Abstract
High-resolution nuclear magnetic resonance (NMR) spectroscopy is introduced for the quality control and authenticity assessment of beer in official food control. Measurements were performed using a 400-MHz NMR spectrometer using flow injection technology for automatic sample changing. Only degassing and addition of buffer (pH 5.6 in D2O for locking and 0.1% TSP for referencing) is required to prepare the beer samples. Differences in the spectral profiles of beers varying in type and origin were studied by principal component analysis (PCA), considering the spectrum to be a characteristic fingerprint. For the first time, the high throughput of a Flow-Injection NMR system allowed a comprehensive database of beer spectra for PCA classification to be established efficiently. Beers made with barley malt could be distinguished from those made with wheat malt. Clustering of beers from the same brewing sites was observed, as well as significant discrimination of beers with deteriorated quality. Using the partial least squares (PLS) method to correlate NMR spectra with results from reference methods, models for calculating the original gravity, ethanol and lactic acid were established. The results obtained suggest that NMR is a useful tool in the quality control of beer samples, since quantitative determination of essential compounds as well as chemometric classification are simultaneously possible. Compared to conventional methods, 1H NMR spectroscopy is faster and requires simpler sample preparation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A range of different analytical methods are used to monitor the quality of beer in breweries or during official food control. The legal parameters “alcohol” and “original gravity” (OG) are usually determined by reference methods like distillation and pycnometry, or by analytical instruments, which combine oscillation-type densimetry and refractometry. In addition to organoleptical and microbiological examination for standard beer analysis, colour and bitterness are assessed by photometry, organic acids are determined using enzymatic analyses or liquid chromatography, and higher alcohols using gas chromatography. The use of these traditional techniques means that beer analysis is time-consuming and expensive.
Nuclear magnetic resonance (NMR), however, enables more rapid and non-invasive characterisation of foods, potentially providing information about a very wide range of different compounds [1]. In brewing science, NMR has mostly been applied to specific problems, like the identification and quantitation of malt and hop constituents, like polyphenols [2], humulones [3] or isohumulones [4]. A low-resolution NMR application was developed to determine the viscosities of the mash and the wort for process and quality control of the brewing and filtering processes [5]. The deuterium distribution in ethanol, determined using site-specific isotope fractionation (SNIF) NMR, was used to characterise the origin of beer [6]. The hyphenation of high-resolution NMR spectroscopy with liquid chromatography and mass spectrometry allowed the identification of oligosaccharides [7, 8] and aromatic compounds [9, 10]. Beer flavour stability and staling mechanisms were evaluated by characterising the reaction of bisulphite with unsaturated aldehydes using 13C and 1H NMR spectroscopy [11].
Duarte et al [12] have recently shown that NMR spectroscopy enables a direct and rapid overview of the chemical composition of beer to be obtained without the need for pre-treatment aside from degassing. Assigning 1- and 2-dimensional NMR spectra from beer samples facilitated the identification of about 30 compounds ranging from organic acids, amino acids, and alcohols to higher molecular weight compounds such as lipids and large aromatic compounds, possibly polyphenolics. However, even using techniques like diffusion-ordered spectroscopy (DOSY) [13], a full assignment of the beer spectra is not possible, mainly due to strong signal overlap. Therefore, an alternative approach to interpreting the NMR spectra using multivariate statistics is required.
In brewing science, multivariate statistical techniques are being increasingly used to evaluate flavour characteristics [14, 15], analytical results [16, 17, 18] or to correlate both [19, 20, 21, 22, 23]. One of the most powerful and widely-used multivariate procedures employed is principal components analysis (PCA) [24]. It is a method for transforming the original measurement variables into new variables called principal components (PC). By plotting the data in a coordinate system defined by the two or three largest principal components, it is possible to identify key relationships in the data, which means to find similarities and differences. This approach has already proven to be of value for the detection and interpretation of NMR spectra, as previously shown for apple juice [25] and grape extracts [26]. The results from 50 beer samples examined by high resolution NMR and PCA suggested that beer distinction is possible, at least in terms of beer types [27].
In this work, based on previous research by Duarte et al [12, 27], a fast routine NMR method was established that used flow injection technology to change samples. For the first time, NMR and PCA are applied to a group of 80 beer samples, and partial least squares (PLS) regression is used to determine the original gravity of the beers from NMR spectra. In addition, substances relevant to the quality like ethanol and lactic acid were simultaneously quantified. The use of NMR spectroscopy in combination with multivariate statistic techniques is assessed for its use in the quality control of beer.
Materials and methods
Beer samples
Eighty beer samples submitted by local authorities to the CVUA Karlsruhe were analysed using NMR. The sample collective comprises a wide range of different beer styles, including 56 bottom-fermented (such as Pilsener, Lager and Export type) and 24 top-fermented (like wheat, Alt, ale type) beers. 40 beer samples were light in colour, and 40 were dark or black. Sixty-six samples were standard “Voll beers” (original gravity 11–16%), and 14 samples were “Stark beer” or “Bock beer” type (original gravity >16%). In all cases, there was a full organoleptical and chemical examination, which included the determination of relative density, ethanol and original gravity using refractometry and oscillation-type densimetry, the determination of lactic acid using enzymatic analysis (R-Biopharm, Darmstadt, Germany), as well as the microbiological determination of beer-spoilage bacteria.
Sample preparation
For sample preparation, the beer samples were degassed by filtration through fluted filter paper and subsequent ultrasonication for 10 min. Prior to NMR analysis, the samples were filtered again through a filter of 0.2-μm pore size to remove suspended particles (for instance yeast). After that, 1.8 ml of the beer sample, 0.2 ml of buffer (pH 5.6 in D2O containing 0.1% of TSP (sodium salt of 3-(trimethylsilyl)-propionate acid-d4), and 0.013% of sodium azide) were placed into an autosampler vial.
NMR measurements
Measurements were performed at a BEST system with a Bruker Avance 400 Spectrometer, using flow injection technology for sample changing (Bruker Biospin, Rheinstetten, Germany). A Gilson Liquid Handler is used to prepare a so-called “sample train” for transfer to the probe. The sample train consists of push solvent, leading and trailing gaps, and the sample itself. The leading and trailing gaps were composed of nitrogen gas gaps and different solvent gaps. The exact set-up is described in Fig. 1 and Table 1. Transfer to the probe is achieved with a dispensing speed of 2 ml/min. After the measurements the sample train is blown out with a pulsed gas stream.
Sample train for fully-automated flow injection of beer samples into the NMR spectrometer. For exact volumes see Table 1
A 4-mm flow injection probe (inverse 1H/13C with z-gradient) was used with an active volume of 120 μl. The following two different pulse sequences were applied to acquire the proton spectra:
-
1.
Presaturation on the water signal with a 1-D presaturation sequence and an additional selective suppression of the C13-satellites of the CH3-group of ethanol at 16.8 ppm. The C13-satellites overlap on the low field site with the doublet of lactate. This experiment allows, therefore, the measurement of lactate.
-
2.
Triple presaturation experiment to suppress the dominating signals of the H2O, CH3- and CH2-groups of ethanol with a modulated shape pulse (100 ms) in the NOESY 1-D presaturation sequence. NOESY mixing time was 100 ms.
General parameters for both experiments: Time domain: 16 k, 32 scans, 4 dummy scans, relaxation delay 10 s, acquisition time 1.02 s, sweep width 8012 Hz (20 ppm). The 2H-signal from D2O was used for the automatic log. Spectra were referenced using the TSP signal at 0 ppm. The data were acquired automatically under control of the BEST-software, requiring about 12 min per sample. Assignment of the spin systems and separation of the components of the complex patterns in the 1-D overlapped proton spectra were made according to Duarte et al [12].
Chemometrics
The free induction decay (FID) files were Fourier transformed (with 0.3 Hz line broadening), and the spectra were phased, baseline-corrected, and calibrated by the TSP signal at 0.0 ppm. The resulting spectra were analysed by PCA and PLS using the software package AMIX 3.2.3 (Bruker Biospin, Rheinstetten, Germany). An input for the PCA, a bucket table, was generated from the complete set of spectra. Ranges, exclusions and bucket widths for the different PCA and PLS analyses are given in Table 2. Scaling to total intensity was applied. The technique of cross-validation was applied to determine the number of principal components (PCs) needed. During cross-validation, one sample at a time (of N samples) is left out, and the prediction ability is tested on the sample omitted. This is repeated N times, resulting in N models, and this will give an estimate of the average prediction ability for the N models. This is then used to select the number of PCs needed. By plotting the data in a coordinate system defined by the two or three largest principal components, it is possible to identify key relationships in the data; in other words, to find similarities and differences between the spectra from the beers.
Using PLS regression, the NMR spectra were correlated with results from reference methods (refractometry and oscillation type densimetry, enzymatic analysis) to establish a model for calculating the original gravity and ethanol as well as the lactic acid contents of the beer samples. Differently from PCA, PLS studies and models the relationship between two data tables, table X and Y. In the context of our study, the X table corresponds to the bucket table calculated from the NMR spectra (the one which is used in PCA as well). Both tables have the same number of lines, one for each sample. While each line of the bucket table contains the bucket version of the NMR spectrum of a sample, a line in the Y table contains extra information on the sample, such as concentrations of certain chemical constituents of the sample. This means that, while the number of columns in the X-table is determined by the number of buckets, the number of columns in the Y table depends on the number of extra variables.
The purpose of PLS regression is to find a small number of relevant factors, which are predictive for Y on the basis of X. For example, if one would like to circumvent the need for a time-consuming wet chemistry concentration determination of a specific constituent of some samples, one might derive a model that catches the relation between NMR spectra and those concentrations. This model would then be used to predict the respective concentration values directly from the spectra without doing wet chemistry. Again, model validation is done via cross-validation, in order to study the predictive power of the PLS-model, for example via comparison of predicted and measured Y-table entries.
Results and discussion
Optimization of sample preparation and flow-injection NMR
In the only previous study by Duarte et al [27], which systematically examined a large number of beer samples by NMR, no sample preparation was performed except degassing (which is an integral part of each beer analysis anyway), and the addition of D2O and TSP. Although the spectra were referenced to the same chemical shift value (δ 0.000 ppm) using the TSP signal, sugar signals showed small shifts (Δδ 0.002–0.003 ppm) and organic acids showed shifts of up to 0.06 ppm in all spectra. The shifts arose from differences in sample pH, to which, in particular, the signals from organic acids in the aliphatic region are very sensitive. These shifts lowered the ability of the multivariate data analysis to classify and its stability. Therefore in this work, the pH was buffered in all samples, so that the differences correspond to true differences in the compositions of the samples and not to pH effects. Before buffer addition, the pH-value varied over a wide range between 4.0 and 4.8 (mean 4.5±0.3). After buffer addition, the pH values lay between 5.2 and 5.5 (mean 5.4±0.07), so that no significant chemical shifts were observable. Additionally, sodium azide was added to preserve the samples. This pre-treatment proved to be advantageous, so that even longer storage of the samples at room temperature led to no significant change in the spectrum.
It was ascertained that two NMR experiments are sufficient for each sample. The C13-satellites of the CH3-group of ethanol overlap on the low field site with the doublet of lactate. Therefore, in the first experiment, the selective C13 suppression at 16.9 ppm in combination with a standard presaturation of the water signal allows the lactate in the proton spectrum to be measured as well as ethanol and all other signals simultaneously. To make the multivariate analysis more sensitive, a second experiment using triple presaturation was employed. This allows us to suppress the dominant signals of H2O and ethanol at 4.7 ppm (H2O), 3.66 ppm and 1.18 ppm (CH3- and CH2-group of ethanol). The analysis time for both experiments is only 12 min per sample. A sample throughput in the context of a screening procedure is thereby ensured.
NMR spectra
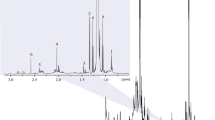
Figure 2 shows the 1-D 1H spectrum of a Pilsener beer recorded at 400 MHz with some assignments indicated. The three strongest peaks in the spectrum reflect the two major components, water and ethanol. A partial assignment of the signals observed, carried out by both 2-D NMR and LC NMR/MS, has already been reported in recent publications [8, 10, 12], enabling us to characterise the beer composition in detail. In the aliphatic region of the spectra (0–3 ppm), peaks from higher alcohols, organic acids, amino acids and fatty acids are observable. The mid-field region (3–6 ppm) shows the contribution of beer carbohydrates, which normally comprise fermentable sugars and dextrins [27]. The signals in the aromatic region (6–10 ppm) show the presence of aromatic amino acids, nucleosides, aromatic alcohols and polyphenolic compounds [10, 12]. However, the spectra also show extensive signal overlap, especially in the aliphatic and aromatic regions (Fig. 2). Individual assignment requires extensive use of 2-D NMR experiments, but even then it was found that many difficulties still occur due to the large spectral complexity and overlap, even in the 2-D displays [12]. Therefore, in this work another approach to interpretating these complex spectra is chosen, which involves the correlation of the overall spectral profile, without requiring individual and complete assignments, using chemometric methods such as PCA.
Principal components analysis
Although the beers differ significantly in their organoleptic properties (for example Pilsener and wheat beers), there are no single substances to which these differences can be attributed. If 1H NMR spectra of different beer types are compared, only marginal differences are obvious at first sight. Both the low intensities and the strong overlaps between the signals also hinder the full interpretation. Properties such as beer type, production process and origin are hidden inside the high spectral complexity. In the following, these properties are uncovered from the NMR spectra using multivariate statistics.
Discrimination of conspicuous beers
PCA was first performed on the complete NMR spectra (excluding only water and ethanol signals). The scatter plot of the first two PCs, which together express 95.5% of the total variability, is shown in Fig. 3. Two groups of samples are noted: a large group comprising 76 samples and a small group of four conspicuous samples. Using the PCA loadings profile, the bucket 4.20–4.24 ppm was identified as cause of the variance in the four samples. This chemical shift can be assigned to lactic acid (quartet at 4.19 of α-CH group). A high content of lactic acid in the four beer samples caused the deviation in PC1.
Utilizing the results from the conventional beer analysis, it was found that the NMR classification is correct. The deviating samples were organoleptically conspicuous, with sour off-flavours, and beer-spoilage lactic acid bacteria were determined in microbiological analysis. Interestingly, these four samples could be assigned to small home- or micro-breweries. During manufacturing controls, deficiencies in work hygiene could be found, which led to recontamination of the beer after wort boiling.
Differentiation between malt types
PCA was also performed on the bucket tables constructed from specific NMR regions in order to account for the contributions of minor components to the variability within the beers analysed. When only the aromatic regions of the spectra are considered for PCA, the PC1 versus PC2 scatter plot of scores suggests that the samples can be separated into two groups (Fig. 4). Wheat beers cluster in a region of negative PC1, whereas the beers made with barley malt (Pilsener, Export and Alt beers) give scores of positive PC1 or PC1 around zero. The separation can be attributed to the malt type. Therefore, this PCA model could be used to check that the beers comply with German beer purity laws [28]. Many beer companies advertise the purity of their products in other countries too, although they misrepresent their products by adding ingredients not traditionally used when brewing beer. Pure beer is expected to be made only from water, malted barley, hops and yeast unless clearly labelled otherwise, as in wheat beer. PCA classification of the aromatic regions of NMR spectra provides, for the first time, a simple method for controlling beer purity.
Allocation of brewing sites
The score scatter plot of PC1 versus PC2 obtained when only aliphatic regions of the spectra are considered for PCA shows considerable dispersion, and so do plots of subsequent PCs. If the beers that originate from the same brewery are tagged, however, their corresponding points occur close together on the plot (see Fig. 5). Each particular brewing process seems to have a greater influence on the aliphatic regions of the spectra than the aromatic regions. Possible causes of this clustering according to breweries may be the influence of the particular brewing water used, the origin of the malt, or the strain of the yeast. The possibility of distinguishing the source brewery of a particular beer may be useful if the brand of beer was uncertain. For example, beers are often poured by bar staff behind a bar and then served to the customer; in order to check that customers are receiving the brands that they actually order and not inferior brands, we could use our scatter plot test. Therefore 1H-NMR can now be used as a complimentary method in addition to organoleptic evidence or complex SNIF-NMR techniques [6] to determine and monitor the origin of beer.
Partial least squares regression
Besides the qualitative determination of beer purity, type and origin, in order to perform quality control on beer we also need to quantify the parameters original gravity and ethanol at the very least. In German beer legislation, there are beer categories that depend on the original gravity. Standard beers have an original gravity of at least 11%. Beer with an original gravity of below 11% must be labelled. Beers named Starkbier or Bockbier are required to have an original gravity of 16% or more. In addition, beer is taxed upon its original gravity, so the determination of this parameter is very important to official food control. Original gravity is the concentration of solids in the unfermented wort from which the beer is made. It is traditionally calculated from real extract and the alcohol content of the beer. In the NMR spectrum, single substances cannot be used to monitor original gravity, because the malt extract contains a complex range of ingredients. Therefore, in order to determine original gravity, the bucket table of the entire NMR range (10.0–0.3 ppm), including ethanol (experiment 1), was correlated to the wet chemical analysis using PLS. The PLS model, verified through cross-validation, showed an excellent correlation (R=0.998) between the standard laboratory values and the NMR prediction. The slopes and intercepts of the regression lines show that there is no constant or proportional difference between the two procedures (Table 3).
The second parameter that must be quantified for official food monitoring is the ethanol concentration. A European Union directive specifies the maximum tolerances for the alcoholic strength written on the beer’s label. For beers with an alcoholic strength that does not exceed 5.5% vol, the tolerance is 0.5% vol, whereas for beers above 5.5% vol the tolerance is 1% vol [29]. The PLS model for ethanol also showed a good correlation (R=0.985). None of the beers gave a false positive out of the tolerance. The NMR prediction method is therefore able to efficiently check that the legal tolerances of ethanol are being complied with.
Lactic acid, which is produced by beer spoilage bacteria, can be used as an indicator of production hygiene. Beer normally contains up to 200 mg/l of lactic acid [30], and higher concentrations suggest the presence of lactic acid bacteria as Lactobacillus or Pediococcus. In the PCA, lactic acid was the discriminating factor for beers with deteriorated quality. By using PLS regression it was possible, furthermore, to predict the lactic acid concentration from the NMR spectra. The correlation with the enzymatic analysis (R=0.954) is adequate for a screening procedure. The NMR procedure allows us to select conspicuous samples which may be selectively analysed for beer spoilage by microbiological analysis.
Conclusion
Increasing numbers of requirements and cost-pressures are pushing governmental and commercial food-testing laboratories to replace traditional reference methods by faster and more economical methods. For this purpose, screening methods, which ensure a very high sample throughput, seem to be the most advantageous.
Modern NMR methods, in combination with chemometric analysis, provide the ability to uncover adulteration and thus consumer deception in all kinds of foods. In contrast to conventional methods, which most often focus on the analysis of few specific components as markers of food adulteration, NMR spectroscopy enables one to record, within a certain range, more or less all of the constituents of the foodstuff in a single experiment.
It was shown that high resolution spectra of beer simultaneously contain both quantitative information about the levels of alcohol and original gravity in the beer, as well as compositional properties like authenticity, quality and origin. Obviously, the combination of spectroscopy and PCA achieves a much higher level of discrimination than simple spectroscopic quantification of the individual peaks.
The rapidity with which information can be obtained about a large number of compounds within the beer, and the minimal sample preparation required indicates that high-field proton NMR is unique in its ability to comprehensively survey a large number of samples. Therefore, there are considerable advantages to using this method in comparison with conventional methods of analysis; in particular, it allows us to focus complex and labour-intensive reference analytics on beers already screened and found to be suspicious by the NMR method.
Due to its ability to deliver high sample throughput, its use of standard NMR equipment in combination with lab automation technology, and its suitability as a cost-efficient screening procedure, NMR-based monitoring techniques will attain increasing importance as methods routinely used in food control in the future. By building a comprehensive database of beer samples, our method will provide the ability to distinguish between types of malt or to quantify the barley proportion in the beer.
References
Belton PS, Delgadillo I, Holmes E, Nicholls A, Nicholson JK, Spraul M (1996) J Agr Food Chem 44:1483–1487
Friedrich W, Galensa R (2002) Eur Food Res Technol 214:388–393
Pusecker K, Holtzel A, Albert K, Bayer E, Wildenauer M, Rust U (1997) Monatsschr Brauwiss 50:70–74
Nord LI, Sorensen SB, Duus JO (2003) Magn Reson Chem 41:660–670
Götz J, Schneider J, Först P, Weisser H (2003) J Am Soc Brew Chem 61:37–47
Martin GJ, Benbernou M, Lantier F (1985) J Inst Brew 91:242–249
Vinogradov E, Bock K (1998) Carbohydr Res 309:57–64
Duarte IF, Godejohann M, Braumann U, Spraul M, Gil AM (2003) J Agr Food Chem 51:4847–4852
Duarte IF, Spraul M, Godejohann M, Braumann U, Gil AM (2003) Application of NMR and hyphenated NMR spectroscopy for the study of beer components. In: Belton PS, Gil AM, Webb GA, Rutledge D (eds) Magnetic resonance in food science: latest developments. Royal Society of Chemistry, Cambridge, UK, pp 151–157
Gil AM, Le Duarte IF, Godejohann M, Braumann U, Maraschin M, Spraul M (2003) Anal Chim Acta 488:35–51
Dufour JP, Leus M, Baxter AJ, Hayman AR, Rezac J, Cepicka J, Viden I (1999) J Am Soc Brew Chem 57:138–144
Duarte I, Barros A, Belton PS, Righelato R, Spraul M, Humpfer E, Gil AM (2002) J Agr Food Chem 50:2475–2481
Gil AM, Duarte I, Cabrita E, Goodfellow BJ, Spraul M, Kerssebaum R (2004) Anal Chim Acta 506:215–223
Mawatari M, Nagashima Y, Aoki T, Hirota T, Yamada M (1991) J Am Soc Brew Chem 49:59–64
Daems V, Delvaux F (1997) Food Qual Prefer 8:373–380
Siebert KJ, Stenroos LE (1989) J Am Soc Brew Chem 47:93–101
Lonkhuijsen HJv, Douma AC, Angelino SAGF (1998) J Am Soc Brew Chem 56:7–11
Siebert KJ (1999) Food Qual Prefer 10:129–137
Harayama K, Hayase F, Kato H (1991) Agric Biol Chem 55:393–398
Harayama K, Hayase F, Kato H (1994) Biosci Biotechnol Biochem 58:1595–1598
King BM, Duineveld CAA (1999) Food Qual Prefer 10:315–324
Foster RT, Samp EJ, Patino H (2001) J Am Soc Brew Chem 59:201–210
Vind-Larsen O, Williams I, Lillelund AC, Aastrup S, Byrne DV (2003) Techn Q Master Brew Assoc Am 40:193–198
Peppard T (1988) The use of principal components analysis in monitoring the quality of beer. In: Linskens HF, Jackson JF (eds) Beer analysis. Springer, Berlin Heidelberg New York, pp 264–279
Belton PS, Colquhoun IJ, Kemsley EK, Delgadillo I, Roma P, Dennis MJ, Sharman M, Holmes E, Nicholson JK, Spraul M (1998) Food Chem 61:207–213
Forveille L, Vercauteren J, Rutledge DN (1996) Food Chem 57:441–450
Duarte IF, Barros A, Almeida C, Spraul M, Gil AM (2004) J Agr Food Chem 52:1031–1038
Donhauser S (1988) German beer purity law and its influences on the properties and analysis of beer. In: Linskens HF, Jackson JF (eds) Beer analysis. Springer, Berlin Heidelberg, New York, pp 280–296
EC (1987) Commission Directive (87/250/EEC) on the indication of alcoholic strength by volume in the labelling of alcoholic beverages for sale to the ultimate consumer. Off J Eur Comm L113:57–58
Uhlig R, Gerstenberg H (1993) Brauwelt 133:280–286
Acknowledgments
The authors thank S. Gonzalez, H. Heger and G. Hofman for excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lachenmeier, D.W., Frank, W., Humpfer, E. et al. Quality control of beer using high-resolution nuclear magnetic resonance spectroscopy and multivariate analysis. Eur Food Res Technol 220, 215–221 (2005). https://doi.org/10.1007/s00217-004-1070-7
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-004-1070-7