Abstract
A real-time PCR assay for the simultaneous detection of Mallard and Muscovy duck is described. Species-specific primers were designed for Mallard or Muscovy duck using the mitochondrial cytochrome b gene sequence. These primer sets were multiplexed with a single duck probe to produce a simple, rapid and robust real-time PCR assay. This assay was shown to be specific for duck compared to a wide range of commercially important meat species and was used for the successful detection of duck meat in complex food matrices. This is the first report of an assay that will detect all species of commercially available duck in commercial products using real-time PCR.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
For many years duck meat has been one of the most expensive commercially available meat types, consumption of which was reserved for special occasions. There has, however, been a recent move towards large-scale production of duck using modern husbandry techniques, which has resulted in a greater availability of duck meat and a proliferation of processed products containing duck meat. Additonally, the range of duck species used commercially has been extended from mainly Mallard ducks (Anas platyrhynchos) to include the subspecies Gressingham and Allesbury, and the Muscovy ducks (Cairina moschate).
There is now a need for a sensitive and robust assay for the detection of all species of commercial duck in heat and pressure-processed products. Heat and pressure processing are known to denature protein and fragment DNA, however DNA-based analyses remain the best option for detection since DNA survives heat and pressure processing relatively intact when compared to proteins [1]. There are few reports on the successful detection of duck species in commercial products. Partis et al. [2] used PCR-RFLP for the detection of duck, however they found that pork preferentially amplified over all other species, such that the method was not applicable for mixtures of meats [2]. Cavalo et al. [3] outlined a method using RAPD for duck detection, but found that similar bands were produced by amplification of both duck and pork, making interpretation of the banding patterns subjective [3]. Both of these reports did however demonstrate the utility of a DNA-based approach, and more recent reports have outlined methods based upon species specific PCR systems, which circumvent co-amplification in mixtures [4–6]. Rodriguez et al. [4, 5] used conventional PCR for the detection of Mule duck, a cross between Mallard and Muscovy duck (Anas platyrhynchos × Cairina moschate), using separate primer sets for two genes: 12S ribosomal RNA and α-actin genes [4, 5]. Rodriguez et al. [6] went on to develop a real-time PCR assay for the detection of Mule duck. However, this assay was based on Mallard DNA sequence and would not detect wild-type Muscovy duck [6], a duck species now commonly used for human consumption.
We report the development of a real-time PCR assay for the detection of both Mallard and Muscovy duck species. Species-specific primers were designed using the mitochondrial cytochrome b gene, which when multiplexed with a generic duck probe, were used for the successful detection of duck meat in complex food matrices which had been heat and pressure processed.
Materials and methods
Meat and meat product samples
Samples of fresh raw meat from Mallard, Gressingham, Allesbury and Muscovy duck species, pig, deer, pheasant, duck, goose, guinea fowl, grouse, pigeon, quail, cow, partridge, sheep, turkey, chicken and commercial products were obtained from local suppliers in the United Kingdom. All samples were stored at 4 °C until DNA extraction.
DNA extraction from raw meat
DNA was extracted from raw meat using a modified version of the GenElute Mammalian Genomic DNA Extraction Kit (Sigma). Briefly, 2 g of tissue was finely minced with a scalpel blade, prior to incubation at 65 °C in 8 ml CTAB buffer (0.055 M CTAB, 1.4 M NaCl, 0.1 M Tris and 0.02 M EDTA, pH 8) containing 50 μg/ml proteinase K (Sigma). After 1 h the samples were cooled briefly and 400 μl of the supernatant used to extract the DNA exactly according to the manufacturers instructions. The final elution step was repeated once and the eluates pooled. DNA was stored short term at 4 °C or long term at –20 °C.
DNA extraction from complex meat matrices
DNA was extracted from complex meat matrices using a modified version of the Wizard DNA cleanup system (Promega). Briefly, 5 g of each sample was incubated at 65 °C in 10 ml CTAB buffer containing 80 μg/ml proteinase K (Sigma). After 2 h the samples were vortexed for 10 s and centrifuged for 10 min at 4,000×g. One millilitre of the supernatant was combined with 800 μl chloroform, vortexed briefly and centrifuged for 10 min at 16000×g. Five hundred microlitres of the aqueous phase was combined with 1 ml Wizard DNA resin and incubated for 5 min at room temperature. The DNA was then purified exactly according to the manufacturers instructions and eluted with 100 μl Tris–EDTA buffer.
Real-time PCR primer and probe set design
Mallard and Muscovy duck specific primers and probe sets were designed using the mitochondrial cytochrome b gene DNA sequence (GenBank Accession numbers AFO59O81 and LO8385 respectively) using Primer Express software (Applied Biosystems) (Table 1). Primers were designed to mismatch all other commercially important species at the 3′ position [7] and were purchased from Sigma Genosys. The probe was labelled with 5′ carboxyfluorescein (FAM) and 3′ minor groove binding (MGB) fluorescent quencher and was purchased from Applied Biosystems.
Real-time PCR reaction set up
Each real-time PCR reaction contained 1 × TaqMan Buffer A, 25 mM MgCl2 solution, 0.625 Units AmpliTaq Gold DNA Polymerase (Applied Biosystems) and 0.5 μM of each deoxynucleotide triphosphate (Sigma) in 25 μl. Optimised primer and probe concentrations are shown in Table 1. DNA template was diluted 1:4 DNA:water and 5 μl of either DNA or water, as a negative control, was added per reaction. Reactions were assembled in Axygen thin walled 96 well plates with optical caps and run on ABI Prism 7700 or 9700 sequence detection systems with the following thermal cycling protocol: 50 °C for 2 min, 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min.
Results and discussion
Primer design
There are two commercially important species of duck: Mallard and Muscovy. These species are of the same family but diverge at the genus level and it is widely considered that all domestic and commercially important duck species, with the exception of the Muscovy, have been bred from the Mallard.
Comparison of the mitochondrial cytochrome b gene sequences for these species revealed that although closely related, there is significant sequence heterogeneity between the species (Fig. 1). We designed an assay that would identify both duck species using a probe common to all duck species combined with species-specific primers. Sequence data from the NCBI database for Mallard and Muscovy ducks and other commercially popular species were aligned and areas of mismatch identified. Using Primer Express software, primer sets were designed to have mismatches to all other species at the 3′ position of both sense and antisense primers for Mallard and Muscovy ducks. The probe was designed to the Muscovy sequence and had only a single base mismatch for the Mallard sequence (Fig. 1).
Efficacy and specificity
The aim of the study was to develop an assay which would identify the presence of duck in commercial products. The identification of the type of duck was less important than the detection of duck per se. Therefore, the primer sets for Mallard and Muscovy were developed separately and then multiplexed to form a duck group assay.
The Mallard and Muscovy assays were found to be highly sensitive on their matching template (Ct values 14.68 and 13.48 for Mallard and Muscovy DNA template using Muscovy and Mallard assays respectively). After primer limitation and multiplexing, the duck group assay was compared to the single assays for amplification efficiency on either Mallard or Muscovy duck template DNA (Fig. 2). There was no significant difference in the Ct values of the single or multiplexed assays for either template. The single assays were found to cross-amplify the alternate template with lower sensitivity (Ct values 28.27 and 30.09 for Muscovy and Mallard DNA template respectively). Cross-amplification at these levels equates to approximately 10,000-fold less amplification efficiency on the alternate template compared to the matching template. However, since there is a common probe for both primer sets, the cross amplification of the alternate template presented no problem for the sensitivity of the assay.
The specificity of the duck group assay was then assessed using DNA from a wide variety of species, including pheasant, chicken, partridge, goose, guinea fowl, pigeon, quail, grouse, turkey, cow, pig, horse, donkey, red deer and sheep, and two varieties of duck originating from the Mallard: Gressingham and Allesbury (Table 2). It was found that there was no cross-amplification of non-duck species (Ct value of 40 equates to no amplification), but good amplification of all types of ducks, demonstrating complete specificity of the duck group assay for duck DNA.
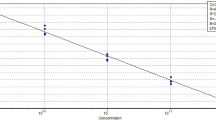
Duck group assay efficiency and limit of detection
Efficiency and limit of detection for the duck group assay were determined using 10-fold dilutions of Mallard duck DNA diluted in water. Primer and probe sets which are working at 100% efficiency will require approximately an additional 3.3 cycles (3.3 Ct) for a 10-fold dilution of template. The limit of detection is the DNA dilution where the Ct values are still reproducibly produced, but beyond which the assay fails. The detection of duck for the duck group assay extended to a 1,000,000-fold dilution of the duck DNA in water (average Ct 33.65) (Fig. 3). Additionally, over this range of dilutions the response was linear, and the slope of the Ct values plotted against the log of the DNA dilution was −3.5, indicating an assay working at 106% efficiency. The limit of detection relates to the detection of 0.0001% (w/w) duck meat in a sample. Although this limit of detection was calculated from dilutions of DNA in water, with detection at these orders of magnitude, the assays could be used as a basis for enforcing accurate product labelling and provide reassurance to consumers concerned about even the smallest levels of contamination of duck meat in products.
Application of assay to commercial samples
The duck group assay was then used to test DNA extracted from a range of commercial samples where either duck or game had been included in the ingredients list (Table 3). Duck was successfully detected in all samples. In samples of roasted and smoked duck and of game casserole, the DNA did not show evidence of significant fragmentation by low Ct values. Similarly, a low Ct value was returned for duck pate which contained not only duck but also pork, pork fat and milk. Higher Ct values were returned for samples which were highly processed or long life: the cat foods. The level of duck in these samples (4%) was only quoted as a minimum since the labelling requirements for pet foods are different from those for human consumption, however we would expect a Ct value of approximately 15 for a sample containing 4% duck. The greater Ct values returned for these samples indicated a high degree of DNA fragmentation. An anomaly for the pet food results was given by the duck biscuits, which returned a relatively low Ct value. This indicated that the duck meat in the biscuit was not as highly processed as in the other pet foods analysed. The presence of duck was therefore detected in all samples analysed, where duck had been listed in the ingredients. Additionally, there was specific detection of duck in complex food matrices, even in the presence of pork, a confounding factor for RFLP and RAPD based assays [2, 3].
Conclusions
A real-time PCR assay for the detection of duck has been developed. The optimised assay is specific, highly sensitive and applicable to complex food matrices. This is the first real-time PCR based assay for the detection of commercial duck species to be published and as such, can now be recommended to food control laboratories.
References
Meyer R, Candrian U, Luthy J (1993) Mitt Gebiete Lebensm Hyg 84:112–121
Partis L, Croan D, Guo Z, Clark R, Coldham T, Murby J (2000) Meat Sci 54:369–376
Cavalo J, Zaragoza, Osta R (2001) Poult Sci 80:522–524
Rodriguez M, Garcia T, Gonzalez I, Asensio L, Mayoral B, Lopez-Calleja I, Hernandez P, Martin R (2003a) J Agric Food Chem 51:1524–1529
Rodriguez M, Garcia T, Gonzalez I, Asensio L, Mayoral B, Lopez-Calleja I, Hernandez P, Martin R (2003b) Meat Sci 65:1257–1263
Rodriguez M, Garcia T, Gonzalez I, Asensio L, Hernandez P, Martin R (2004) J Agric Food Chem 52:1478–1483
Hird H, Hill M, Goodier R (2003) Meat Sci 65:1117–1123
Acknowledgments
This study was funded under the food authenticity programme of the Food Standards Agency of the United Kingdom
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hird, H., Chisholm, J. & Brown, J. The detection of commercial duck species in food using a single probe-multiple species-specific primer real-time PCR assay. Eur Food Res Technol 221, 559–563 (2005). https://doi.org/10.1007/s00217-005-1197-1
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-005-1197-1