) was suitable for PCR amplification.
Reliability and specificity
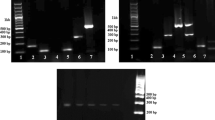
To confirm that all extracts contained amplifiable mtDNA, extracted mtDNA samples were submitted to PCR amplification using the internal reference primers to target the mitochondrial 16S rRNA gene. The result showed that each specimen produced a specific amplified band (Fig. 2a). To further assess the specificity of the designed primers, each set of primers (UP, forward, and reverse primers) were assayed by PCR amplification with its corresponding mtDNA (Fig. 2b) or a mixture of eight animal mtDNA (Fig. 2c). The results clearly indicated that all of the selected primers could amplify specific fragments from the target animal specimen. Although primers for chicken and pig mtDNA-amplified nonspecific bands that were less than 100 bp (Fig. 2c) in mixed templates, these nonspecific bands had no effect on the determination of test results because the target bands were between 181 bp and 678 bp. Therefore, these screened primers could be further applied to the UP-M-PCR system.
M Marker 100 bp, 0: negative control (reagents with primers without DNAs). a The bands amplified by internal reference primers. PCR products from dog: 241 bp, chicken: 249 bp, cattle: 234 bp, pig: 239 bp, horse: 240 bp, donkey: 239 bp, fox: 240 bp, rabbit: 241 bp. b, c Simplex PCR and specificity of simplex assay of mtDNA from raw meat. b Template is its corresponding DNA. c Templates are a mixture of eight animal mtDNA. PCR products from dog:181 bp, chicken: 229 bp, cattle: 287 bp, pig: 412 bp, horse: 451 bp, donkey: 510 bp, fox: 570 bp, rabbit: 678 bp
Optimization of UP-M-PCR system
Mitochondrial DNAs extracted from eight types of meats were mixed and used as templates in the same amount (10 ng each). Validation of multiplex PCR condition was performed with single, double, and eight types of templates, respectively. Each primer was mixed in a specific concentration (as mentioned in the UP-M-PCR section). Figure 3 shows 2% agarose gel electrophoresis of multiplex PCR products amplified from eight types of meat. Figure 3a–d shows the amplification bands resulting from single template, two templates, or eight templates, respectively. UP-M-PCR resulted in a specific band of target size from eight meat species and no fragment produced by non-specific amplification between 181 and 678 bp.
Octuplex PCR results for assay validation. M: Marker 100 bp, 0: negative control (reagents with primers without DNAs). a Lanes 1–8 represent PCR products from dog, chicken, cattle, pig, horse, donkey, fox and rabbit, respectively. b Lanes 1–7 represent PCR products from dog and chicken, dog and cattle, dog and pig, dog and horse, dog and donkey, dog and fox, dog and rabbit, respectively. Lanes 8–13 represent PCR products from chicken and cattle, chicken and pig, chicken and horse, chicken and donkey, chicken and fox, chicken and rabbit, respectively. Lanes 14–18 represent PCR products from cattle and pig, cattle and horse, cattle and donkey, cattle and fox, cattle and rabbit, respectively. Lanes 19–22 represent PCR products from pig and horse, pig and donkey, pig and fox, pig and rabbit, respectively. c Lanes 1–6 represent PCR products from horse and donkey, horse and fox, horse and rabbit, donkey and fox, donkey and rabbit, fox and rabbit, respectively. d Lanes 1–4 represent the results of four repeated experiments with the lanes being amplified from dog, chicken, cattle, pig, horse, donkey, fox and rabbit meat
Sensitivity test
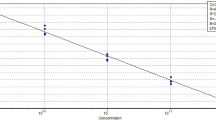
The UP-M-PCR assay was used to amplify five different concentrations of mtDNA to determine the assay sensitivity—the minimum mtDNA concentration of each target meat species that could still be detected. The detectable mtDNA concentration for each target species was different (Fig. 4). The PCR products of chicken, horse, and fox were detected from 0.1 ng/μL mtDNA. The PCR products of dog, cattle, pig, donkey, and rabbit were detected from as low as 0.05 ng/μL mtDNA.
Application to commercial food products
The UP-M-PCR assay was used for the detection of target animal mtDNA in 103 commercial meat products labeled as dog, chicken, cattle, pig, horse, donkey, fox, and rabbit meat, respectively. The results showed that mutton, beef, and donkey meat products may be adulterated by pork or chicken meat. Meat balls and sausages were detected to be mixed with chicken meat. Meat inspection in the market demonstrated the practical application of this method. The detailed adulteration of commercial meat products is summarized in Table 2. Positive results were sent to sequencing and the results were consistent with the multiplex PCR results.
Discussion
The present study focused on developing a rapid, sensitive, specific, and cost-effective method to examine different types of commercial meat products for their authenticity. The use of mtDNA in this method is mainly based on the following considerations. First, genes on mtDNA contain high DNA variations among different species and low DNA variation among individuals of the same species, providing high confidence in the determination of meat species [20, 21]. In other words, the short amplicon does not compromise the specificity. Second, the approximately 3500 mitochondrial copies in a single skeletal muscle cell make the assay very robust and sensitive [22]. Lastly, the sequence data from Cytb and rRNA genes are available on DNA databases for many species, which enables additional comparison of amplified sequences if needed. These are the main reasons targeting on the mtDNAs for the determination of animal species. To improve the efficiency and accuracy for practical use, a pair of control primers for the 16S rRNA gene was introduced in multiplex PCR as a positive control to assess possible amplification problems in DNA quality, equipment, PCR program, or reagents.
The design of primers, especially universal primer, was very important on multiplex PCR techniques. The universal primer sequences were not matched with genomic DNA and mitochondrion DNA sequences of dog, chicken, cattle, pig, horse, donkey, fox, and rabbit by sequence alignment. The PCR experiments confirmed that universal primers did not produce amplification products with DNA and mtDNA of these species (Data not shown). Meanwhile, to circumvent the possible problem that the DNA fragments of processed foods are mainly destroyed, all the amplified products are designed below 700 bp in size. After a series of optimization, UP-M-PCR amplification with universal primers was performed with the appropriate cycling conditions and annealing temperature. In the first ten cycles, the annealing temperature was 70 °C, much higher than the annealing temperature of the universal primer. Amplification products with universal primers were carried out from the specific reverse primers, while universal primers did not participate in the amplification in the first ten cycles. In the latter 25 cycles, the annealing temperature was set at 60 °C, and the universal primers could be amplified using the products obtained from the first 10 cycles as templates.
Primer specificity in the entire DNA of a target species was examined by single PCR using a mixture of three types of primers (UP, forward, and reverse primers). The results clearly indicated that all of the primers could amplify specific fragments ranging between 181 and 678 bp from the target animal specimen (Fig. 2a, b). In the multiplex PCR system, non-specific amplification is easy to occur among primers and between primers and templates because primers and templates are in the same reaction system. The UP-M-PCR resulted in specific bands of target size from eight meat species and no non-specific fragments were produced between 181 and 678 bp (Fig. 3). At the same time, the amplification efficiency of each pair of specific primers must be considered in addition to the consistency of Tm for each pair of primers in the multiplex PCR system. To maintain the consistency of the amplification of the species-specific primers, the primer concentration was adjusted.
The cross-reactivity between multiple primers is one of the main factors affecting the practical use of multiplex PCR. However, the use of universal primers can reduce the concentration of species-specific primers and thus reduce the cross-reactivity between primers. In this study, when the concentration of species-specific primers in the UP-M-PCR system was diluted to 0.00132 μM, the specific DNA fragments still appeared clearly, which made the concentration of primers more than 100 times lower than that of traditional multiplex PCR. By the novel UP-M-PCR method, the PCR products of chicken, horse, and fox were detected from 0.1 ng/μL mtDNA. The PCR products of dog, cattle, pig, donkey, and rabbit meat were detected from as low as 0.05 ng/μL mtDNA (Fig. 4). This sensitivity could be readily used in food inspection. To test the applicability of the developed UP-M-PCR, 103 commercial meat products from market were examined and analyzed. The results suggested that this assay was a simple and rapid technology for identifying meant source from multiple species. The detected positive results were confirmed by Sanger sequencing.
In conclusion, this report developed a UP-M-PCR method that can not only effectively reduce the concentration of species-specific primers but also increase the detection flux through the use of universal primers. Based on the fact that mitochondrial genes are significant different among different species, the established UP-M-PCR system could identify eight different meat sources in a multiplex PCR by the different size of the amplified fragments. The specificity, sensitivity, and efficiency of the cost-effective assay developed on the conventional PCR platform make it possess great popularization and application value in the field of food inspection.
References
Costa ND (2010) Reducing the meat and livestock industry’s environmental footprint. Nutr Diet 64:185–191
Chen G, Liu S, Yang Z, Wang D (2015) Characteristics Chinese meat consumption in 2020 and forecast analysis of Chinese rural economy. Chin Rural Econ 2:76–82
Ballin NZ, Vogensen FK, Karlsson AH (2009) Species determination—Can we detect and quantify meat adulteration? Meat Sci 83:165–174
Agneta H, Hanna L, Lande B, Thorsdottir I (2013) Protein intake from 0 to 18 years of age and its relation to health: a systematic literature review for the 5th Nordic Nutrition Recommendations. Food Nutr Res 57:88–97
Al-Jowder O, Kemsley EK, Wilson RH (2002) Detection of adulteration in cooked meat products by mid-infrared spectroscopy. J Agric Food Chem 50:1325–1329
Skouridou V, Tomaso S, Rau J (2019) Duplex PCR-ELONA for the detection of pork adulteration in meat products. Food Chem 287:354–362
Asensio L, Isabel G, Teresa G (2008) Determination of food authenticity by enzyme-linked immunosorbent assay (ELISA). Food Control 19:0–8
Mamani-Linares LW, Gallo C, Alomar D (2011) Identification of cattle, llama and horse meat by near infrared reflectance or transflectance spectroscopy. Meat Sci 90:378–385
Chung GS, Lee MH, Kim JM (1998) Differentiation the species of origin of meats on the basis of the contents of histidine dipeptides in muscle. J Vet Sci 40:1–6
Bargen CV, Brockmeyer J, Humpf HU (2014) Meat authentication: a new HPLC − MS/MS based method for the fast and sensitive detection of horse and pork in highly processed food. J Agric Food Chem 62:9428–9435
Nurjuliana M, Cheman YB, Mathashim D (2011) Rapid identification of pork for halal authentication using the electronic nose and gas chromatography mass spectrometer with headspace analyzer. Meat Sci 88:638–644
Giaretta N, Di Giuseppe A, Lipper M (2013) Myoglobin as marker in meat adulteration: a UPLC method for determining the presence of pork meat in raw beef burger. Food Chem 141:1814–1820
Roy S, Rahman IA, Santos JH, Ahmed MU (2016) Meat species identification using DNA-redox electrostatic interactions and non-specific adsorption on graphene biochips. Food Control 61:70–78
Deb R, Sengar GS, Singh U (2017) LAMP assay for rapid diagnosis of cow DNA in goat milk and meat samples. Iran J Vet Res 18:134–137
Quinto CA, Tinoco R, Hellberg RS (2016) DNA barcoding reveals mislabeling of game meat species on the US commercial market. Food Control 59:386–392
Xu R, Wei S, Zhou G (2018) Multiplex TaqMan locked nucleic acid real-time PCR for the differential identification of various meat and meat products. Meat Sci 137:41–46
Qin P, Qu W, Xu J (2019) A sensitive multiplex PCR protocol for simultaneous detection of chicken, duck, and pork in beef samples. J Food Sci Technol (Mysore) 56:1266–1274
Jin P, Ding H, Li P (2013) Analysis of meat products adulterated in Suzhou area in 2013. Chin J Food Hyg 26:168–172
Li N, Wang J, Shen Q (2014) Survey of duck, chicken and pork component in lamb or beef slices sold in Beijing. Chin J Food Hyg 26:227–232
Linacre A (2006) Application of mitochondrial DNA technologies in wildlife investigations-species identification. Forensic Sci Rev 18:1–8
Tobe SS, Kitchener AC, Linacre AM (2010) Reconstructing mammalian phylogenies: a detailed comparison of the cytochrome B and cytochrome oxidase subunit I mitochondrial genes. PLoS One 5:e14156
Miller FJ, Rosenfeldt FL, Zhang C, Linnane AW, Nagley P (2003) Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: lack of change of copy number with age. Nucleic Acids Res 31:e61
Ali ME, Razzak MA, Hamid SB et al (2015) Multiplex PCR assay for the detection of five meat species forbidden in Islamic foods[J]. Food Chem 177:214–224
Matsunaga T, Chikuni K, Tanabe R et al (1999) A quick and simple method for the identification of meat species and meat products by PCR assay. Meat Sci 51(2):143–148
Acknowledgements
This work was supported by Suzhou Key Technologies for Prevention and Control of Major Diseases and Infectious Diseases (Project no: Gwzx201606), as well as the Suzhou People’s Livelihood Science and Technology (Project nos: SS201660, SS201843).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflicts of interest.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, W., Tao, J., Xue, M. et al. A multiplex PCR method mediated by universal primers for the identification of eight meat ingredients in food products. Eur Food Res Technol 245, 2385–2392 (2019). https://doi.org/10.1007/s00217-019-03350-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-019-03350-9