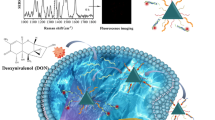
Abstract
The fluorescent nanoprobes for reduced thiol compounds (represented by glutathione, GSH) are constructed based on the aggregation-induced emission (AIE) luminescence mechanism and endosome escape technology. First, a DNA sequence was designed with the decoration of biotin at the 5′-end, disulfide bound in the internal portion, and amino at the 3′-end. The aptamer of the MCF-7 cell was also one of the most important structures in our DNA sequence for the selectivity of MCF-7 cells. We modified streptavidin-modified magnetic beads (MB) with biotin-modified influenza virus hemagglutinin peptide (HA) and biotin-DNA-amino to form MB/DNA/HA. Carboxyl-modified tetraphenylethylene (TPE), an iconic AIE fluorogen, was bonded with amino-modified DNA by covalent interactions (TPE/DNA). Then, the TPE molecule was attached on the outer layer of MB via biotin-modified TPE/DNA to form MB/DNA/HA/TPE. Compared with traditional AIE/biomolecule conjugates, the nanoprobe had an enhanced endosome escape function, due to the assembly of HA. This construction made the intracellular fluorescence response more accurate. In the presence of reduced thiol compounds (take GSH, for example), the disulfide bond on the DNA was reduced by thiol-disulfide exchange reactions and the TPE molecule was released into the solution. The shedding TPE molecule was more hydrophobic than TPE/DNA and the conversion of TPE/DNA to shedding TPE could lead to the aggregation of the TPE fluorogen. Thus, its fluorescence was enhanced. Under the optimized condition, the fluorescence intensity increased with the increase in concentration of GSH‚ ranging from 1.0 × 10−9 M to 1.0 × 10−5 M‚ and the detection limit was 1.0 × 10−9 M. The relative standard deviation (RSD) was calculated to be 3.6%. The recovery in cell homogenate was from 94.5 to 102.7%. The nanoprobe provided a way for the detection of reduced thiol compounds in MCF-7 cells. We envision that, in the near future, our strategy of DNA-instructed AIE could be widely applied for biosensing and bioimaging in vitro and even in vivo with dramatically enhanced sensitivity.

Graphical Abstract
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glutathione (GSH) is a tripeptide compound composed of glutamic acid (Glu), cysteine (Cys), and glycine (Gly). It is very common in cells and mainly plays an antioxidant role [1]. The redox state in cells plays a role in regulating apoptosis, which directly affects the occurrence of human diseases, and the level of glutathione was closely related to the stable state of the redox state [2, 3]. When cells are diseased, intracellular GSH is often overexpressed. This is because cytopathic diseases can cause excessive cell proliferation, and a large amount of GSH assistance is needed to complete energy conversion during cell proliferation. The content of GSH was less in normal cells, which can avoid apoptosis, but GSH has a higher content in cancer cells [4, 5]. Therefore, accurate and effective monitoring of intracellular GSH expression levels was of great significance for the diagnosis and treatment of diseases.
Fluorescent biosensors are important tools for the analysis and detection of biomolecules in living cells, and there are currently many fluorescent probes used to detect GSH [6,7,8]. At this stage, the solubility of most fluorophores in aqueous solutions is very low. Although many organic dyes with fluorescent “on/off” effects are designed to be ionic in order to increase the solubility in aqueous solutions, they are still prone to aggregation-caused quenching (ACQ) [9, 10]. At the beginning of this century, Tang et al. discovered and proposed the concept of aggregation-induced emission (AIE) [11]. With the further study of the mechanism of AIE, researchers find that the mechanism of action of AIE and ACQ is opposite. When the AIE fluorescent molecular group is well dispersed, the dynamic rotation and vibration of intramolecular motion will reduce the excitation energy, and then reduce the fluorescence intensity. When AIE clusters are aggregated, intramolecular movement will be restricted, preventing the dissipation of excitation energy, which will increase the fluorescence intensity [12]. In recent years, more and more AIE-effect fluorophores have been discovered, and because of their advanced characteristics (such as superabsorption, high luminosity, and high anti-whitening), this type of fluorescent group is widely used in the fields of biosensing and bioimaging [13,14,15]. When used in intracellular detection, the accuracy of detection may be reduced due to aggregation quenching. Studies have found that fluorescent substances with AIE characteristics can effectively solve the traditional fluorescence quenching problem. Some prior studies have demonstrated the feasibility of using AIE/peptide or AIE/nucleic acid or AIE/protein conjugates to exhibit significant potential for biomedical applications such as intracellular imaging and drug delivery [16, 17]. Ouyang et al. synthesized terminal AIEgen-labeled nucleotides that could serve as substrates for some polymerases and applied them into the sequencing of small DNA fragments [18]. Tian et al. reported that human papillomavirus capsid protein was assembled with the complex of DNA and AIE fluorogen. The co-assembled nanoparticles showed enhanced fluorescence, improved biocompatibility, and commendable protection, ensuring virtual brighter imaging in HeLa cells [19]. A supramolecular AIE dot as the drug nanocarrier via the host-guest interaction between PEG-peptide and α-cyclodextrins realized long blood circulation time [20]. Such supramolecular AIE dots could simultaneously achieved a stealth surface during blood circulation and enhanced cellular uptake in the tumor via the removal of the shielding sequence and the exposure of the cell-penetrating sequence by overexpressed matrix metalloproteinase in the tumor site. Such active bioconjugates can be readily degraded, which severely reduces the bioavailability of the imaging and drug delivery. Endosomal escape has therefore been regarded as a critical “bottleneck” for on-demand intracellular applications.
Here, we proposed a sensitive reduced thiol compound detection method. Based on streptavidin-modified magnetic beads (MB), we modified TPE to the surface of streptavidin-coated MB by DNA containing disulfide bonds. At the same time, the influenza virus hemagglutinin peptide (HA) was modified on the surface of MB to improve the endosome/lysosome escape effect of fluorescent probes. Under the action of reduced thiol compounds, the disulfide bond was broken, and TPE could be released into the cell to generate aggregate fluorescence. The detection method has high specificity and high sensitivity, and was suitable for directly detecting reduced thiol compounds in cells.
Experimental section
Material and instrument
Streptavidin-modified magnetic beads (MB) were purchased from Beijing Tianenze Biotechnology Co., Ltd. (Beijing, China). The DNA sequences used in the experiments (see Table S1 in Electronic Supplementary Material, ESM) were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). DMSO, EDC, and NHS were purchased from Aladdin. Influenza virus hemagglutinin peptide (HA) was provided by Kingsray Biotechnology Co., Ltd. 1-(4-Carboxybenzene)-1,2,2-tristyrene was purchased from Shanghai Dibo Chemicals Technology Co., Ltd. Fetal bovine serum was provided by Thermo Fisher Scientific. L02 cells were purchased from Silver Amethyst Biotech Co., Ltd. and MCF-7 human breast cancer cells were purchased from Shanghai Zeye Biotechnology Co., Ltd.
The UV absorption spectrum was measured using NanoDrop 2000 UV-Vis spectrometer (NanoDrop, USA); transmission electron microscopy images were obtained using a JEM-2100 transmission electron microscope (JEOL Ltd., Japan); Nano-ZS90 zeta potentiometer (Malvern Instrument Ltd., Britain) was used to measure the particle size of the nanomaterials; F-4600 fluorescence spectrophotometer (Hitachi, Japan) was used to measure fluorescence; Nikon laser confocal microscope (Nikon, Japan) was used to obtain confocal fluorescence images of the cells.
Synthesis of MB/DNA/HA/TPE
Two hundred microliters of streptavidin-modified magnetic bead solution (MB) were rinsed twice with PBS, and then mixed with 70 μL of biotinylated DNA (1 × 10−4 M) and 30 μL biotinylated HA (1 × 10−4 M) [21], incubating for 2 h at 25 °C. The free DNA and HA were then removed by magnetic separation to obtain nanoparticles MB/DNA/HA. One hundred microliters of TPE-COOH (10 mM) with DMSO as the solvent was mixed with 10 μL of EDC (0.5 mM) aqueous solution and 10 μL of NHS (0.5 mM) aqueous solution, and shaken for 30 min at 25 °C. The activated TPE-COOH was then transferred to a centrifuge tube containing nanoparticle MB/DNA/HA and mixed uniformly, then reacted for 12 h at 25 °C. The nanoprobe MB/DNA/HA/TPE was obtained by magnetic separation.
Fluorescence detection
We selected 100 μL of GSH solutions of different concentrations (10−9 M, 10−8 M, 10−7 M, 10−6 M, 10−5 M) as research objects. Two hundred microliters of MB/DNA/HA/TPE were used to incubate with the GSH solution. After incubation, fluorescence spectrophotometer was performed for fluorescence detections.
Cell culture experiment
MCF-7 breast cancer cells and L02 normal cells involved in all experiments were incubated in a 37 °C constant temperature and humidity incubator with a CO2 concentration of 5% and an air humidity of 95%. The cell incubation medium was added with 10% fetal bovine serum and 1% double antibody (penicillin-streptomycin) in DMEM.
Cell colocalization imaging experiment
We used L02 cells and MCF-7 cells to detect the localization analysis of the nanoprobe in the cells. First, we pre-seeded MCF-7 cells and L02 cells in a 35-mm diameter glass bottom cell culture dish. The cell seeding density was about 10,000 cells. The cell culture dishes were placed in the cell culture incubator at constant temperature and humidity for 12 h. After adhering to the wall, the cells were incubated with DAPI, nanoprobes, and LysoTracker Red in sequence under the same condition. After incubation, the cells were washed 3 times with PBS, and then detected using laser confocal imaging.
Results and discussion
MB/DNA/HA/TPE probe design
Here, streptavidin-modified nanomagnetic beads (MB) were modified with the DNA (5′-biotin) and influenza virus hemagglutinin peptide (HA) based on the specific binding between streptavidin and biotin (Fig. 1). The carboxyl-modified TPE (TPE-COOH) was then modified to the other end of the DNA (-NH2-3′), based on the coupling reaction, eventually forming MB/DNA/HA/TPE probe. In order to improve the specificity of the probe, aptamer AS1411 sequence was chosen as a part of DNA sequence. -S-S-NH2 was modified at the 3′-end of DNA to form a site for a simple thiol-disulfide exchange. The presence of HA achieved the endosomal escape effect of the probe in the cell. When the probe entered the target cell (MCF-7), the intracellular GSH could act on the disulfide bonds, causing the disulfide bonds to break and the TPE molecule falling off. In the biological environment, the solubility of TPE is very low. So the detached TPE molecule could aggregate and emit strong fluorescence. This design provided a new way for accurately detecting the expression level of GSH in cells, and a reasonable theory for disease surveillance.
We optimized experiment conditions. We tested the effect of pH. As shown in Fig. S1 in ESM, the results showed that the best pH was 7.5. As the temperature increasing, the fluorescence intensity enhanced. When the temperature exceeded 25 °C, the fluorescence intensity was changed little. So we chose 25 °C as the experimental temperature. Besides, in order to study the response rate of nanoprobe to GSH, we incubated nanoprobe with 10−7 M GSH at 25 °C and performed continuous-time fluorescence detection. As shown in Fig. S2A in ESM, the fluorescence signal reached the platform after 2 h. Then, we detected the fluorescence signals of TPE, MB/DNA/HA/TPE, and MB/DNA/HA/TPE + GSH. The positions of fluorescence peaks of TPE are in good agreement with MB/DNA/HA/TPE + GSH, indicating that there was no FRET effect (see Fig. S2B in ESM).
Then, we did the DLS and TEM characterizations of the probe before and after GSH stimulation. As shown in Fig. S3 in ESM, the MB was 97.3 ± 2 nm, and MB/DNA/HA/TPE was 101.2 ± 2 nm. And the particle size of the probe did not change significantly before and after GSH stimulation.
MB/DNA/HA/TPE probe for GSH detection
We took GSH as an example for reduced thiol compounds. In order to study the detection capability of the MB/DNA/HA/TPE probe for GSH, GSH solutions with different concentrations were used as the detection sample. After incubation, the solution was subjected to fluorescence detection. According to the experimental results, as the GSH concentration increases, the fluorescence signal was enhanced (Fig. 2). The proposed strategy allowed the detection of GSH with a linear range from 1 × 10−9 M to 1 × 10−5 M and the detection limit was 1.0 × 10−9 M. Compared with those sensitive techniques listed in Table S4 in ESM, our proposed method has been the most sensitive tool for the determination of GSH, to the best of our knowledge. The linear equation of fluorescence is Y = 6.35X + 65.77 (Y represents the relative fluorescence intensity, X represents the logarithm of concentration of GSH, M, R = 0.998). The RSD was 3.6%, which makes it clear that this developed method has a good reproducibility. The calculated quantum yield was 0.49.
In order to study the reliability of the probe detection, we chose the commonly used GSH kit (Dojindo China Co., Ltd., Shanghai, China) on the market to compare with the fluorescent detection effect of this work. We selected GSH-diluted samples of different concentrations as the detection target, and used GSH kit and the fluorescent probe for quantitative detection. Figure S4A and Table S2A in ESM show the comparison of the detection results of the proposed method and the GSH kit method. We found that the two detection methods have a relatively good linear relationship with R = 0.998. Figure S4B and Table S2B in ESM show the comparison of the detection results of the proposed method and disulfide-bound molecular beacon method [22]. We found that the two detection methods have a relatively good linear relationship with R = 0.999. Here, the kit used was Solarbio Reduced Glutathione (GSH) Assay Kit, and the student work used disulfide-bound molecular beacon.
In order to detect the detection sensitivity and detection ability of the probe in biological samples, we selected MCF-7 cell disruption solutions (100 cells to 10,000 cells) of different amount as target samples for quantitative analysis and detection. As shown in Fig. S5 in ESM, as the number of cells increases, the fluorescent signal also gradually increases. The change of fluorescence intensity based on the variation in cell number from 100 cells to 10,000 cells satisfied the regression equation, Y = 7.28X − 9.57 (Y represents the fluorescence intensity; X represents the number of cells, R = 0.997). At the same time, we tested the recovery in cell homogenate. The test results showed that the probe can effectively detect biological samples (see Table S3 in ESM). The above results further indicate that the probe has great development potential in biological detection.
Selectivity to reduced thiol compounds
In order to detect the selectivity of MB/DNA/HA/TPE, we evaluated the response of reduced thiol to the probe. As shown in Fig. 3, MB/DNA/HA/TPE probe was treated to GSH, Cys, Hcy, HSA, N-acetylcysteine, glucose, His, methionine, and Ca2+ separately. We found that the fluorescence signals of GSH, Cys, Hcy, HAS, and N-acetylcysteine were significantly higher than other substances because of the disulfide bonds cleaved by reduced thiol, resulting in TPE molecule released. So this probe used to determine reduced thiol compounds has satisfactory selectivity.
In order to verify the reaction of the probe with intracellular thiol, we introduced the thiol scavenger N-acetylmaleimide (NEM) to incubate the cells [23]. One group of L02 cells and two groups of MCF-7 cells were treated separately. One group of MCF-7 cell was pretreated with NEM, and the other group of MCF-7 cells and L02 cells were not pretreated with NEM. Then, three groups of cells were crushed using ultrasound. The three groups of cell crushed liquids were treated with the fluorescent probe, and fluorescence detection was performed using a fluorescence spectrophotometer. As can be seen from Fig. 4A, when the cells that have not been treated with NEM, the fluorescence intensity was the strongest. On the contrary, the fluorescence signal of the cells treated with NEM was basically the same as that of the probe, indicating that under the action of NEM, the thiol compounds in the cell were shielded and the probe cannot respond fluorescently. At the same time, we performed flow fluorescence detection on the cells (Fig. 4B). MCF-7 cells were used as assay cells and divided the cells into three groups. One group was pretreated cells with no NEM + MB/DNA/HA/TPE. The other group was pretreated with no NEM + no MB/DNA/HA/TPE. The third group was pretreated with NEM + MB/DNA/HA/TPE. Then, we used flow cytometry to detect the intracellular fluorescence signal. The results showed that in the NEM-treated cells, the probe had almost no fluorescence response, while in the non-NEM-treated MCF-7 cells, the probe was detected to a strong fluorescent signal. These all suggested that the probe has good selectivity for GSH and can recognize and monitor intracellular thiol.
A Fluorescence detection of GSH in cells. (a) MCF-7 cells + NEM + MB/DNA/HA/TPE, (b) MCF-7 cells + no NEM + MB/DNA/HA/TPE, (c) no MCF-7 cells + no NEM + MB/DNA/HA/TPE. The RSD was 3.3%. B Flow fluorescence intensity of the fluorescent probe MB/DNA/HA/TPE in the cell. (a) MCF-7 cells + no NEM + MB/DNA/HA/TPE, (b) MCF-7 cells + no NEM + no MB/DNA/HA/TPE, (c) MCF-7 cells + NEM + MB/DNA/HA/TPE
Endosome escaping of MB/DNA/HA /TPE
In order to further study the fluorescence imaging performance of probe in cells, we have done confocal imaging experiments with and without probes. As shown in Fig. S6 in ESM, under the same conditions, cell showed a significant fluorescence signal after interaction with the nanoprobe by comparison with the cell without the probe. The results indicated that the background intensity of cell autofluorescence was not enough to interfere with this method.
In order to study the feasibility of this method, we selected MCF-7 cells and L02 cells as the experimental models (Fig. 5). First, MCF-7 cells and L02 cells were pre-seeded in small cell culture dishes. Then, the cells were sequentially incubated with DAPI, MB/DNA/HA/TPE (or MB/DNA/TPE), and LysoTracker Red under the same conditions. After incubation, the cells were washed 3 times with PBS. MB/DNA/HA/TPE and MB/DNA/TPE were localized in cells, respectively. The red spots labeled with LysoTracker Red dye were lysosomes. The blue spots labeled with DAPI dye were the nucleus. And the localization of TPE was indicated with green fluorescence. After incubating with MB/DNA/HA/TPE for 2 h, MCF-7 cells were no obvious green fluorescence. After incubating with MB/DNA/TPE for 2 h, MCF-7 cells were some green fluorescence indicating the probe was inside the cell. All these results indicated that the modification of HA to this probe can prompt the probe to endosome escaping. After incubation with MB/DNA/TPE for 2 h, L02 cells were no obvious green fluorescence. The comparisons showed that probe can efficiently target MCF-7 cells due to the specificity of aptamer. In summary, the MB/DNA/HA/TPE probes have both the ability to escape from lysosomes and the targeting ability of MCF-7 cells.
Conclusion
In summary, we designed and synthesized a new type of fluorescent nanoprobe MB/DNA/HA/TPE for the detection of reduced thiol compounds (GSH as an example). This nanoprobe can effectively detect GSH in solution. The vitro experiments showed that the probe can response to MCF-7 cells rather than L02 cells, due to the selectivity of the aptamer. Besides, the nanoprobe displayed the advantage of lysosome escape effect in cells, making more accurate reduced thiol compound-induced fluorescent signal in cells. We believe that the nanoprobes based on lysosome escape and AIE mechanism have great potential in the study of biological processes in cells due to its excellent selectivity and good sensitivity.
References
Yin C, Xiong K, Huo F, Salamanca JC, Strongin RM. Fluorescent probes with multiple binding sites for the discrimination of Cys, Hcy, and GSH. Angew Chem Int Ed. 2017;56(43):13188–98.
Qin Y, Fan J, Yang W, Shen B, Yang Y, Zhou Q, et al. Endogenous Cys-assisted GSH@AgNCs-rGO nanoprobe for real-time monitoring of dynamic change in GSH levels regulated by natural drug. Anal Chem. 2020;92:1988–96.
Han X, Song X, Yu F, Chen L. A ratiometric fluorescent probe for imaging and quantifying anti-apoptotic effects of GSH under temperature stress. Chem Sci. 2017;8:6991–7002.
Gong F, Cheng L, Yang N, Jin Q, Tian L, Wang M, et al. Bimetallic oxide MnMoOX nanorods for in vivo photoacoustic imaging of GSH and tumor-specific photothermal therapy. Nano Lett. 2018;18:6037–44.
Guo X, Wang L, Duval K, Fan J, Zhou S, Chen Z. Dimeric drug polymeric micelles with acid-active tumor targeting and FRET-traceable drug release. Adv Mater. 2018;30:1870020.
Yue Y, Huo F, Cheng F, Zhu X, Mafireyi T, Strongin RM, et al. Functional synthetic probes for selective targeting and multi-analyte detection and imaging. Chem Soc Rev. 2019;48:4155–77.
Song A, Feng T, Shen X, Gai S, Zhai Y, Chen H. Fluorescence detection of glutathione S-transferases in a low GSH level environment. Chem Commun. 2019;55:7219–22.
Xiong K, Huo F, Chao J, Zhang Y, Yin C. Colorimetric and NIR fluorescence probe with multiple binding sites for distinguishing detection of Cys/Hcy and GSH in vivo. Anal Chem. 2019;91(2):1472–8.
Mei J, Huang Y, Tian H. Progress and trends in AIE-based bioprobes: a brief overview. ACS Appl Mater Interfaces. 2018;10:12217–61.
Xia F, Wu J, Wu X, Hu Q, Dai J, Lou X. Modular design of peptide- or DNA-modified AIEgen probes for biosensing applications. Accounts Chem Res. 2019;52:3064–74.
Luo J, Xie Z, Lam JWY, Cheng L, Chen H, Qiu C, et al. Aggregation-induced emission of 1-methyl-1,2,3,4,5-pentaphenylsilole. Chem Commun. 2001:1740–1.
Liu J, Su H, Meng L, Zhao Y, Deng C, Ng JCY, et al. What makes efficient circularly polarised luminescence in the condensed phase: aggregation-induced circular dichroism and light emission. Chem Sci. 2012;3(9):2737–47.
Qian J, Tang B. AIE luminogens for bioimaging and theranostics: from organelles to animals. Chem. 2017;3(1):56–91.
Gao Y, He Z, He X, Zhang H, Weng J, Yang X, et al. Dual-color emissive AIEgen for specific and label-free double-stranded DNA recognition and single-nucleotide polymorphisms detection. J Am Chem Soc. 2019;141:20097–106.
Wang X, Dai J, Min X, Yu Z, Cheng Y, Huang K, et al. DNA-conjugated amphiphilic aggregation-induced emission probe for cancer tissue imaging and prognosis analysis. Anal Chem. 2018;90:8162–9.
Min X, Zhuang Y, Zhang Z, Jia Y, Hakeem A, Zheng F, et al. Lab in a tube: sensitive detection of microRNAs in urine samples from bladder cancer patients using a single-label DNA probe with AIEgens. ACS Appl Mater Interfaces. 2015;7:16813–8.
Ding Y, Shi L, Wei H. A “turn on” fluorescent probe for heparin and its oversulfated chondroitin sulfate contaminant. Chem Sci. 2015;6:6361–6.
Sun F, Zhao S, Peng M, Fu Q, Gao H, Jia Y, et al. Sequencing of small DNA fragments with aggregated-induced-emission molecule-labeled nucleotides. Anal Chem. 2020. https://doi.org/10.1021/acs.analchem.0c00707.
Jing J, Xue Y, Liu Y, Xu B, Li H, Liu L, et al. Co-assembly of HPV capsid proteins and aggregation-induced emission fluorogens for improved cell imaging. Nanoscale. 2020;12:5501–6.
Chen X, Gao H, Deng Y, Jin Q, Ji J, Ding D. Supramolecular aggregation-induced emission nanodots with programmed tumor microenvironment responsiveness for image-guided orthotopic pancreatic cancer therapy. ACS Nano. 2020;14:5121–34.
Sun X, Ying N, Ju C, Li Z, Xu N, Qu G, et al. Modified beacon probe assisted dual signal amplification for visual detection of microRNA. Anal Biochem. 2018;550:68–71.
Guo Y, Wang H, Sun Y, Qu B. A disulfide bound-molecular beacon as a fluorescent probe for the detection of reduced glutathione and its application in cells. Chem Commun. 2012;48:3221–3.
Wi Y, Le H, Verwilst K, Sunwoo K, Kim S, Song J, et al. Modulating the GSH/Trx selectivity of a fluorogenic disulfide-based thiol sensor to reveal diminished GSH levels under ER stress. Chem Commun. 2018;54:8897–900.
Funding
This work was supported by the Development Plan of Youth Innovation Team in Colleges and Universities of Shandong Province (2020KJC003), the Natural Science Foundation of Shandong Province of China (ZR2016JL010), and the Primary Research and Development Plan of Shandong Province (2018GSF118172).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 738 kb)
Rights and permissions
About this article
Cite this article
Hu, Y., Cao, X., Guo, Y. et al. An aggregation-induced emission fluorogen/DNA probe carrying an endosome escaping pass for tracking reduced thiol compounds in cells. Anal Bioanal Chem 412, 7811–7817 (2020). https://doi.org/10.1007/s00216-020-02909-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-02909-w