Abstract
A single-probe strip test for the rapid and sensitive detection of miRNA-21 mimics is reported herein. Highly specific structurally responsive bi-functional, thiol and biotin, DNA/LNA oligonucleotide probes (molecular beacons-MB) were designed and conjugated with gold nanoparticles (AuNPs) (i.e. biotin-MB-AuNPs). The proposed design had the ability to modulate the accessibility of the biotin group as a function of the presence of a miRNA target allowing the interaction of the boilable with the streptavidin test zone only in the presence of the miRNA-21 mimics. For quantitative evaluation, images of the strip tests were recorded using a flatbed scanner (Epson Perfection V370 Photo). The colour intensities of the test zones of the strip tests were analysed with the ImageJ software (Scion Corp., USA) and quantified as a function of pixel intensity. The response of the strip test was linear over the range 0.5 to 20 nM miRNA-21 (limit of detection of 115 pM) and showed good reproducibility (intra and inter CVs below 8 %); furthermore, the assay was shown to be highly selective, discriminating other interference miRNAs mimics (e.g. miRNA-221 and miRNA-205). Finally, the proposed strip test was used for detection of miRNA-21 mimics in spiked serum samples, demonstrating its potential for point-of-care clinical applications. Main advantages of the single-probe strip test design are its versatility, simplicity and robustness, which can be easily extended to other miRNA targets by tuning the sequence of the single probe. Furthermore, the use of the structurally responsive single probe is particularly relevant in the case of short-length targets, such as miRNA, whereas a conventional sandwich approach might require a careful control of assay conditions such as hybridization temperature and salt concentration.

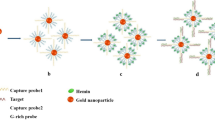
Pictorial representation of the working principle of the proposed structurally responsive oligonucleotide-based single-probe lateral-flow test and example of the test response as a function of increasing concentration of target analyte.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are a class of noncoding RNA molecules, between 17 and 25 nucleotides in length, which are encoded in the genomes of plants and animals. These are involved, upon binding with messenger RNA, in a series of important living process, including early development, cell proliferation, cell apoptosis, fat metabolism and cell differentiation [1]. It is not surprising that, in recent years, the number of reports linking miRNA overexpression or downregulation to diseases and subsequently proposing miRNA detection as an early diagnostic approach has increased enormously. This correlation makes miRNAs good candidates as biomarkers for diagnostic, prognostic and predictive information. For pursuing these applications, it is crucial to develop fast, accurate and sensitive methods for miRNA detection [1]. Among the over 1000 sequences reported, miRNA-21, a mammalian miRNA encoded by the mir-21 gene [2], has been shown to play an important role in the development of heart disease [3] and to be commonly overexpressed in solid tumours of the lung, breast, stomach, prostate, colon, brain, head and neck, oesophagus and pancreas [4]. Several techniques have been reported for detection of miRNA-21 including RT-PCR [5–8], northern blotting [9], Bioluminescence [10], microarray [11] and electrochemical [1, 12], but miRNA detection is still challenging mostly because of its short length and its low concentration (nano to pico Molar).
Each technique has its advantages and its limitations; for instance, northern blotting, currently the “golden standard” in miRNA detection, is very time-consuming and requires large amount of RNA samples and radioactive probes. RT-PCR exhibits high sensitivity, but it is limited by high cost, tedious sample preparation and, even more importantly, by the length of the miRNA sequences [12, 13]. Recently, Yin et al. [1] and Zhou et al. [12] proposed an assay based on the combination of LNA/DNA molecular beacon probes (MB) and electrochemical transduction for the determination of miRNA-21. Despite the high sensitivity, specificity and detection limit that was achieved with the proposed assay design, electrochemical techniques require relatively expensive instrumentation, can be time-consuming and require highly skilled personnel compared to simply strip tests. There are only few reported strip tests for miRNA detection [14, 15]. The work described by Hou et al. [14] adopted a very complicated approach based on the use of a biotinylated bridging probe, streptavidin labeling and nuclease digestion. This approach is very complicated requiring 4 consecutive steps, prior to run the sample onto the strip. All these steps are time-consuming (total assay time of 70 min.), some of them extremely complicated (requiring high control of temperature and pH). The multiple step incubations, mung-bean nuclease digestion and the eventually silver enhancements are all very critical time-dependent steps limiting the real practical applications. The work described by Gao et al. [15] for detection of miRNA-215 is less complex; however, it still required multiple probes hybridisation and a bridging probe for detection. Thus, a simple, direct and inexpensive strip tests for detecting miRNA remains desirable.
Molecular beacons (MBs) are single-stranded oligonucleotide probes with a characteristic stem-and-loop structure [16], where the loop contains 18–30 base pairs complementary to a specific target sequence, and the stem sequence of 5–7 bases, present at both the ends of the loop, is introduced to confer to the MBs its specific structural conformation [17]. A significant property of MBs is its ability to change conformation upon recognition of its specific target; this characteristic allowed the development of reagent/washless assay [18, 19]. MBs have found application in the direct detection of DNA and RNA in living cells [20–24], as reporting probes in real-time PCR for DNA, RNA and pathogenic detection [25–29], as probes in DNA arrays [30–33], as probes to detect UV-induced oligonucleotide damage [34], to distinguish miRNAs that have slight variations in their nucleotide sequence [35] or in electrochemical genosensor [19].
Unfortunately, the majority of the approaches described above are affected by limitations as relatively high equipment cost, long assay time, tedious sample preparation and the requirement for highly trained personnel; all these factors obstruct their practical applications [36]. There is, therefore, a need for the development of an inexpensive, sensitive, quick and simple to use tool in DNA/RNA analysis.
One potential approach is based on coupling MBs with gold nanoparticles (AuNPs) and use this bio-conjugates as a reporter, whereby the resulting optical signal can be easily read with the naked eye without the need for expensive instrumentation and thus providing an attractive point-of-care platform [37]. Among the variety of colorimetric methods available, lateral-flow test strips offer several advantages such as ease of use, a simple colour readout and relatively low cost. They have also been shown to have wide applications for DNA detection [38]. Despite He et al. demonstrated, using synthetic targets, the potentialities [39, 40] of MBs in lateral flow assays, most of the current DNA lateral-flow strip tests are based on sandwich hybridisation approaches that imply the use of quite complicated assay design and test zone construction. Herein, we report on the development of a simple probe lateral-flow strip test, based on immobilised streptavidin in the test zone and MBs-AuNPs conjugates as recognition/transducing element for the rapid, within minutes and sensitive detection miRNA-21. Quantitative analysis of the strip tests can be simply performed by using a home scanner (Epson Perfection V370 Photo) and imaging software (ImageJ).
Experimental
Materials
Streptavidin (52.8 kDa), chloroauric acid (HAuCl4), Tween 20, dithiothreitol (DTT), bovine serum albumin (BSA), sodium hydroxide, sodium chloride, sodium citrate, sodium acetate, fuming acetic acid and Trizma-HCl were purchased from Sigma-Aldrich (USA). Nitrocellulose membranes (HF120), conjugate pad, sample pad and adsorption pad were purchased from Millipore (Billerica, MA). All chemicals used in this study were of analytical reagent grade. All solutions were prepared with ultrapure (18.2 MΩ) water from a Millipore Milli-Q water purification system (Billerica, MA).
The sequence of the DNA/LNA MBs was taken from a previous report [41] with the only difference that a 10 PolyT tail was added at its 5′ end in an attempt to improve, following recognition of the target, the availability of the biotin reported [18]. The presence of a thiol group at the 5′ end of the MB allowed its immobilisation onto the AuNPs. A biotin at the 3′ was used to allow interaction between the AuNP-based biorecognition/transducer element and the streptavidin located in the test line, giving rise in this way to the typical colorimetric response.
Sequence of the miRNA targets was taken from miRBase (http://www.mirbase.org). miRNA-205 and miRNA-221 were chosen as test sequences in the selectivity experiment because they demonstrated to be involved in cancers and cancer development. miRNA-205 has a role in different cancers such as breast [42], prostate [43], lung [6] and bladder [44]. Also, miRNA-221 has shown to be an oncogenic miRNA and to have a potential role in bladder [45] and astrocytic tumours [46].
In order to facilitate the handling of the sample, RNA sequences were replaced by RNA-mimics oligonucleotides; these consist of DNA sequences in which thymine was replaced by desoxy uridine.
RNA-mimics sequences were obtained from Integrated DNA Technologies, Inc. (Coralville, IA, USA) and biomers.net (Germany); LNA-modified oligonucleotide probes were obtained from Exiqon (Denmark):
-
MB:
-
5′-/5BioTEG/GGCCGTCAACATCAGTCTGATAAGCTACGGCCTTTTTTTTTT/ 3ThioMC3-D/-3′ (in bold and italics are the LNA bases)
-
-
Control line DNA probe:
-
5′-GGC CGT AGC TTA TCA GAC TGA TGT TGA CGG CC/3BioTEG/-3′
-
miRNA-21 mimics: 5′-UAG CUU AUC AGA CUG AUG UUG A-3′
-
miRNA-205 mimics: 5′-UCC UUC AUU CCA CCG GAG UCU GU-3′ (homology factor 41 %)
-
miRNA-221 mimics: 5′-AGC UAC AUU GUC UGC UGG GUU UC-3′ (homology factor 18 %)
-
Oligonucleotides stock solutions (100 μM) were prepared by dissolving the lyophilised synthetic sequences in filtered (filter size: 0.2 μm) MilliQ water. All stock solutions were stored at −20 °C. In order to reduce the risks of deactivation of the thiol group, the stock solution of the MB was divided in aliquots that were stored at −20 °C and defrosted only when used. All experiments have been performed, if not stated otherwise, at room temperature (21 °C).
Procedure
Preparation of gold nanoparticles
Gold nanoparticles (AuNPs) were synthesised according to the citrate reduction of HAuCl4 [18]. All glassware and the magnetic stirring bar, stopper and condenser used in this preparation were thoroughly cleaned in aqua regia (Caution: very corrosive solution handle with care), rinsed with copious amount of deionised water and then Millipore-filtered water and oven-dried prior to use.
In a 100-mL round-bottom flask equipped with a condenser, 50 mL of 1 mM HAuCl4 was brought to a rolling boil with vigorous stirring. Rapid addition of 5 mL of 38.8 mM sodium citrate to the vortex of the solution resulted in a colour change from pale yellow to burgundy. Boiling was continued for 10 min; the heating mantle was then removed, and stirring was continued for an additional 15 min. After the solution cooled to room temperature, it was placed in storage at 4 °C. The resulting solution of colloidal particles was characterised by spectrophotometric techniques and using zeta potential analyser (Zetasizer, Malvern Instruments Ltd., Worcestershire, UK).
Preparation of biotin-MB-AuNPs Conjugates
Preparation of biotin-MB-AuNPs conjugate was performed in glass vials; in order to reduce the unspecific adsorption of the MBs and of the biotin-MB-AuNPs conjugate onto the wall of the glass vials, these were treated for 2 h in 12 M NaOH at room temperature [47]. In the rinsed glass vial, MB were added to a 250 μL of the AuNPs (OD 2.3) in 0.1 mM phosphate buffer at pH 7.4 to obtain a final DNA-to-AuNPs ratio of 500:1 [18]; the solution was left to react at room temperature, under gentle mixing, overnight. The biotin-MB-AuNPs mixture was then subjected to an “aging process” consisting of a stepwise increase of the concentration of NaCl up to 0.3 M, prior to an overnight incubation at room temperature under gentle shaking. The biotin-MB-AuNPs conjugates were washed twice by sequential centrifugation (12,000 rpm, 20 min, 21 °C) and resuspension in 0.3 M NaCl and 0.1 mM phosphate buffer at pH 7.4. The final solution of the biotin-MB-AuNPs in a 0.3 M NaCl and 0.1 mM phosphate buffer at pH 7.4 was stored at 4 °C until use.
Quantification of the MBs loading onto the AuNPs was performed by DTT cleavage experiments [48]. The pellet obtained from 150 μL of functionalised AuNPs was resuspended in 75 μL of freshly prepared solution of DTT (10 mM) in water and left to react for 1 h under gentle shaking. Following the 1 h incubation, the supernatant was collected and the concentration of the DNA was measured spectrophotometrically at 260 nm using a NanoDrop 2000 UV–vis Spectrophotometer (Thermo Scientific, Sweden). Concentration of the DNA in the supernatant was calculated by fitting the obtained absorbance value in a previously constructed calibration curves (between 25 nM and 2 μM). The number of DNA strains in the solution was then calculated using the obtained concentration, the initial volume and the Avogadro number. The number of AuNPs in the analysed sample was estimated from value tabulated on BBI website (http://bbisolutions.com) and extrapolated to our sample concentration.
Preparation of reagents for control line
Sixty microliters of 1 mM biotinylated control line DNA probes and 140 μL of 10 mM PBS (pH 7.4) were added to 300 μL of 31.7 μM streptavidin solution, and the mixture was incubated 1 h at RT. The excess DNA probes were removed by centrifugation for 20 min with a centrifugal filter (cutoff 30000, Millipore) at 6500 rpm. The resulting conjugates were washed three times with 0.5 mL of PBS in the same centrifugal filter. Finally, 500 μL of PBS was added into the remaining solution in the filter.
Preparation of lateral-flow strip test
The test zone and control zone of the strip were prepared by dispensing the streptavidin solution (2.38, 4.75 and 9.50 μM) and streptavidin-biotinylated DNA solutions (i.e. 9.50 μM) onto the nitrocellulose membrane using the IsoFlowTM reagent dispensing system with a dispensing rate of 0.1 μL mm−1 (Arista Biologicals Inc, USA). The control zone was coated twice to maximise the signal. The membrane was then dried in a desiccator overnight at room temperature. After drying, the absorption pad was placed at the far end of the nitrocellulose membrane downstream of the flow direction. Then, the sample pad was placed at the front end of the nitrocellulose membrane (i.e. at the origin of sample flow). Subsequently, the nitrocellulose membrane assembled with sample pad and absorption pad was cut into 0.5 cm width strips by using an automated A-Point membrane cutter (Arista Biologicals Inc, USA). The resulting strips were stored in a desiccator at room temperature for future experiments.
Assay procedure
For preparing sample solution, desired amount of target miRNA was mixed with optimised volume of biotin-MB-AuNPs conjugate (2.5 μL, O.D. 2.3 at 520 nm) in optimum buffer (in this work 0.5 × SSC; pH 7) to a final volume of 25 μL and incubated for 5 min to allow the hybridisation between the target miRNA mimics and MBs. Following 20 μL of the solution was applied to the sample application zone of the strip and left to run for 3 min. The assay was completed by a further addition of 20 μL running buffer (0.5 × SSC; 0.05 % v/v Tween 20; 5 mg mL−1 BSA; pH 7) to the application zone. The bands were visualised within 5 min. For quantitative measurements, the strip was placed into a scanner (Epson Perfection V370 Photo), and image recorded with a resolution of 1200 dpi and image type 24-bit colour. The optical intensities of the test line and control line were analysed with ImageJ software (Scion Corp., USA).
Serum samples
Serum was prepared from human whole blood collected from an apparently healthy, consenting blood donor at Linköping University Hospital with ethical approval. In brief, the blood sample was allowed to clot at room temperature for 30 min and centrifuged at 1500 r.c.f. for 15 min at room temperature followed by extraction of the serum from the top layer. The desired different concentrations of miRNA-21 mimics in the serum samples were obtained by spiking 20 μL of it with 2 μL of appropriated miRNA-21 stock solution.
Characterisation
UV–vis measurements were performed using a SHIMADZU UV-2450 spectrophotometer (Shimadzu, Japan) with a 0.5-nm resolution or NanoDrop 2000 (Thermo Scientific, USA). The hydrodynamic size and zeta potentials (surface charge) of the AuNPs and biotin-MB-AuNPs conjugate were measured using a Zetasizer Nano ZS90 (Malvern Instruments Ltd., Worcestershire, UK). Particle size was measured based on dynamic light scattering, measuring the Brownian motion of the particles and converting the data into a size distribution graph using the Stokes-Einstein relationship. Zeta potential was measured based on the laser doppler micro-electrophoresis principle. The electrophoretic mobility (μ) was converted to the zeta-potential (ζ) by using the Smoluchowski relation ζ = μη / ε, where η and ε are the viscosity and permittivity of the solution respectively. The measurements were performed at 25 °C, and the mean zeta potential values were calculated by taking an average of 3 repeated measurements.
Results and discussion
Design of molecular beacon-based AuNPs and lateral-flow strip tests
Scheme 1 illustrates the working principle of the proposed biotin-MB-AuNPs based lateral-flow miRNA test. The biotin-MB-AuNPs and sample mixture are loaded from the upstream of the strip tests (Scheme 1a); if target miRNA-21 mimics is present in the mixture, this will hybridise with the biotin-MB-AuNPs conjugate inducing the opening of the biotin-MBs. This change in biotin-MBs structure results in exposing the biotin group, previously “protected” by the steric hindrance generated by the stem-loop structure, making it available for interaction with the streptavidin present in the test zone. The capture of the miRNA-21 mimics/biotin-MB-AuNPs complex at the test zone will result in the appearance of a visible red band in the test zone. The excess/non-captured biotin-MB-AuNPs will continue to migrate toward the control zone where the biotin-MB-AuNPs will be captured, via hybridisation reaction, by immobilised DNA probes reverse-complementary to the MB (Scheme 1b) generating a visible red colour control line. To read the strip tests, two red bands in both the test zone and control zone are observed in the presence of target miRNA-21 mimics. On the contrary, in the absence of target miRNA-21 mimics, the biotin-MB will retain its stem-loop structure not allowing in this way the interaction between the biotin and the streptavidin at the test line; as a result, a red band will be only visible in the control zone (Scheme 1c). The miRNA mimics, consisting of DNA sequences in which thymine was replaced by desoxy uridine, has the advantage of being more stable compared with miRNA. This allowed performing more consistence and robust optimisations for the development of the strip tests. Despite the DNA-DNA, RNA-RNA and DNA-RNA duplexes that are known to have different thermodynamics and stability, these differences are strongly buffered by a high dA/(U.T) contempt (>50 %) in the sequence [48] and by high concentration of salt in the hybridisation solution [49]. In our studies, all the miRNA mimics sequences contained more than 50 % of dA/(U.T) and all experiments were performed in 0.5x SSC which contains enough salt. Therefore, it is reasonable to suggest that under our assay condition, the duplex stability/formation will be mainly determined by the base-pairing making the obtained finding of great relevance also for future applications to miRNA sample analysis.
A Schematic illustration of the design and configuration of the single probe miRNA strip test. B The principle of the single probe biotin-MB-AuNPs changing structure and exposing the biotin group upon target recognition. C The detection principle for the miRNA strip test indicating positive and negative results
Characterisation of AuNPs and biotin-MB-AuNPs conjugation
The synthesised citrate-capped AuNPs were characterised by dynamic light scattering; these have been shown to have an average hydrodynamic diameter of 29.52 ± 6.34 nm (Fig. S1A in the Electronic Supplementary Material, ESM). The concentration of the stock citrate-capped AuNPs was quantified by UV–Vis measurements at 520 nm with an optical density (O.D.) value of 2.3. Functionalisation of the AuNPs with the biotin-MBs was performed following the protocol described in the Materials and Methods (section “Preparation of biotin-MB-AuNPs Conjugates”). The resulting biotin-MB-AuNPs had an average hydrodynamic diameter of 29.83 ± 7.51 nm (Fig. S1A in the ESM), which is similar to the citrate-capped AuNPs and indicates that the biotin-MB-AuNPs have good colloidal stability without aggregation. AuNPs with size of 30 nm were considered to allow a good compromise between the flow efficiency, bio-receptor loading (especially being this considerably affected by the hair pin structure of the MB) [50, and to maintain a strong red colour intensity, without significant plasmonic blue shift, of the AuNPs allowing in this way both visual determination and optical measurement.
The stability of the biotin-MB-AuNPs was further challenged by measuring UV–vis spectra at high NaCl (0.3 M) concentration (Fig. S1B in the ESM). The presence of a clear plasmonic peak at 527 nm and the absence of particle aggregation (no blue shift was observed) were considered a qualitative confirmation of the successful functionalisation of the AuNPs with the biotin-MBs. To further evaluate the surface properties of the resulting particles, zeta potential measurements of the citrate-capped AuNPs before and after functionalisation with the biotin-MBs were performed. The citrate-capped AuNPs and the corresponding functionalised biotin-MB-AuNPs had average zeta potential values of −26.2 ± 1.7 and −8.58 ± 1.7 mV, respectively. The higher negative value recorded at the citrate-capped AuNPs was the result of the higher surface charge density provided by the highly packed critic acids, due to their small size, when compared to the MB. To be noticed that the zeta potential value recorded for the MB functionalised particles is consistent with those previously reported [51]. DTT cleavage experiments have been used to confirm and quantify the presence of biotin-MBs onto the AuNPs; these experiments were performed using the NanoDrop 2000. Based on the AuNPs O.D. 2.3 and their size (∼30 nm), an average loading of 152 ± 8 MB molecules per AuNP was obtained. The obtained value is below the maximum possible loading reported by Hurst et al. [52]; this result was expected due to the fact that the bulky structure of the closed MBs is not favourable for high loading.
Optimisation of lateral-flow strip tests
Buffer composition is well known to influence the stability of oligonucleotides duplex; for example, a high stringency solution (low salt composition) is well known to improve the selectivity of MBs. For these reasons, different hybridisation and running buffers were evaluated. Various concentration of SSC buffers (4×, 3×, 2×, 1× and 0.5×; pH 7) were investigated as possible hybridisation and running buffers. These were screened by performing a series of strip detection assays (target miRNA concentration 10 nM; pre-hybridisation time 5 min) and comparing the signal obtained to the noise (response in the absence of target). Accordingly to the obtained results, (data not shown) 0.5 × SSC; pH 7 and 0.5 × SSC; 0.05 % v/v Tween 20; BSA 5 mg mL−1; pH 7 were used, respectively, as hybridisation and running buffer.
Two key parameters in the development of a lateral-flow strip tests are the amount of the immobilised streptavidin (capturing probe) in the test line and the amount of biotin-MB-AuNPs (biolabel) used in the test. Fine tuning of these two parameters is very important to ensure the desired cutoff sensitivity and dynamic range of the strip test. As part of this optimisation, three different concentrations of immobilised streptavidin (2.38, 4.75 and 9.50 μM) and three different amounts of biotin-MB-AuNPs (2, 2.5 and 3 μL; O.D. 2.3) were selected and their various combinations were used for the detection of 0, 1 and 5 nM of the targeted miRNA-21 mimics, respectively.
In Fig. 1a, the results obtained for the different experiments are presented. From a close analysis, it was clear that no observable signals were recorded when 2.38 μM of streptavidin was immobilised at the test zone, due to the insufficient amount of biotin-MB-AuNPs captured. On the other hand, when the streptavidin concentration was increased to 4.75 and 9.50 μM, clear visible signals were observed at the test zone. In order to compare the analytical performance of the different experimental conditions, the signal-to-noise (S/N) ratio was plotted (Fig. 1b). As expected, the different results clustered as a function of the amount of streptavidin immobilised onto the test zone, whereas the amount of biotin-MB-AuNPs had less influence on the analytic performance. Interestingly, a higher S/N ratio and subsequently better discrimination was recorded for the lowest concentration of immobilised streptavidin (i.e. 4.75 μM). In order to gain a better understanding of the obtained result, the absolute responses for the different experiments were plotted and compared. As it can be seen in Fig. 1c, the increase in streptavidin concentration in the test line resulted not only, as expected, in an increase in the absolute signal response but also in a significant increase, from 2.1 to 2.9 times, in the absolute blank signal (no target analyte). The significant increase in blank signal reduced the analytical sensitivity of the strip tests for detection of low concentrations of target analyte. Moreover, the use of higher concentrations of the biotin-MB-AuNPs seems to have a saturation effect on the signal response at a lower streptavidin concentration; this could be clearly seen when comparing the results obtained for the strip tests prepared with 4.75 μM streptavidin for 2.5 and 3 μL of the biotin-MB-AuNPs label (Fig. 1b). Therefore, an optimised setup of 4.75 μM immobilised streptavidin and 2.5 μL of biotin-MB-AuNPs (O.D. 2.3) were chosen for future experiments.
A Optimisation on the amount of immobilised streptavidin and biotin-MB-AuNPs for the strip tests. B Comparison on the S/N ratio of strip tests with different settings. C The absolute signal response of strip tests with different settings indicating the absolute blank signals. D Effect on the hybridisation time as a function of the strip test response
The hybridisation time between MB probe and the target miRNA mimics can also significantly influence the efficiency of the assay. As part of the optimisation of the assay condition, the relationship between hybridisation time and signal responses was evaluated. This study was performed by mixing the biotin-MB-AuNPs with 5 nM of the miRNA-21 target (final concentration) and letting them to interact for different times (0, 5, 15 and 30 min, respectively) prior to be introduced in the sample pad. As it can be seen from Fig. 1d, a visible signal can be observed immediately without pre-incubation time (time zero) and the response increased gradually with the time. Five minutes was selected as the optimum hybridisation time providing a good compromise between the assay time, signal response and operational ease.
Analytical performance
In order to evaluate the analytical performance of the developed strip test a series of calibration curves for detection of miRNA-21 mimics (4 repetitions; Fig. S2 in the ESM), in the high pM/low nM region ranging from 0.5 to 40 nM, were tested. This range of concentration has been selected accordingly to previous report concerning quantification of miRNA-21 in biological fluids [8, 53].
Figure 2a presented the typical images of strip tests after addition of standard miRNA-21 mimics showing clearly a significant increase in the observed colour intensity in the test zone as a function of increase in miRNA-21 concentration with a visual cutoff value of 0.5 nM. The plot of the signal intensity corresponding to the peak area (insert of Fig. 2b) recorded at the test lines, obtained accordingly to the instrumental procedure described in tne experimental section (section “Assay Procedure”), against the concentration of miRNA-21is reported in Fig. 2b. As it can be seen from the calibration curve, a linear signal response of the miRNA-21 mimics was recorded in the concentration range from 0.5 to 20 nM (with good correlation coefficient R 2 of 0.999 and a limited of detection of 115 pM (3σ/slope; n = 10). It is also important to notice that our miRNA strip tests show a wide linear detection range compared to the existing miRNA strip tests [14, 15]. The wide liner detection range recorded in our miRNA-21 strip tests could be explained by the advantage of the use of a single probe protocol, having in this way the signal generation governed by a less complicated affinity kinetic with the miRNA/probe hybridisation as the limiting factor.
A Images of the strip tests after applying different concentration of miRNA-21 mimics (0 to 40 nM). B Calibration curve of the test zone colour intensity (peak area) as a function of miRNA concentration (n = 4); insert shows the raw data of peak area. C The stability of the biotin-MB-AuNPs and miRNA-21 strip tests over a period of 28 days upon storage at room temperature
The stability of the biotin-MB-AuNPs and miRNA-21 strip tests was evaluated over a period of 28 days upon storage at room temperature. Results show that the strip tests are relatively stable for detection of miRNA-21 mimics at low (1 nM), medium (5 nM) and high (20 nM) concentration, even if we observed a slightly increase of the absolute signal as a function of time (Fig. 2c). This increase could be mainly due to instability of the biotin-MB-AuNPs probably due to structural changes in the MB layer that result in partial exposure of the biotin with subsequent slightly increase in the non-specific signal.
The intra-assay and inter-assay coefficients of variations (CV) were evaluated to assess the reproducibility of the miRNA-21 strip tests. The intra-assay CV of the miRNA-21 strip tests were determined by performing repeated measurements with same procedures, conditions and instrumentation of same batch of biotin-MB-AuNPs, while the inter-assay CV of the miRNA-21 strip tests was determined by performing repeated measurements with same procedures, conditions and instrumentation of three different batches of biotin-MB-AuNPs. The obtained intra-assay and inter-assay CVs for different analyte concentrations (3 measurements for each analyte concentration) are summarised in Table 1. The intra-assay CV for detection of miRNA-21 mimics with concentration of 1, 5 and 20 nM was 6.59, 4.07 and 3.05 %, respectively. While the inter-assay CV of the mi-RNA strip tests for detection miRNA-21 mimics with concentration of 1, 5 and 20 nM were 7.84, 7.08 and 7.79 %, respectively. The inter-assay CV is slightly higher compared with the intra-assay CV, which is expected due to batch variation. The intra-assay and inter-assay CV of all measurements were below 8 %. These results show that the miRNA-21 strip tests give good reproducibility.
Another important parameter related to the analytical performance of the strip test is its selectivity. This was evaluated by comparing the signal responses of the developed strip test for the target miRNA-21 mimics against two other interfering miRNA mimics (i.e. miRNA-221 mimics and miRNA-205 mimics) at the concentrations of 0, 1 and 5 nM. The selected non-targets miRNAs (i.e. miRNA-221 and miRNA-205) have been also demonstrated to be overexpressed in the presence of different cancers [42, 43].
As it can be seen from Fig. 3, a clear increase in signal response was only observed in the presence of target miRNA-21 mimics. In contrast, no significant signal was observed for miRNA-205 mimics and a small signal was observed for miRNA-221 mimics only at high concentration level of 5 nM. Although a small signal was observed for miRNA-221 mimics alone at high concentration, we did not observe significant influence of this during the detection of miRNA-21 mimics mixed with the other non-target miRNA mimics; this is because the miRNA-21 mimics has higher affinity and selectivity for competitive binding with the biotin-MB-AuNPs. This result confirmed the high selectivity of the biotin-MB-AuNPs and the resulting strip test for detection of miRNA-21 mimics.
Finally, the suitability of the developed test for the detection of miRNA-21 mimics in real sample was demonstrated by testing the strip tests with spiked serum samples. Measured concentration of miRNA-21 mimics in the serum sample was calculated by fitting the peak area obtained for the different sample with the calibration curve presented in Fig. 2b (Peak area = 1444∙CmiRNA (nM) + 4172.5). In Fig. 4, the correlation plot is presented; this was built by plotting the concentration obtained using the strip test (by fitting the reading in the calibrations curve of Fig. 2b) versus the actual known concentration of miRNA-21 mimics in the spiked serum samples. It is noted that the measured zero concentration shows a slightly negative value. This could be explained by the fact that serum has a relatively high protein content (i.e. albumin) which will reduce the non-specific binding of the AuNPs label at the test zone in the real sample measurement resulting in this way in a partial suppression of the background response. The equation showed in the correlation plot has a scope of 0.94 which indicated a good matching between the measured experimental values and the actual concentration of the miRNA-21 mimics in serum sample. The presented strip tests were designed for serum sample which required a simple centrifugation step to separate the serum from blood sample prior to measurement. In the future, we envisage to integrate a blood-separating membrane in the strip to replace the centrifugation procedure.
Conclusion
We have developed a lateral-flow strip test for simple, rapid and selective detection of miRNA-21 mimics based on a single-probe single-step assay design with immobilised streptavidin in the test zone and detection by structure responsive biotin-MB-AuNPs biolabel. The presence of target miRNA induced the structural changes of the biotin-MB on the AuNPs exposing the biotin group which can be simply captured by the generic streptavidin test zone resulting in a visible signal. The developed strip test has been shown to allow detection of miRNA-21 mimics down to 0.5 nM, with a dynamic range between 0.5 and 20 nM, in a fast (ca. 13 min) and easy (1 single step) way. The intra-assay and inter-assay CV of the strip test, below 8 %, show the good reproducibility of this. The developed test was shown to have a high selectivity for miRNA-21 in comparison to other oncological relevant miRNA (miRNA-221 and miRNA-205). Furthermore, we further proved the detection of spiked miRNA-21 mimics in a complex serum sample demonstrating the potential of the strip test for point-of-care clinical applications. The demonstrated design approach is envisaged to be extended to the detection of other miRNAs by simple tuning of the sequence of the MB foreseeing the future application of it as a simple, cost-effective distributed biosensor for decentralised screening of miRNA. Besides the versatility, the use of a single probe is envisaged to improve the assay simplicity and robustness. The moving away from conventional sandwich approach makes the assay less sensitive to the experimental condition (i.e. salt concentration and temperature); this is particularly important in the case of short targets as are miRNA.
References
Zhou Y, Zhang Z, Xu Z, Yin H, Ai S. MicroRNA-21 detection based on molecular switching by amperometry. New J Chem. 2012;36(10):1985–91.
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–8.
Small EM, Frost RJA, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121(8):1022–32.
Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27(31):4373–9.
Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DSB. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57(1):84–91.
Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non–small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54(10):1696–704.
Wei J, Gao W, Zhu C-J, Liu Y-Q, Mei Z, Cheng T, et al. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non–small cell lung cancer. Chin J Cancer. 2011;30(6):407–14.
Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang SL, Dai B, et al. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011;71(3):326–31.
Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, et al. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res. 2010;38(7), e98.
Cissell KA, Rahimi Y, Shrestha S, Hunt EA, Deo SK. Bioluminescence-based detection of microRNA, miR21 in breast cancer cells. Anal Chem. 2008;80(7):2319–25.
Wang ZX, Bian HB, Wang JR, Cheng ZX, Wang KM, De W, et al. Prognostic significance of serum miRNA-21 expression in human non-small cell lung cancer. Oncology. 2011;104(7):847–51.
Yin H, Zhou Y, Zhang H, Meng X, Ai S. Electrochemical determination of microRNA-21 based on graphene, LNA integrated molecular beacon, AuNPs and biotin multifunctional bio bar codes and enzymatic assay system. Biosens Bioelectron. 2012;33(1):247–53.
Li W, Ruan K. MicroRNA detection by microarray. Anal Bioanal Chem. 2009;394(4):1117–24.
Hou SY, Hsiao YL, Lin MS, Yen CC, Chang CS. MicroRNA detection using lateral flow nucleic acid strips with gold nanoparticles. Talanta. 2012;99:375–9.
Gao X, Xu H, Baloda M, Gurung AS, Xu LP, Wang T, et al. Visual detection of microRNA with lateral flow nucleic acid biosensor. Biosens Bioelectron. 2014;54:578–84.
Fang X, Liu X, Schuster S, Tan W. Designing a novel molecular beacon for surface-immobilized DNA hybridization studies. J Am Chem Soc. 1999;121(12):2921–2.
Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14(3):303–8.
Beni V, Hayes K, Lerga TM, O’Sullivan CK. Development of a gold nano-particle-based fluorescent molecular beacon for detection of cystic fibrosis associated mutation. Biosens Bioelectron. 2010;26(2):307–13.
Nasef H, Beni V, O’Sullivan CK. Electrochemical molecular beacon DNA biosensor for the detection and discrimination of the DF508 cystic fibrosis mutation. J Electroanal Chem. 2011;662(2):322–7.
Alexander JC, Pandit A, Bao G, Connolly D, Rochev Y. Monitoring mRNA in living cells in a 3D in vitro model using TAT-peptide linked molecular beacons. Lab Chip. 2011;11(22):3908–14.
Bratu DP, Catrina IE, Marras SAE. Tiny molecular beacons for in vivo mRNA detection. In: Gerst JE, Editors. RNA Detection and Visualization. Humana Press; 2011. p. 141–157.
Catrina IE, Marras SAE, Bratu DP. Tiny molecular beacons: LNA/2′-O-methyl RNA chimeric probes for imaging dynamic mRNA processes in living cells. ACS Chem Biol. 2012;7(9):1586–95.
Chen AK, Behlke MA, Tsourkas A. Avoiding false-positive signals with nucleasevulnerable molecular beacons in single living cells. Nucleic Acids Res. 2007;35(16), e105.
van den Bogaard PC, Tyagi S. Using molecular beacons to study dispersal of mRNPs from the gene locus. In: Hancock R, editors. The Nucleus. Humana Press; 2008. p. 91–103.
Alsmadi O, Al-Rubeaan K, Wakil SM, Imtiaz F, Mohamed G, Al-Saud H, et al. Genetic study of Saudi diabetes (GSSD): significant association of the KCNJ11 E23K polymorphism with type 2 diabetes. Diabetes Metab Res Rev. 2008;24(2):137–40.
Bhagwat A, Patel J, Chua T, Chan A, Cruz S, Aguilar GG. Detection of Salmonella species in foodstuffs. In: Marx A, Seitz O, editors. Molecular beacons: signalling nucleic acid probes, methods, and protocols. Humana Press; 2008. p. 33–43.
Blackard JT, Komurian-Pradel F, Perret M, Sodoyer M, Smeaton L, Clair JBS, et al. Intrahepatic cytokine expression is downregulated during HCV/HIV co‐infection. J Med Virol. 2006;78(2):202–7.
Chen W, Martinez G, Mulchandani A. Molecular beacons: a real-time polymerase chain reaction assay for detecting Salmonella. Anal Biochem. 2000;280(1):166–72.
Hadjinicolaou A, Farcas G, Demetriou V, Mazzulli T, Poutanen S, Willey B, et al. Development of a molecular-beacon-based multi-allelic real-time RT-PCR assay for the detection of human coronavirus causing severe acute respiratory syndrome (SARS-CoV): a general methodology for detecting rapidly mutating viruses. Arch Virol. 2011;156(4):671–80.
Culha M, Stokes DL, Griffin GD, Vo-Dinh T. Application of a miniature biochip using the molecular beacon probe in breast cancer gene BRCA1 detection. Biosens Bioelectron. 2004;19(9):1007–12.
Epstein JR, Leung APK, Lee K-H, Walt DR. High-density, microsphere-based fiber optic DNA microarrays. Biosens Bioelectron. 2003;18(5–6):541–6.
Kim H, Kane MD, Kim S, Dominguez W, Applegate BM, Savikhin S. A molecular beacon DNA microarray system for rapid detection of E. coli O157: H7 that eliminates the risk of a false negative signal. Biosens Bioelectron. 2007;22(6):1041–7.
Martinez K, Estevez MC, Wu Y, Phillips JA, Medley CD, Tan W. Locked nucleic acid based beacons for surface interaction studies and biosensor development. Anal Chem. 2009;81(9):3448–54.
El-Yazbi AF, Loppnow GR. Detecting UV-induced nucleic-acid damage. TrAC Trends Anal Chem. 2014;61:83–91.
Baker MB, Bao G, Searles CD. In vitro quantification of specific microRNA using molecular beacons. Nucleic Acids Res. 2012;40(2), e13.
Mao X, Xu H, Zeng Q, Zeng L, Liu G. Molecular beacon-functionalized gold nanoparticles as probes in dry-reagent strip biosensor for DNA analysis. Chem Commun. 2009;(21): 3065–3067.
Guo Z, Duan J, Yang F, Li M, Hao T, Wang S, et al. A test strip platform based on DNA-functionalized gold nanoparticles for on-site detection of mercury (II) ions. Talanta. 2012;93:49–54.
Xu H, Mao X, Zeng Q, Wang S, Kawde A-N, Liu G. Aptamer-functionalized gold nanoparticles as probes in a dry-reagent strip biosensor for protein analysis. Anal Chem. 2008;81(2):669–75.
He Y, Zeng K, Gurung AS, Baloda M, Xu H, Zhang X, et al. Visual detection of single-nucleotide polymorphism with hairpin oligonucleotide-functionalized gold nanoparticles. Anal Chem. 2010;82(17):7169–77.
He Y, Zhang X, Zhang S, Kris MK, Man FC, Kawde AN, et al. Visual detection of single-base mismatches in DNA using hairpin oligonucleotide with double-target DNA binding sequences and gold nanoparticles. Biosens Bioelectron. 2012;34(1):37–43.
Meng X, Xu M, Zhu J, Yin H, Ai S. Fabrication of DNA electrochemical biosensor based on gold nanoparticles, locked nucleic acid modified hairpin DNA and enzymatic signal amplification. Electrochim Acta. 2012;71:233–8.
Greene SB, Herschkowitz JI, Rosen JM. The ups and downs of miR-205: identifying the roles of miR-205 in mammary gland development and breast cancer. RNA Biol. 2010;7(3):300–4.
Majid S, Dar AA, Saini S, Yamamura S, Hirata H, Tanaka Y, et al. MicroRNA‐205–directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. 2010;116(24):5637–49.
Wiklund ED, Bramsen JB, Hulf T, Dyrskjøt L, Ramanathan R, Hansen TB, et al. Coordinated epigenetic repression of the miR‐200 family and miR‐205 in invasive bladder cancer. Int J Cancer. 2011;128(6):1327–34.
Lu Q, Lu C, Zhou G-P, Zhang W, Xiao H, Wang X-R. MicroRNA-221 silencing predisposed human bladder cancer cells to undergo apoptosis induced by TRAIL. Urol Oncol: Sem Orig Investig. 2010;28(6):635–41.
Conti A, Aguennouz MH, La Torre D, Tomasello C, Cardali S, Angileri F, et al. miR-21 and 221 upregulation and miR-181b downregulation in human grade II–IV astrocytic tumors. J Neurooncol. 2009;93(3):325–32.
Liu J, Lu Y. Preparation of aptamer-linked gold nanoparticle purple aggregates for colorimetric sensing of analytes. Nat Protocols. 2006;1(1):246–52.
Lesnik EA, Freier SM. Relative thermodynamic stability of DNA, RNA, and DNA:RNA hybrid duplexes: relationship with base composition and structure. Biochemistry. 1995;34(34):10807–15.
Lang BE, Schwarz FP. Thermodynamic dependence of DNA/DNA and DNA/RNA hybridization reactions on temperature and ionic strength. Biophys Chem. 2007;131(1–3):96–104.
Beni V, Zewdu T, Joda H, Katakis I, O’Sullivan CK. Gold nanoparticle fluorescent molecular beacon for low-resolution DQ2 gene HLA typing. Anal Bioanal Chem. 2012;402(3):1001–9.
Chen T, Wu CS, Jimenez E, Zhu Z, Dajac JG, You M, et al. DNA micelle flares for intracellular mRNA imaging and gene therapy. Angew Chem. 2013;125:2066–70.
Hurst SJ, Lytton-Jean AKR, Mirkin CA. Maximizing DNA loading on a range of gold nanoparticle sizes. Anal Chem. 2006;78(24):8313–8.
Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13(5):358–69.
Acknowledgments
Kamalodin Kor is grateful to the Ministry of Science, Research and Technology of Iran for financial support (project ref. 214140) to carry out this project at Linköping University, Sweden.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicting interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 2020 kb)
Rights and permissions
About this article
Cite this article
Kor, K., Turner, A.P.F., Zarei, K. et al. Structurally responsive oligonucleotide-based single-probe lateral-flow test for detection of miRNA-21 mimics. Anal Bioanal Chem 408, 1475–1485 (2016). https://doi.org/10.1007/s00216-015-9250-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-015-9250-9