Abstract
Staphylococcal enterotoxin (SE) B is one of the most common serotype of SEs to cause staphylococcal food poisoning. To ensure food safety, rapid and low-cost methods have been constantly developed and applied to detect SEB worldwide. In the present investigation, a panel of aptamers of single-stranded DNA molecules against SEB was obtained by optimizing the procedure of systematic evolution of ligands by exponential enrichment, and five of them were selected for further analyzing the characteristics of sequences and second structures. Afterward, an aptamer-based colorimetric method of SEB detection was carried out by employing unmodified gold nanoparticle probes (AuNPs). Results showed that the main second structures of these aptamers were stem loop and hairpin forms. In addition, applying one of these aptamers (No. 15-1), SEB could be detected at the concentration of 10 ng/mL by AuNPs-based colorimetric method. Moreover, the aptamer also possessed a good selectivity toward SEB and SEC1. Our work demonstrated that aptamers had their potential applications as a bioprobe for the detection of SEs in food products.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Staphylococcal food poisoning is one of the most commonly erupted foodborne illness that results from the consumption of foods containing staphylococcal enterotoxins (SEs) produced by enterotoxinogenic strains of Staphylococcus aureus [1–4]. SEs, with a lot of serotypes, can bring various types of disease symptoms [5]. Besides, they are potent gastrointestinal exotoxins resistant to proteolytic enzymes, high temperature (at 100 °C), and extreme pH values because of their compact tertiary structures, hence retaining their activities in the digestive tract after ingestion [6–8]. Among the SEs, SEB is one of the most commonly presented serotypes in staphylococcal food-poisoning cases [9, 10], and rapid, accurate, and reliable detection of SEB is necessary and significant to protect public health against SEB.
Currently, immuno-based assays [11, 12], surface plasmon resonance (SPR) assay [13], and biomolecular interaction mass spectrometry [14] are the most important methods to identify SEs. As is well known, the availability of specific and high-affinity antibodies is of paramount importance in an immuno-detection system [15]. However, these antibodies are sensitive to temperature and have limited lifetimes; moreover, there are still several other problems in the production of antitoxic protein antibodies, such as identical antisera cannot be prepared constantly or maintaining hybridoma involves high costs [16].
Aptamers, generated through the systematic evolution of ligands by exponential enrichment (SELEX), are random single-stranded nucleic acid oligomers (ssDNA or RNA) with a specific and complex three-dimensional shape [17]. As an alternative strategy, aptamers possessing high recognition ability toward specific molecular targets have a strong potential application as bioprobes for targeting drug, developing new drugs, and biosensing [18, 19]. What’s more, aptamers have many advantages over antibodies, such as more stable, easier modification, easier synthesis, and higher affinity, and they can be fluorescently labeled and do not require experimental animals for synthesis [20, 21]. Due to these properties, a variety of aptamer-based analytical methods including electrochemistry, fluorescence, atomic force microscope, and quartz crystal microbalance have been developed for molecular recognition and detection [22–25]. Moreover, the aptamer-based colorimetric method applying gold nanoparticles (AuNPs) as sensing elements is attracting more and more attention because of the potential to eliminate the use of analytical instruments and observe results by naked eyes [26–28].
In this study, SELEX technique was applied to select ssDNA aptamers specific for SEB. And then, an unmodified AuNPs-based colorimetric method employing the selected aptamers was attempted to detect SEB. To the best of our knowledge, this is the first time the abovementioned method is applied for testing SEB. Compared to the aptamers reported for SEB detection [15, 29], this study provided another rapid and cheap way, and the aptamers acquired could be applied for simultaneous detection of multifarious SEs.
Materials and methods
Purified SEA, SEB, and SEC1 were kindly provided by Dr. Jiang from Academy of Military Medical Sciences (Beijing, China). Taq DNA polymerase and dNTPs were obtained from Zomanbio Co., (Beijing, China), and pEASY-T1 cloning vector was purchased from TransGen Biotech Co., (Beijing, China). All other reagents were of analytical purity.
ssDNA original random library and primers
The ssDNA original library containing a random sequence of 40 nucleotides was flanked by two constant regions (5′-ACCGACCGTGCTGGACTCT-(N)40-AGTATGAGCGAGCGTTGCG-3′). A forward primer (P1: ACCGACCGTGCTGGACTCT) and a reverse primer (P2: CGCAACGCTCGCTCATACT) were used to generate double-strand DNA, and a stem loop primer (P3: GCTAACGCGGTGGGACTTCCTAGTCCCACCGCGCGCAACGCTCGCTCATACT) was used for single-strand DNA (ssDNA) generation together with the primer P1. All oligonucleotides were synthesized and purified with polyacrylamide gel electrophoresis (Genscrip Biological Co., Nanjing, China).
Preparation of ASIAN library by unequal-length strand PCR
In the SELEX process, the original ssDNA library was applied as the starting pool for the first selection round. After the first selection round, every other selection round needs the last selection round product as “starting pool,” which was called ASIAN library in this study. Herewith, we employed the method of unequal-length strand PCR to prepare ssDNA of ASIAN library.
With primers of P1 and P3, the unequal length of ssDNA molecules was prepared by PCR as follows: 25 μL of PCR mixture contains 2.5 μL of 10× PCR buffer, 2 μL of 2.5 mM dNTPs, 10 pmol each primer, 1 pmol template, and 1 U Taq DNA polymerase. The mixture was thermally cycled 30 times through 94 °C for 45 s, 60 °C for 90 s, and followed by 5 min extension at 60 °C. The PCR products were analyzed by 8 % polyacrylamide gel electrophoresis (PAGE) with 7 M urea, and the lower band DNAs of interest were purified from the gel for the next round of selection.
The amount of separated ssDNA was measured by urea denatured PAGE silver staining. Five different concentrations of origin ssDNA with the same volume and equivoluminal DNA loading buffer (8 M Urea, 20 mM EDTA, 5 mM Tris–HCl (pH 7.5), 3.7 mM bromophenol blue) were subjected to urea denatured 8 % PAGE according to Laemmli [30]. The gels were stained with silver and then analyzed by the software Quantity One [31].
In vitro selection of DNA aptamers with specific binding to SEB
The SELEX procedure was applied with reference to Wen et al. [32]. For the initial selection, 2 nmol ssDNA original library was heated to 100 °C in 1 mL binding buffer (20 mM HEPES-Na, 120 mM NaCl, 5 mM KCl, 1 mM CaCl2, 2 mM MgCl2, pH 7.35) for 10 min and cooled at 4 °C immediately for 5 min to facilitate the equilibration of different conformations. A certain amount of SEB was then added and incubated at 37 °C for 60 min with shaking. SEB·ssDNA complexes were separated from free ssDNA by pressured filtration with mixed cellulose membrane treated by 0.4 M KOH previously. After that, the mixed cellulose membrane was washed by 1 mL binding buffer for 5 times and cut into pieces; 100 μL of elution buffer (7 M urea, 0.5 M NH4Ac, 7 mM SDS, 1 mM EDTA, pH 8.0) was added to the pieces and heated at 100 °C for 10 min and then centrifuged at 7,500 rpm/min for 10 min at 4 °C, and the supernatant was collected. The SEB·ssDNA was extracted by chloroform and then ethanol precipitated. The precipitated ssDNA was dissolved in 100 μL of deionized water and amplified by unequal-length strand PCR using the forward primer and step loop primer as described in “Preparation of ASIAN library by unequal-length strand PCR” section.
Cloning and sequencing
After 15th round of aptamer selection, the collected ssDNA was amplified by PCR with primers P1 and P2. Each 25 μL of parallel PCR mixtures contained 2.5 μL of 10× PCR buffer, 2 μL of 2.5 mM dNTPs, 10 pmol each primer, 1 U Taq DNA polymerase, and an appropriate amount of template ssDNA. PCR conditions employed were as follows: an initial heat activation step at 94 °C for 5 min and 10 cycles of 94 °C for 45 s, 68 °C for 45 s, and 72 °C for 45 s, and followed by 5 min extension at 72 °C. The products were purified with Miniquick Purification Kit (ZomanBio, Beijing, China) and subsequently cloned into Escherichia coli (DH5α) using the pEASY-T1 vector system. Ten clones were picked and sequenced. Their secondary structures were investigated by RNA structure [33].
Preparation of AuNPs and detection of SEB by aptamer-based colorimetric analysis
AuNPs (~12.74 nm in diameter) were prepared by the citrate reduction of HAuCl4 [34]. Aptamer-based colorimetric detection of SEB was performed as follows: First, 120 μL of AuNPs was mixed with 2 μL of 132 μM aptamer No.15-1 (aptamer No.15-1 was selected from the other four according to simulation results between aptamers and SEB by Visual Molecular Dynamics software, data not shown), and then, the solution was allowed to react for 10 min at room temperature. Secondly, different concentrations of SEB in deionized water were added to the prepared solution, and the solution was allowed to react for another 15 min at room temperature. Finally, 80 μL of 2.75 M NaCl was added to develop color. The absorbance spectrum was measured using UV–VIS spectrometer (UV1700, SHIMADZU). The concentration of SEB was quantified by the absorption ratio (A620/A520) [35]. The specification of aptamer was confirmed by adding BSA, SEA, and SEC1 in a similar way [36].
Results and discussion
Preparation of ASIAN library using special stem loop primers
Generally, specific aptamers can be obtained through several rounds of SELEX screening, and the success of the SELEX screening depends on the preparation of high purity ssDNA of ASIAN library. Asymmetric PCR and biotin–streptavidin separation are usually employed to prepare ssDNA, but the products acquired appear as a smear of band and not of high purity [37]. By contrast, unequal-length strand PCR using special stem loop primer is cheap, does not need any special modification of primers, and yields good-quality products [38, 39].
Herewith, the stem loop primer P3 contained two parts: the longer 5′ reverse repeat sequence with the ability to form stem loop structures and the 3′ complement sequence of the template (Fig. 1a). Because of the higher T m of the stem loop primer, advanced structures still remained the same when the Taq DNA polymerase worked at the temperature range of 55–65 °C and thus prevented the elongation of the (+) strand. As was shown, PCR products had two bands with different migration rates in 7 M urea denatured 8 % PAGE (Fig. 1b). The lower band, which would be employed as the single-strand template, was 33nt shorter than the upper one because of the formation of stem loop structure. Just as implied by Fig. 1b, a large quantity of ssDNA with high purity could be recovered, and the ASIAN library could be applied for the next round of screening.
Unequal-length strand PCR for the preparation of ASIAN library ssDNA using a stem loop primer. a Schematic of PCR product using the P1 primer and the stem loop primer. In the 5′ terminus of (−) primer, there is a GC-rich reverse repeat sequence that can form stem loop structures and prevent the (+) strand elongation. b Strand separation analysis in a urea denatured PAGE. Lane 1, DL2000 DNA Marker; lane 2, the ssDNA of the native library; lane 3, the unequal-length ssDNA product
In vitro selection of ssDNA with specific binding to SEB
Selection processes of ssDNA aptamers binding to SEB were as described in “In vitro selection of DNA aptamers with specific binding to SEB”. The starting pool of ssDNA (2 nmol, approximately 1014 sequences) contained a 40-base central random region and two constant regions. A total of fifteen rounds of SELEX were performed. The original ssDNA was diluted to different concentration in the same proportion and analyzed by 7 M urea denatured 8 % PAGE. And with the gradient reduction in the molar volume of original ssDNA, the color of ssDNA bands faded and the width became narrow gradually (Fig. 2a). The gray value of each lane was measured by Quantity One software, and correlation between molar volume and gray value was implemented by software Origin 8.0 (Fig. 2b). The results revealed that there was a good linear relationship and high consistency between them, which further confirmed the possibility of this method for quantification of ssDNA.
Sequence and structure analysis of the selected aptamers
High-affinity ssDNA were obtained by the 15th round of selection, and ssDNA from the 15th round were amplified to dsDNA and then cloned. Afterward, ten clones were further selected and sequenced. Among them, sequences of five clones (Table 1) were consistent with the original library which contained a random sequence of 40 nucleotides flanked by two constant regions. RNA structure results showed that the second structures of aptamers were mainly stem loop and hairpin. Stem loop might be the structural basis for aptamers binding to target molecules [40, 41]. The stem might play a stabilizing role, and the loop was directly bound to ligands. In aptamer No. 15-1 (Fig. 3), G·T mismatch can be found, and this mismatch often occurred in the normal double helix at the turning point of stacked base pairs, which easily lead to the closure of rings.
Detection of SEB by aptamer-based colorimetric analysis using AuNPs as probe
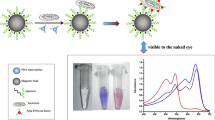
It has been reported that unfolded short ssDNA can adsorb onto the surface of AuNPs without any modification, and such ssDNA-treated AuNPs are more stable than untreated ones in the presence of a given high concentration of salt [35]. Based on this, a colorimetric method using AuNPs aggregation was developed to detect target molecules [42]. The mechanism of the colorimetric method to detect SEB was as follows. Because of the negative capping agent’s electrostatic repulsion against van der Waals attraction between AuNPs [43], the AuNPs were dispersed and stable. However, the addition of enough salt could lead to the aggregation of the AuNPs, which brought the color of AuNPs solution from red to blue. After adding aptamers into AuNPs, aptamers would adsorb onto the surface of the AuNPs, so there is no color change and aggregation of AuNPs after adding high concentration salt. But in the presence of SEB, SEB could bind with aptamers in competition with AuNPs. This could in turn bring the color change of AuNPs solution when adding salt. As shown in Fig. 4a, in the absence and presence of SEB in AuNPs solution treated with aptamer (No. 15-1), once 2 M NaCl was added, the UV–Vis spectrum of AuNPs had a redshifted. The peak at 520 nm (the dispersed AuNPs) decreased, while the peak at 620 nm increased (the aggregated AuNPs). And the absorption ratio (A620/A520) increased with the increasing SEB concentration and the binding time of the aptamer and SEB (Fig. 4b), the absorption ratio reached a maximum at 10 min and then reached a constant value. Applying the measured absorption spectra, the absorption ratio at 620 and 520 nm was found to correlate with the concentration of SEB (Fig. 4c). The linear regression equation is y = 0.0038x + 0.0765, and the R 2 is 0.9874. According to the linear range of the curve, the working range to detect SEB was assigned to a concentration between 10 and 50 ng/mL. Because the colorimetric analysis is simple, cost-effective, and conjugated easily with biomaterials, it will be a promising method to detect target molecules.
a UV–Vis absorption spectra of AuNPs in the absence and presence of SEB. b Plot of changes of absorption ratio (A620/A520) in the presence of various amounts of SEB. c Plot of A620/A520 versus the concentration of SEB. d The ratio of A620/A520 of different interferents to that of SEB in the presence of each interferent at a concentration of 50 ng/mL
The selectivity of aptamer (No. 15-1) to SEB was evaluated by measuring the absorption ratio value, A620/A520 to some common interferents such as BSA, SEA, and SEC1 at a concentration of 50 ng/mL. The data derived from Fig. 4d showed that the absorption ratio at 620 and 520 nm in the presence of SEB was larger than those of blank or other proteins. However, it also revealed that the aptamers obtained had an obvious cross-reaction with SEA and SEC1, indicating that the aptamers selected had potential applications as a bioprobe for the detection of SEs in food products.
Conclusions
In summary, aptamers toward SEB were successfully obtained by employing SELEX system, and second structures of the aptamers were mainly stem loop and hairpin according to RNA structure web server. What’s more, with the aptamer-based colorimetric method applying AuNPs as probe, SEB could be easily detected. The results also implied the good selectivity of aptamers toward SEA and SEC1. Obviously, the colorimetric method would provide a new possibility for the detection of SEs in food products.
Abbreviations
- AuNPs:
-
Gold nanoparticle probes
- BSA:
-
Bovine serum albumin
- PAGE:
-
Polyacrylamide gel electrophoresis
- PCR:
-
Polymerase chain reaction
- SE:
-
Staphylococcal enterotoxin
- SELEX:
-
Systematic evolution of ligands by exponential enrichment
- SPR:
-
Surface plasmon resonance
- ssDNA:
-
Single-stranded DNA
References
Omoe K, Imanishi K, Hu DL, Kato H, Takahashi OH, Nakane A, Uchiyama T, Shinagawa K (2004) Biological properties of staphylococcal enterotoxin-like toxin type R. Infect Immun 72:3664–3667
Sandel MK, McKillip JL (2004) Virulence and recovery of Staphylococcus aureus relevant to the food industry using improvements on traditional approaches. Food Control 15:5–10
Gisch K, Gehrke N, Bros M, Priesmeyer C, Knop J, Reske-Kunz AB, Suduwe S (2007) Formalin-fixed Staphylococcus aureus particles prevent allergic sensitization in a murine model of type I allergy. Int Arch Allergy Immunol 144:183–196
Hu SK, Liu SY, Hu WF, Zheng TL, Xu JG (2013) Molecular biological characteristics of Staphylococcus aureus isolated from food. Eur Food Res Tech 236:285–291
Balaban N, Rasooly A (2000) Staphylococcal enterotoxins. Int J Food Microbiol 61:1–10
Bergdoll MS (1983) In: Easmon CSF, Adlam C (eds) Enterotoxins. Academic Press, New York
Mantis NJ (2005) Vaccines against the category B toxins: staphylococcal enterotoxin B, epsilon toxin and ricin. Adv Drug Deliv Rev 57:1424–1439
Argudín MÁ, Mendoza MC, Rodicio MR (2010) Food Poisoning and Staphylococcus aureus Enterotoxins. Toxins 2:1751–1773
Omonigho SE, Ikenebomeh MJ (2002) Microbiological quality assessment of garri with reference to thermonuclease activity. Food Control 13:535–541
Umoh VJ, Odoba MB (1999) Safety and quality evaluation of street foods sold in Zaria, Nigeria. Food Control 10:9–14
Ewald S, Christensen S (1987) Detection of enterotoxin production by Staphylococcus aureus from aviation catering meals by the ELISA and the microslide immunodiffusion test. Int J Food Microbiol 5:87–91
Ewald S (1988) Evaluation of enzyme-linked immunosorbent assay (ELISA) for detection of staphylococcal enterotoxin in foods. Int J Food Microbiol 6:141–153
Rassoly A (2001) Surface plasmon resonance analysis of staphylococcal enterotoxin B in food. J Food Prot 64:37–43
Nedelkov D, Nelson RW (2003) Detection of staphylococcal enterotoxin B via biomolecular interaction analysis mass spectrometry. Appl Environ Microbiol 69:5212–5215
DeGrasse JA (2012) A Single-stranded DNA aptamer that selectively binds to Staphylococcus aureus enterotoxin B. PLoS ONE 7(3):e33410. doi:10.1371/journal.pone.0033410
Urushibata YJ, Itoh K, Ohshima M, Seto Y (2010) Generation of Fab fragment-like molecular recognition proteins against Staphylococcal enterotoxin B by phage display technology. Clin Vaccine Immunol 17:1708–1717
Stoltenburg R, Reinemann C, Strehlitz B (2007) SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng 24:381–403
Ng EWM, Shima DT, Calias P, Cunningham ET, Guyer DR, Adamis AP (2006) Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov 5:123–132
Tuerk C (1997) In: White BA (ed) Using the SELEX combinatorial chemistry process to find high affinity nucleic acid ligands to target molecules. Humana Press, Totowa
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510
Mosing RK, Bowser MT (2007) Microfluidic selection and applications of aptamers. J Sep Sci 30:1420–1426
Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D, Sullenger BA (2002) RNA aptamers as reversible antagonists of coagulation factor IXa. Nature 419:90–94
Remmele M (2003) Nucleic acid aptamers as tools and drugs: recent developments. ChemBioChem 4:963–971
White RR, Sullenger BA, Rusconi CP (2000) Developing aptamers into therapeutics. J Clin Invest 106:929–934
Fialová M, Kypr J, Vorlíčková M (2006) The thrombin binding aptamer GGTTGGTGTGGTTGG forms a bimolecular guanine tetraplex. Biochem Biophys Res Commun 34:50–54
Li Y, Lee HJ, Corn RM (2006) Fabrication and characterization of RNA aptamer microarrays for the study of protein-aptamer interactions with SPR imaging. Nucleic Acids Res 34:6416–6424
Huang CC, Huang YF, Cao ZH, Tan WH, Chang HT (2005) Aptamer-modified gold nanoparticles for colorimetric determination of platelet-derived growth factors and their receptors. Anal Chem 77:5735–5741
Rhouati A, Paniel N, Meraihi Z, Marty JL (2011) Development of an oligosorbent for detection of ochratoxin A. Food Control 22:1790–1796
John GB, Johnathan LK (2002) Use of magnetic beads in selection and detection of biotoxin aptamers by electrochemiluminescence and enzymatic methods. Biotechniques 32:178–183
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 22:7680–7685
Bian S, Wang Y, Yu T, Ding XS, Wu WT, Ye BP, Xi T (2004) Application of silver staining of polyacrylamide gels in semi-quantitative RT-PCR. J China Pharm Univ 35:178–182
Wen JD, Gray CW, Gray DM (2001) SELEX selection of high-affinity oligonucleotides for bacteriophage Ff gene 5 protein. Biochemistry 40:9300–9310
Seetin MG, Mathews DH (2012) RNA structure prediction: an overview of methods. Methods Mol Biol 905:99–122
Ye Y, Zhou YX, Mo ZZ, Cheng W, Yang S, Wang XH, Chen FS (2010) Rapid detection of aflatoxin B1 on membrane by dot-immunogold filtration assay. Talanta 81:792–798
Song KM, Cho M, Jo H, Min K, Jeon SH, Kim T, Han MS, Ku JK, Ban C (2011) Gold nanoparticle-based colorimetric detection of kanamycin using a DNA aptamer. Anal Biochem 415:175–181
Li L, Li BX, Qi YY, Jin Y (2009) Label-free aptamer-based colorimetric detection of mercury ions in aqueous media using unmodified gold nanoparticles as colorimetric probe. Anal Bioanal Chem 393:2051–2057
Erdogan F, Kirchner R, Mann W, Ropers HH, Nuber U (2001) Detection of mitochondrial single nucleotide polymorphisms using a primer elongation reaction on oligonucleotide microarrays. Nucleic Acids Res 29:e36
Williams KP, Bartel DP (1995) PCR product with strands of unequal length. Nucleic Acids Res 23:4220–4221
Cao XX, Li SH, Chen LC, Ding HM, Xu H, Huang YP, Li J, Liu NL, Cao WH, Zhu YJ, Shen BF, Shao NS (2009) Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Res 37:4621–4628
Davis KA, Abrams B, Lin Y, Jayasena SD (1998) Staining of cell surface human CD4 with 2’-F-pyrimidine-containing RNA aptamers for flow cytometry. Nucleic Acids Res 26:3915–3924
Fukusaki E, Kato T, Maeda H, Kawazoe N, Ito Y, Okazawa A, Kajiyama S, Kobayashi A (2000) DNA aptamers that bind to chitin. Bioorg Med 10:423–425
Li F, Zhang J, Cao XN, Wang LH, Li D, Song SP, Ye BC, Fan CH (2009) Adenosine detection by using gold nanoparticles and designed aptamer sequences. Analyst 134:1355–1360
Li HX, Rothberg L (2004) Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc Natl Acad Sci USA 101:10436–10439
Acknowledgments
This research was supported by the Project of the National High Technology Research and Development Program of China (863 Program, No. 2012AA101601) and National Science Foundation of China (No. 31271876).
Conflict of interest
None.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Aiping Liu and Yongxia Zhang contributed equally to the research.
Rights and permissions
About this article
Cite this article
Liu, A., Zhang, Y., Chen, W. et al. Gold nanoparticle-based colorimetric detection of staphylococcal enterotoxin B using ssDNA aptamers. Eur Food Res Technol 237, 323–329 (2013). https://doi.org/10.1007/s00217-013-1995-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-013-1995-9