Abstract
Angiotensin-I-converting enzyme (ACE)-inhibitory peptides are encrypted in milk protein sequences. They may express an antihypertensive effect if they are released by proteolysis in foods and/or during gastrointestinal digestion. A bioinformatic, in silico, approach was developed to evaluate how systematic initial proteolysis, i.e. cleavage after one specific type of amino acid (C-end) at a time in milk proteins, influence the formation of ACE-inhibitory peptides by subsequent gastrointestinal proteolysis. Computer simulation was done and a peptide QSAR model was used to estimate the combined ACE inhibition by digested proteins. Initial proteolytic cleavage at the C-end of amino acids isoleucine and proline gave, based on calculations, increased effect of ACE-inhibitory peptides after gastrointestinal proteolysis of milk proteins. Cleavage after most other amino acid residues had little or no effect. Results indicate that initial proteolysis in foods have to be specific in order to increase formation of bioavailable ACE-inhibitory peptides during gastrointestinal digestion.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Release of bioactive peptides occurs during proteolysis of milk proteins. Some of the bioactive peptides encrypted in the protein sequences can have health related/physiological effects; research in this area has been extensively reviewed [1, 2]. One of the bioactive properties of peptides derived from proteolysis that has been investigated much is the inhibition of angiotensin-I-converting enzyme (ACE). A number of ACE-inhibitory peptides have been isolated and characterised from foods and protein hydrolysates [3], facilitating improved understanding of their structure-activity relationship [4]. However, isolation and characterisation of ACE-inhibitory peptides in vitro is no guarantee for physiological effect in vivo [5] making it necessary to take their bioavailability into account [6].
Generally speaking, there are two strategies for increased effect of bioactive peptides in dairy products. Either bioactive peptide preparations can be added, or food proteins can undergo specific proteolysis to promote the release of encrypted bioactive peptides. It is important then to take into account that extensive proteolysis also occurs during gastrointestinal digestion. Potential bioactive compounds from the diet, including bioactive peptides, can be broken down during gastrointestinal digestion before they are absorbed and transported into the blood stream to give a physiological effect. This has to be taken into account if dairy products shall be developed with health claims due to ACE-inhibitory peptides. If proteolysis of proteins in foods shall be used as a strategy to increase the effect of bioactive peptides, it must be done, so that the biological activity of peptides (combined effect of concentration and specific activity) after gastrointestinal digestion becomes higher than that obtained naturally by gastrointestinal digestion of intact proteins.
To evaluate food proteins as potential precursors of bioactive peptides, a bioinformatics approach has been to screen protein sequences and compare with bioactive peptide sequences in databases [7]. This approach has been applied to predict possible release of bioactive peptides from plant proteins using proteolytic enzymes with different specificity [8] and to explore release of ACE-inhibitory peptides from pea and milk proteins during gastrointestinal digestion [9]. Besides comparing characterised peptide sequences in databases, peptide quantitative structure-activity relationship (QSAR) models could be used. QSAR models are mathematical functions that describe relationship between activity and chemical structure expressed by variables. Such models are used both to predict activity of untested chemical structures and to predict the chemical structure of compounds with specific activity. A QSAR model for ACE-inhibitory peptides derived from milk proteins have recently been presented [4].
Our objective was to theoretically quantify how initial proteolysis of milk proteins affects the formation of ACE-inhibitory peptides during gastrointestinal proteolysis.
Materials and methods
In silico gastrointestinal proteolysis
Protein sequences (Table 1) were obtained from the Swiss-Prot/TrEMBL database at the ExPASy Molecular Biology Server (http://www.expasy.org). The molecular masses used are those from the revision by Farrell et al. [10] on nomenclature of milk proteins.
In silico gastrointestinal proteolysis of protein and peptide sequences was performed with the software PeptideCutter [11] (http://www.expasy.org/tools/peptidecutter) using the combination of pepsin (pH 1.3), trypsin and chymotrypsin—low specificity without including the more sophisticated model on cleavage probability of chymotrypsin and trypsin. A detailed procedure for performing in silico simulation of gastrointestinal proteolysis was described by Vermeirssen et al. [9].
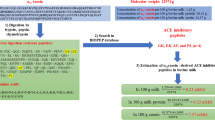
To evaluate the effect of systematic proteolysis on milk proteins before gastrointestinal proteolysis, the protein sequence was first cleaved at the C-end of a given type of amino acid and the obtained fragments then underwent in silico gastrointestinal proteolysis.
Estimation of ACE inhibition by protein hydrolysate
Estimation of ACE inhibition expressed as log IC50 (μmol l−1) was done by the QSAR model reported by Pripp et al. [4]
where x 1 is a measure of side-chain hydrophobicity and x 2 is positively charged side chain for amino acid in C-terminal position and x 3 is van der Waals volume of the amino acid next to the C-terminal position. IC50 is the concentration (μmol l−1) needed to inhibit ACE by 50%.
In order to estimate the ACE inhibition for a protein hydrolysate, the combined IC50 value (μmol l−1) from peptides (i=1, 2,..., p) in protein hydrolysate \(({IC_{\overline {50}}})\) was expressed as
where p is the number of peptides obtained after hydrolysis of a protein sequence, IC50 i the QSAR model estimated IC50 (μmol l−1) for peptide i from hydrolysis of protein sequence, where i=1, 2,..., p. The combined IC50 can be understood as the IC50 value of a given single peptide, which if it occurred in same total concentration as all peptides in the hydrolysate of interest, would give exactly the same IC50 as that found for the hydrolysate.
Corresponding protein concentration (mg/ml) to give a hydrolysate that inhibit 50% activity of ACE was then
where M w is the molecular mass of protein sequence (g/mol) (Table 1).
A spreadsheet in Microsoft Office Excel 2003 was developed with formulas and database of physico-chemical variables. It was able to estimate IC50 for protein hydrolysates directly from the one-letter coded peptide sequences obtained by in silico gastrointestinal proteolysis.
Results and discussion
Hydrolysing food proteins with combinations of pepsin, trypsin and α-chymotrypsin has been used to simulate experimentally the formation of ACE-inhibitory peptides during gastrointestinal proteolysis. Using the outlined bioinformatic approach, it was possible to theoretically estimate IC50 values for inhibition of ACE after gastrointestinal proteolysis of milk proteins. The major whey proteins α-lactalbumin and β-lactoglobulin gave IC50 values (Eq. 3) of 0.047 mg/ml and 0.034 mg/ml, respectively, and αs1-casein, αs2-casein, β-casein and κ-casein gave IC50 values of 0.025, 0.045, 0.060 and 0.047 mg/ml, respectively. Vermeirssen et al. [12] found, by experimentally simulating gastrointestinal proteolysis in vitro using pepsin, trypsin and α-chymotrypsin, ACE inhibitor activities of IC50 0.076 mg/ml for pea and 0.048 mg/ml for whey protein hydrolysates. An experimental IC50 value of 0.048 mg/ml for digested whey proteins (in batch simulated gastrointestinal proteolysis) is consistent with the theoretical estimation for α-lactalbumin and β-lactoglobulin. Theoretical estimation of IC50 values for some major pea proteins after digestion was also examined and gave mainly IC50 values in the range 0.03–0.06 mg/ml. Difference between experimental findings for in vitro gastrointestinal proteolysis and theoretical estimation for pea proteins might be due to the presence of protease inhibitors in peas [13]. This could affect experimental findings, but are not taken into account in the theoretical estimations. In the study by Vermeirssen et al. [9] on in silico digestion, β-lactoglobulin and β-casein were found to be very promising sources of ACE-inhibitory peptides. Only small differences between the milk proteins with no initial proteolysis were found in the present study. Dziuba [8] examined also bioinformatically release possibilities of bioactive peptides from plant proteins with different proteolytic enzymes. Gastrointestinal proteolytic enzymes such as chymotrypsin and trypsin gave a lower probability of release compared to those of several other enzymes examined, emphasising the relevance of gastrointestinal proteolysis on formation of bioactive peptides. A major difference between these studies is that a database for ACE-inhibitory peptides was used by Vermeirssen et al. [9] and Dziuba et al. [8], while the present work is based on a peptide QSAR model. An advantage with the present approach is that the ACE inhibition after in silico gastrointestinal proteolysis is expressed as IC50 (mg protein/ml) and that estimation of ACE inhibition of a specific peptide sequence is not limited to whether it is characterised and reported in a database.
Specific additional, controlled proteolysis in dairy products could be an approach to increase formation of bioavailable ACE-inhibitory peptides after gastrointestinal digestion compared to digestion of untreated milk proteins. Microbial fermentations have been shown to influence ACE-inhibitor activity of digested proteins in vitro [14] and in vivo [15]. However, proteolysis in food products could also enable gastrointestinal enzymes to degrade potential ACE-inhibitory peptides more efficiently into free amino acids, and thereby decrease the physiological ACE-inhibitory effect. Modelling how initial proteolytic activities in food influence ACE-inhibitory activity after gastrointestinal digestion could be a tool to find such proteolytic activity that could increase the physiological ACE-inhibitory effect. The approach chosen in this study was to assume cleavage at the C-end of a given type of amino acid in the protein sequence, perform in silico gastrointestinal proteolysis on the fragments and estimate ACE inhibition for the resulting hydrolysate (Eq. 3) using a peptide QSAR model. This was done for each of the 20 different amino acids in the sequences. Calculations for the major milk proteins and for milk (based on relative protein distribution in Table 1) are shown in Fig. 1. However, some of the resulting peptides after in silico proteolysis might be too large to cross the intestinal barrier, enter the blood stream and thus be bioavailable. ACE inhibition by only taking di- and tri-peptides into account was therefore also calculated separately (Fig. 1), since smaller peptides are more likely absorbed by the intestinal peptide transport system [16].
Calculated, in silico, ACE inhibition expressed as 1/IC50 for protein hydrolysate (Eq. 3) of the six major milk proteins and of milk (relative protein distribution cf. Table 1) when all peptides (■) or only di- and tri-peptides (□) (assumed bioavailable) were taken into account. Initial proteolysis was by cleavage at the C-end of a given amino acid type (indicated by one-letter code) in protein sequence followed by simulated gastrointestinal proteolysis. Results for simulated gastrointestinal proteolysis of untreated protein or milk, i.e. no initial proteolysis, are indicated by the denotation “Untr” in the diagrams
Among the different treatments, initial cleavage at the C-end of isoleucine contributed to a significantly increased theoretical ACE inhibition from all major milk proteins, except β-lactoglobulin, compared to gastrointestinal proteolysis of the same proteins without any initial proteolysis. Cleavage at the C-end of aspartic acid resulted in a clearly increased ACE inhibition from α-lactalbumin, and cleavage after valine resulted in clearly increased ACE inhibition from β-casein, κ-casein and α-lactalbumin. A similar pattern was found if only di- and tri-peptides (assumed bioavailable) were taken into account. Cleavage at the C-end of isoleucine gave clearly increased “bioavailable” ACE-inhibitor activity from all proteins, except β-casein, and cleavage after valine more than doubled increased “bioavailable” ACE inhibition from β-casein and α-lactalbumin. Investigation of structure-activity relationship for ACE-inhibitory peptides indicates that binding to ACE is influenced by the C-terminal tri-peptide region of the substrate and that peptides containing hydrophobic amino acid residues in the C-terminal region display the highest inhibitor potential [2, 4]. Cleavage of milk proteins at the C-end of hydrophobic amino acids like isoleucine promoted, according to these theoretical estimations, the ACE inhibition from several milk proteins by increased formation of highly inhibitory peptide structures after gastrointestinal digestion. Many identified ACE-inhibitory peptides contain proline in the C-terminal position, and initial cleavage after proline residues could therefore increase the production of such ACE-inhibitory peptides from milk proteins after gastrointestinal digestion. This was also found to be the case for all four caseins. β-casein has an especially high content of proline residues, and the effect of cleavage at the C-end of proline gives theoretically more than three-fold increase of ACE inhibition and a ten-fold increase in “bioavailable” ACE inhibition (only di and tri-peptides taken into account). If the relative distribution of these six proteins in milk (Table 1) is used to estimate effect of initial proteolysis in milk, cleavage at the C-end of isoleucine or proline is also the most promising way to produce high ACE-inhibitor activity from milk during gastrointestinal digestion (Fig. 1). Initial proteolytic treatment of milk with prolyl endopeptidase could then be a useful approach to increase formation of ACE-inhibitory peptides during digestion, since this enzyme prefers to cleave protein fragments at the C-end of prolyl residues. Dziuba [8] found also that prolyl endopeptidase gave a high release probability of bioactive peptides from plant proteins, even though the additional influence of gastrointestinal proteolysis was not studied. Cleavage at the C-end of other amino acid residues produces, according to the theoretical estimations, little or no increased ACE-inhibitor activity compared to that obtained by gastrointestinal digestion of untreated milk proteins.
Bioinformatic, in silico, research on the effect of proteolysis of food proteins on formation of bioactive peptides during gastrointestinal digestion could be an approach to predict biotechnological treatments to develop foods with specific health effects (functional foods). Based on the present findings, additional, controlled proteolysis in dairy foods should be rather specific in order to obtain increased ACE-inhibition from peptides obtained during gastrointestinal digestion compared to that obtained by gastrointestinal proteolysis of untreated milk proteins. It might therefore be speculated if the effect of initial proteolysis in foods is too limited to make functional foods with claimed health effects due to ACE inhibition. Enriching foods with preparations containing bioavailable ACE- inhibitory peptides may be needed to produce a functional food product with claimed blood-pressure reducing effect. Further experiments using in silico, in vitro and in vivo studies are needed for verification. However, the bioinformatic approach using in silico gastrointestinal proteolysis and peptide QSAR model provide tools to approach the complexity of proteolysis in foods, digestion and formation of bioactive peptides.
References
Gobbetti M, Stepaniak L, De Angelis M, Corsetti A, Di Cagno R (2002) Crit Rev Food Sci 42:223–239
FitzGerald RJ, Meisel A (2003) Milk protein hydrolysates and bioactive peptides. In: Fox PF, McSweeney PLH (eds) Advanced dairy chemistry, vol 1, proteins, 3rd edn. Kluwer Academic/Plenum Publishers, New York, pp 675–698
Dziuba J, Minkiewicz P, Nalecz D, Iwaniak A (1999) Nahrung 43:190–195
Pripp AH, Isaksson T, Stepaniak L, Sørhaug T (2004) Eur Food Res Technol 219:579–583, DOI 10.1007/s00217-004-1004-4
FitzGerald RJ, Murray BA, Walsh DJ (2004) J Nutr 134:980S–988S
Vermeirssen V, Van Camp J, Verstraete W (2004) Br J Nutr 92:357–366, DOI 10.1079/BJN20041189
Dziuba J, Iwaniak A, Minkiewicz P (2003) Polimery 48:50–53
Dziuba J, Niklewicz M, Iwaniak A, Darewicz M, Minkiewicz P (2004) Acta Aliment Hung 33:227–235
Vermeirssen V, van der Bent A, Van Camp J, van Amerongen A, Verstraete W (2004) Biochimie 86:231–239, DOI 10.1016/j.biochi.2004.01.003
Farrell HM, Jimenez-Flores R, Bleck GT, Brown EM, Butler JE, Creamer LK, Hicks CL, Hollar CM, Ng-Kwai-Hang KF, Swaisgood HE (2004) J Dairy Sci 87:1641–1674
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein Identification and Analysis Tools on the ExPASy Server. In Walker JM (ed) The Proteomics Protocols Handbook, Humana Press, Totowa, NJ, pp 571–607
Vermeirssen V, Van Camp J, Devos L, Verstraete W (2003) J Agric Food Chem 51:5680–5687, DOI 10.1021/jf034097v
Wang XF, Warkentin TD, Briggs CJ, Olmah BD, Campbell CG, Woods S (1998) J Agric Food Chem 46:2620–2623, DOI 10.1021/jf971007b
Vermeirssen V, Van Camp J, Decroos K, Van Wijmelbeke L, Verstraete W (2003) J Dairy Sci 86:429–438
Seppo L, Jauhiainen T, Poussa T, Korpela R (2003) Am J Clin Nutr 77:326–330
Yang CY, Dantzig AH, Pidgeon C (1999) Pharm Res 16:1331–1343, DOI 10.1023/A:1018982505021
Acknowledgement
Professor Terje Sørhaug, Department of Chemistry, Biotechnology and Food Science, Norwegian University of Life Sciences, Ås, Norway is acknowledged for critical reading of the manuscript
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pripp, A.H. Initial proteolysis of milk proteins and its effect on formation of ACE-inhibitory peptides during gastrointestinal proteolysis: a bioinformatic, in silico, approach. Eur Food Res Technol 221, 712–716 (2005). https://doi.org/10.1007/s00217-005-0083-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-005-0083-1