Abstract
Pancreatic lipase plays essential roles in the digestion, transport, and processing of dietary lipids in humans. Inhibition of pancreatic lipase leading to the decrease of lipid absorption may be used for treating obesity. In the present study, a new approach of ultrafiltration coupled with high-performance liquid chromatography and quadrupole-time-of-flight mass spectrometry was established for rapidly detecting lipase binders from different extracts of medicinal plants. Rutin, a model inhibitor of lipase, was selected to optimize the screening conditions, including ion strength, temperature, pH, and incubation time. Meanwhile, the specificity of the approach was investigated by using denatured lipase and inactive compound emodin. The optimal screening conditions were as follows: ion strength 75 mM, temperature 37 °C, pH 7.4, and incubation time 10 min. Furthermore, linearity, accuracy, precision, and matrix effect of the approach were well validated. Finally, lipase binders were screened from different extracts of Dendrobium officinale by applying the established approach and were subsequently subjected to traditional lipase inhibitory assay. Eleven lipase inhibitors were identified, eight of which, namely naringenine, vicenin II, schaftoside, isoschaftoside, isoquercetrin, kaempferol 3-O-β-d-glucopyranoside, vitexin 2″-O-glucoside, and vitexin 2″-O-rhamnoside, were reported for the first time. In addition, docking experiments were performed to determine the preferred binding sites of these new lipase inhibitors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lipases, a subclass of the esterases, are the enzymes that catalyze the hydrolysis of fats. For humans, lipases play essential roles in the digestion, transport, and processing of dietary lipids (e.g., triglycerides, fats, oils). Human pancreatic lipase (HPL), the main enzyme that breaks down dietary fats in the human digestive system, converts triglyceride substrates found in ingested oils to monoglycerides and two fatty acids. Actually, HPL serves to process dietary lipids into simpler forms that can be more easily absorbed and transported throughout the body. Therefore, inhibition of pancreatic lipase leading to the decrease of the absorption of lipids may be used for treating obesity [1].
A plethora of plant extracts were reported to have potential inhibitory activities of pancreatic lipase. For instance, acteoside, a phenolic compound and one of the most abundant compounds in Chinese herbal tea kudingcha, showed a significant inhibitory effect against lipase and might have contribution to the antiobesity effect of kudingcha [2]. Similarly, polyphenols in oolong tea were identified to possess potential lipase inhibitory activities, including (−)-epigallocatechin, 3,5-di-O-gallate oolonghomo-bisflavan A, and theaflavin 3,3′-O-gallate [3]. In our previous work, three flavonoids from lotus leaf were identified to be lipase inhibitors and might be responsible for the weight-reducing effect of lotus leaf [4]. To the best of our knowledge, most of the natural lipase inhibitors were polyphenols, saponins, and terpenes. As the part of our continuing search for biologically active antiobesity agents from medicinal plants, a variety of plants from traditional Chinese medicine had been screened for their antilipase activity. Dendrobium officinale, which belongs to Orchidaceae, is well known for improving the symptoms of diabetes [5–7]. It was reported that the methanolic extract exhibited a pleiotropic effect on obesity-induced parameters and exerted a renoprotective effect in high-fat diet-fed mice [8]. Polysaccharides and phenols are the main constituents identified in D. officinale [9]. Therefore, it was hypothesized that the antilipase agents might exist in the plant.
Ultrafiltration, as a type of membrane filtration, has the ability to separate and concentrate target macromolecules. It has been widely applied in chemical and pharmaceutical manufacturing, food and beverage processing, and waste water treatment, and so far, particularly in the investigation of the binding degrees of biomacromolecule (i.e., human serum albumin or drug targets) because the binding of the agents to the target macromolecules is the prerequisite for the bioactivities of the agents. High-throughput affinity selection-mass spectrometry screening (i.e., ALIS system) has been successfully applied in screening inhibitors of the lipid phosphatase SHIP2 and ERK from combinatorial libraries [10, 11]. Meanwhile, Zhu et al. [12] established an ultrafiltration-based approach to screen ligands binding to cyclooxygenase-2 from radix Aconiti extracts and 25 ligands were identified. Munigunti et al. [13] selected PfTrxR as target and, finally, identified nine structurally diverse natural compounds with PfTrxR-inhibiting activity. Moreover, Wang et al. [14] developed an ultrafiltration-based method to screen xanthine oxidase inhibitors from S elaginella tamariscina. As a result, three ligands were validated to be real inhibitors. In summary, the advantage of the ultrafiltration-based approach lies in that it rapidly provides the binding information between the drug target and compounds. Meanwhile, the synergistic or antagonistic effects of two or more compounds can also be investigated by using such an approach.
In the present study, a new ultrafiltration-based approach was established to screen and identify antiobesity agents from the extracts of medicinal plants. First, the screening conditions (i.e., ion strength, incubation time, temperature, pH) were optimized. Second, the specificity, linearity, precision, accuracy, and matrix effect of the established approach were all validated. Third, the established approach was applied to screen antiobesity agents from the extracts of different organs (i.e., stem, leaf, and flower) of D. officinale. Finally, an in silico approach, which applied the docking method to interrogate the binding sites of ligands, was also employed to elucidate the intermolecular interaction between the ligand and enzyme.
Experimental
Chemicals and reagents
Lipase (porcine pancreas) and 4-methylumbelliferyl oleate were purchased from Sigma Chemical Co. Centrifugal ultrafiltration filters (Microcon YM-10, 10 kDa cutoff) were obtained from Millipore Co. HPLC-grade acetonitrile was obtained from Merck. Vicenin II, schaftoside, isoschaftoside, isoquercetrin, and trans-N-feruloyltyramine were purchased from Sichuan Weikeqi Biological Technology Co. Rutin, naringenine, kaempferol 3-O-β-d-glucopyranoside, vitexin 2″-O-glucoside, and vitexin 2″-O-rhamnoside were purchased from Shanghai Yuanye Bio-Technology Co. Syringoside, linolenic acid, and palmitic acid were purchased from Chengdu Must Biological Technology Co. All solutions and dilutions were prepared with ultrapure water from a Milli-Q water purification system.
Apparatus
The analytical apparatus included a Shimadzu HPLC system and a Q-TOF 5600-plus mass spectrometer equipped with Turbo V sources and a TurboIonSpray interface. Instrument control and data processing were conducted with Analyst software (version 1.5.2, AB Sciex) and PeakView software (version 1.2, AB Sciex).
Preparation of D. officinale extracts
The stem, leaf, and flower of D. officinale were collected from Yandang Mountain in Yueqing, Wenzhou City, Zhejiang Province, and authorized by an expert in the field. Voucher specimens have been deposited in the herbarium of Jiangsu Key Laboratory of Chinese Medicine Processing, Nanjing University of Chinese Medicine (No. DO131101). The sample preparation procedures were as follows: first, each organ (stem, leaf, or flower) of D. officinale was pulverized into powder and sieved (60 mesh). Second, an aliquot (1.0 g) of the powder was accurately weighed and refluxed in 50 mL of water at 100 °C for 2 h and then filtered. The filtrate was evaporated to dryness in vacuo and the residue was dissolved in 2 mL of water. Third, the solution was centrifuged at 13,400 rpm for 10 min and the supernatant was stored at −20 °C for the next experiment.
Optimization conditions for ligand screening
One known compound, i.e., rutin, was selected as a model inhibitor to optimize the screening conditions [15]. The screening procedures were as follows: test compound (20 μL in phosphate-buffered saline (PBS) buffer, 0.1 mg mL−1) and 160 μL of assay buffer consisting of 75 mM PBS buffer (pH 7.4) were placed into an Eppendorf (EP) tube and incubated for 2 min at 25 °C. The binding assay was initiated by the addition of lipase (20 μL in PBS, 1 mg mL−1) and incubated further for 60 min at 25 °C. After incubation, the binding mixture was filtered through an ultramembrane filter (Microcon YM-10) according to the modified method of Yang et al. [16] and centrifuged at 13,400 rpm for 20 min at 4 °C.
Four parameters were investigated in the current study to optimize the screening conditions. First, a gradient concentration (10, 30, 50, 75, and 100 mM) and a gradient pH (pH 5.7, 6.2, 6.8, 7.4, and 8.0) of phosphate buffers were used to study the influence of pH and ion strength on the binding experiments. Second, different incubation temperatures (21, 30, 35, 37, and 40 °C) and incubation times (10, 20, 30, 40, and 50 min) were evaluated. All the binding assays were performed in duplicate and analyzed as described above.
Validation of the established approach
Specificity
To assess the specificity of the established method, a quality control sample solution containing rutin and emodin (each 0.1 mg mL−1) was prepared and then subjected to the screening procedure as described above in the section “Optimization conditions for ligand screening.” The filtrates were sent for HPLC analysis to evaluate the potential interferences from other inactive constituents. Denatured lipase (boiled at 100 °C for 5 min) was used to incubate with the quality control (QC) sample to avoid the unspecific binding effect.
Linearity and lower limit of quantification
Calibration standards were prepared at eight concentrations (the final concentrations of rutin were 1.36, 2.72, 5.43, 10.86, 21.72, 43.44, 86.88, and 173.76 μM). The lower limits of quantification were determined by injecting a low concentration of standard solutions which generated S/N ratios of about 10 [17].
Accuracy, precision, and matrix effect
The QC sample (0.1 mg mL−1 for rutin) was prepared and analyzed for five times in the same day (intraday precision) and in three separate days (interday precision). Accuracy was expressed as the percentage deviation of the mean detected concentrations from the nominal concentrations and described as relative error (RE). The precision of the established approach at each QC concentration was determined as the relative standard deviation (RSD). The suitability of the precision and accuracy was assessed by the following criteria: the RSD should not exceed 15 % and the accuracy should be within 15 % of the actual values for the QC sample [17].
The matrix effect was evaluated by comparing the peak area of buffer solution spiked with standard solution of rutin to that of different organ extracts (stem, leaf, and flower) of D. officinale spiked with the standard solutions of rutin at three QC levels [17].
Screening for potential ligands from D. officinale
Twenty micrograms of powder from the extract of each organ (stem, leaf, or flower) was dissolved in 1 mL water and sonicated for 10 min. After that, the solution was centrifuged at 13,400 rpm for 10 min. The supernatant of the solution was transferred to a new EP tube. The incubation procedures were as follows: Each 20 μL of the supernatant was added into one of the two EP tubes, separately. Then, 20 μL lipase solution (1 mg mL−1) and 160 μL PBS solution (pH 7.4) were added successively to one EP tube and set as the experimental group. The other EP tube was added with 180 μL of PBS solution (pH 7.4) and set as the control group. Two EP tubes were incubated at 37 °C for 10 min. After that, the solutions in the tubes were transferred to ultramembrane filters and centrifuged at 13,400 rpm for 20 min. Finally, the ultrafiltrates were collected for high-performance liquid chromatography and quadrupole-time-of-flight mass spectrometry (HPLC-Q-TOF-MS) analysis. The binding capacities of constituents to lipase were determined by calculating the ratio (A 0 − A t)/A 0 × 100 %. A t and A 0 represented the total ion chromatogram (TIC) peak areas of the constituents in the experimental group and control group, respectively.
Experiments for characterization of constituents in the stem, leaf, and flower of D. officinale were performed by using high-performance liquid chromatography coupled with Q-TOF 5600-plus mass spectrometer. The HPLC analysis was performed on XBridge-C18 column (250 mm × 4.6 mm, 5 μm, Waters) and the HPLC conditions were as follows: mobile phase A: 0.1 % aqueous formic acid; mobile phase B: acetonitrile; gradient elution 0 min, 5 % B; 25 min, 25 % B; 30 min, 25 % B; 80 min, 100 % B; flow rate 1 mL min−1; and injection volume 10 μL.
The acquisition parameters for quadrupole-time-of-flight mass spectrometry were as follows: collision energy, −35 eV; ion spray voltage, −4.5 kV; nebulizer gas (gas 1), 55 psi; declustering potential, −60 V; heater gas (gas 2), 55 psi; turbo spray temperature, 550 °C; and curtain gas, 35 psi. A full MS-IDA-MS/MS scan was acquired on the samples, in which a full mass scan was also recorded, and the eight most abundant ions were selected for the MS2 spectra acquisition. The Q-TOF/MS scan range was set as m/z 100–1500.
Competitive binding experiments between high concentration of rutin and plant extracts on lipase
To verify the result of the ultrafiltration-based approach, competitive experiments were performed using the known ligand rutin. Rutin was added to the naringenine solution (0.1 mg mL−1) or incubation mixture of different extracts of D. officinale as a competitive ligand with a final concentration of 0.1 mg mL−1. Other incubation conditions were the same as described above.
Measurement of lipase inhibitory effects
Lipase inhibitory activity was measured according to the method previously described with slight modifications [4]. A substrate solution with a concentration of 0.1 M was prepared by sonication of 4-methylumbelliferyl oleate suspended in PBS buffer (pH 6.8). Samples were dissolved separately in PBS buffer to make 0.2 mg mL−1 solutions and then diluted to yield gradient concentrations of solutions. After that, the lipase (25 μL) and sample solutions (25 μL) in microplate wells were preincubated for 5 min at 25 °C. Then, 50 μL of substrate solution was added to each reaction mixture and incubated for 20 min at 25 °C. Finally, 100 μL of 0.1 M sodium citrate (pH 4.2) was added to stop the reaction. The amount of 4-methylumbelliferone released by lipase was measured with EnSpire® Multimode Plate Reader at an excitation wavelength of 320 nm and an emission wavelength of 450 nm. Orlistat was used as the positive control.
Docking study
Docking experiments were implemented using Autodock Vina (ver.1.1.2). The structures of constituents identified in D. officinale were drawn and sent to be visualized in chem3D Ultra 10.0 (Cambridge software). Semiempirical MM method was applied to minimize molecule energy in chem3D. After that, the molecule was converted to pdbqt format by our written script in-house. The crystal structure of porcine pancreatic lipase complexed with 3,6,9,12-tetraoxaicosan-1-ol (code ID: 1ETH) was retrieved from the RCSB Protein Data Bank [2]. Moreover, the cocrystallized ligand and water molecules were removed and the protein was outputted in pdbqt format. The center coordination of the docking box was 19.693, 0.054, and 17.628, and the box size was restricted to 16 × 16 × 16 with the spacing of 0.375 Å. Other parameters were left as default. Finally, the binding affinities of the constituents in D. officinale were calculated. The conformations with the most favorable free energy of binding were selected for analyzing the interactions between lipase and the constituents.
Results and discussion
Optimization conditions for ligand screening
Ionic strength is one of the main characteristics of a buffer, which will affect the distribution of charge on the exterior surfaces of the enzyme. As displayed in Fig. 1A, with the increase in the PBS concentration from 10 to 100 mM, the amount of binding rutin arose first and then reached the highest level at 75 mM. This phenomenon can be explained by the fact that a certain concentration of ions may assist in the binding of rutin to lipase, but if the concentration of ions exceeds the threshold (i.e., 75 mM), the effect of ions will be reversed, which will neutralize the surface charge of lipase and weaken the electrostatic interaction between rutin and lipase.
Effects of incubation parameters on interaction between rutin and lipase. (A) Ion strength varied from 10 to 100 mM, incubation temperature 37 °C, incubation time 30 min, pH value 6.8; (B) incubation temperature varied from 21 to 40 °C, ion strength 10 mM, incubation time 30 min, pH value 6.8; (C) pH values varied from 5.7 to 8.0, incubation temperature 37 °C, ion strength 10 mM, incubation time 30 min; and (D) incubation time varied from 10 to 50 min, pH value 7.4, incubation temperature 37 °C, ion strength 10 mM
The pH of the buffer solution is another important factor affecting the surface charge and activity of the enzyme. Generally, each enzyme has its optimum pH to catalyze its substrate. The optimum pH of porcine pancreatic lipase is 7.4. The curve in Fig. 1B represents the effect of pH on the binding of rutin to lipase. It clearly showed that the binding degree of rutin was relatively higher at pH around 6.2 or 7.4 than that at other pH values. The isoelectric point (pI) of lipase is about 6.44, and when the solution pH value is above the pI, the lipase has a net negative surface charge, which favors electrostatic interaction with rutin (pKa 6.8) that has less negative charge at pH 7.4 than at pH 8.0.
The length of incubation time is a factor affecting the binding degree. As shown in Fig. 1C, the highest binding degree was achieved after incubation for 10 min. An incubation time of 10 min was sufficient for the interaction between lipase and rutin. The increasing incubation time from 20 to 50 min exerted no significant effect on the interaction between lipase and rutin.
The incubation temperature was also investigated by varied temperatures ranging from 21 to 40 °C. As shown in Fig. 1D, 37 °C is the most suitable temperature for the incubation due to the largest binding degree at the temperature. Our results were in agreement with the report that the optimal reaction temperature was 37 °C for soluble lipase. In summary, the optimal conditions for screening were as follows: ion concentration 75 mM, pH 7.4, incubation time 10 min, and incubation temperature 37 °C.
Validation of the established approach
Specificity
Rutin, which belongs to flavonoid-O-diglycoside, is a well-known lipase inhibitor. Emodin, which is an anthraquinone, shows a negative lipase inhibitory effect. Therefore, rutin and emodin were chosen to investigate the specificity of the established approach and the result was shown in Fig. 2. After incubation with the lipase, the peak area of rutin was decreased, while the peak area of emodin remained unchanged. It revealed that the established approach had good specificity with no interference of the inactive compound emodin. To avoid the nonspecific binding, denatured lipase was used to be incubated with the mixture of rutin and emodin. The peak areas of rutin and emodin remained unchanged, which indicated that none of the unspecific binding to the denatured lipase was observed.
(A) HPLC chromatograms of mixture of rutin and emodin before (black) and after (red) interaction with pancreatic lipase (PL). (B) HPLC chromatograms of mixture of rutin and emodin before (black) and after (red) interaction with denatured pancreatic lipase. HPLC conditions were as follows: reversed-phase XBridge-C18 column (250 mm × 4.6 mm, 5 μm, Waters); mobile phase consisting of solvent A (0.1 % formic acid in water) and solvent B (acetonitrile), which was programmed as follows: 0–10 min, 10–35 % solvent B; 10–25 min, 35–100 % solvent B; 25–30 min, 100 % solvent B; flow rate 1 mL min−1; column temperature 30 °C; injection volume 10 μL
Linearity and lower limits of quantitation (LLOQs)
Linear ranges, regression equations, limit of quantification, and correlation coefficients obtained from typical calibration curves were as follows: 1.36–173.76 μM, y = 6156.4x − 3457.2, 1.36 μM, and 0.9997. The standard curve of rutin showed good linearity with coefficients of correlation (r) of 0.9997. The LLOQs were appropriate for quantitative detection of analytes in the enzymatic studies.
Precision and accuracy
The intra- and interday precisions of rutin were 0.39 and 0.40 %, respectively, and the accuracies ranged from 6.8 to 7.6 %, which fulfilled the requirements of an analytical assay.
Matrix effect
The relative standard deviations for rutin spiked into D. officinale extract were listed in Table 1. The recoveries of D. officinale extract spiked with standard solutions were in the ranges of 87 to 110 %. The results confirmed that the established approach was free from any matrix effect.
Characterization of constituents in D. officinale using HPLC-Q-TOF-MS
The total ion current chromatograms in negative electrospray ionization (ESI) mode of the stem, leaf, and flower were displayed in Fig. 3. A total of 53 compounds were identified and their detailed information was shown in Table 2. The retention time and fragmentation information of compounds 18, 22, 27, 31, 32, 33, 34, 38, 45, 46, and 52 were compared with those of standard compounds. The major constituents of D. officinale were phenolics, flavonoids, pyrrole derivatives, and unsaturated fatty acids.
TIC chromatograms of extracts of the stem (A), leaf (B), and flower (C) of Dendrobium officinale before (black) and after (red) interaction with pancreatic lipase (PL). HPLC conditions were as follows: reversed-phase XBridge-C18 column (250 mm × 4.6 mm, 5 μm, Waters); mobile phase consisting of solvent A (0.1 % aqueous formic acid) and solvent B (acetonitrile), which was programmed as follows: 0–25 min, 5–25 % solvent B; 25–30 min, 25 % solvent B; 30–80 min, 25–100 % solvent B; flow rate 1 mL min−1; column temperature 30 °C; injection volume 10 μL
Phenolic compounds are the major bioactive constituents in D. officinale. Fifteen phenolic compounds were identified in this study (compounds 2–10, 13, 30, 41–43, 45). Fragmentations with loss of 162 Da (hexose) or 294 Da (hexose and pentose) were the characteristic ions of these phenolic compounds. As shown in Fig. S1A in the Electronic Supplementary Material (ESM), compound 4 at the retention time of 7.00 min displayed [M−H]− ion at m/z 329. Fragment ion at m/z 167 in MS2 spectrum was produced by losing one molecule of hexose. In comparison with the reference [18], compound 4 was unambiguously deduced as vanillic acid glucoside. Similarly, compounds 2, 5, 7, and 9 shared the same neutral loss of 162 Da, which indicated that a hexose group was conjugated to their skeleton. By comparing with related references, these compounds were deduced as vanilloloside [19], leonuriside A [20], glucosyringic acid [21], and koaburside [22]. Compound 6 showed the [M−H]− ion at m/z 331 and neutral loss of 152 Da in MS2 spectrum, which was consistent with the fragmentation pathway of koaburaside [23]. Compounds 8 and 10 showed the [M−H]− ions at m/z 447 and m/z 491, respectively. The MS2 spectra of compounds 8 and 10 were shown in Fig. S1B and S1C in the ESM. Both of them exhibited the same neutral loss of 294 and 338 Da, which corresponded to losses of the hexose-pentose group and carboxyl group. Compared with the literature [24], compounds 8 and 10 were identified as 2,6-dihydroxybenzoic acid 2-O-β-d-apiofuranosyl-(1 → 2)-β-d-glucopyranoside and 2,3-dimethoxybenzoic acid 4-O-β-d-apiofuranosyl-(1 → 2)-β-d-glucopyranoside, respectively. Similarly, compound 13 showed the [M−H]− ions at m/z 417 and produced ion at m/z 152 with a neutral loss of 265 Da in MS2 spectrum, which indicated that a pentose-pentose group was conjugated to the skeleton. The retention time of compound 13 was a little later than that of compound 8. Therefore, compound 13 was deduced as 2,6-dihydroxybenzoic acid 2-O-β-d-apiofuranosyl-(1 → 2)-β-d-xylopyranoside. The five remaining compounds (i.e., 30, 41–43, 45) were deduced by comparing with the fragmentation information in the literature [25–28].
Flavonoids, especially flavone C-glycosides, are the second major active constituents of D. officinale, such as vicenin 1, schaftoside, vitexin 2″-O-rhamnoside, and so on [29]. Eleven flavone C-glycosides and four flavone O-glycosides were identified in this study (compounds 18, 21–23, 25, 27, 29, 31–35, 37, 38, 52). Flavone C-glycoside usually produces neutral loss of 120 Da, which results from the cross-ring cleavage of hexose at 0, 2 positions in negative ESI mode. As presented in Fig. S2A in the ESM, compound 18 produced [M−H]− ion at m/z 593 and fragment ions at m/z 473, m/z 383, and m/z 353 in MS2 spectrum. The ion at m/z 473 was a result of the cross-ring cleavage at 0, 2 positions of one hexose group, and m/z 353 was the suggestion of the cross-ring cleavage at 0, 2 positions of two hexose groups simultaneously. The cross-ring cleavage at 0, 2 sites and 0, 3 sites of two hexose groups yielded the product ion at m/z 383. Compared with the standard, compound 18 was unambiguously assigned as vicenin 2. Similarly, as presented in the ESM in Fig. S2B and S2C, compounds 22 and 27 displayed the [M−H]− ion at m/z 563 (C26H28O14), as well as the characteristic neutral loss of 180 Da yielding product ion at m/z 383. The typical neutral losses of 60 Da (m/z 503) and 90 Da (m/z 473) corresponded to the cross-ring cleavage at 0, 3 sites of a pentose and hexose, respectively. Compared with the standards, compounds 22 and 27 were unambiguously assigned as isoschaftoside and schaftoside, respectively. Similarly, compounds 21, 23, 25, 35, and 37 showed the same [M−H]− ions at m/z 563 (C26H28O14). In comparison with the references, these compounds were tentatively deduced as vicenin 1, isoschaftoside isomer, isoschaftoside isomer, vicenin 3, and neoschaftoside, respectively. Compounds 31 and 32 were unambiguously assigned as vitexin-2″-O-glucoside and vitexin 2″-O-rhamnoside by comparing with the standards. Compounds 29 and 31 shared the same [M−H]− ion at m/z 593 and product ions at m/z 413 and m/z 293. Therefore, compound 29 was tentatively deduced as vitexin-2″-O-glucoside isomer.
Compounds 33 and 34 were assigned as quercetin glycosides by their product ions at m/z 300 and m/z 301 in negative MS2 spectra. The kinds of sugars conjugated with flavonoid aglycone are easily assigned by their fragment ion information in MS2 spectrum, which result from the losses of 162 Da for a hexose (a glucose or a galactose), 146 Da for a rhamnose, and 132 Da for a pentose (an apiose, an arabinose, or a xylose). Compared with the standards, compounds 33 and 34 were unambiguously assigned as quercetin-3-O-rutinoside (rutin) and quercetin 3-O-β-d-glucoside (isoquercetrin), respectively. Similarly, compounds 38 and 52 were unambiguously deduced as kaempferol 3-O-β-d-glucopyranoside and naringenine by comparing with the standards.
Nitrogen-containing compounds were rarely found in the Dendrobium species, such as trans-N-coumaroyltyramine and trans-N-feruloyltyramine [30]. According to the nitrogen rule, seven compounds were identified to be nitrogen-containing compounds (compounds 1, 19, 24, 44, 46, 47, and 50). Compound 46 showed the ion [M−H]− at m/z 312 and product ions at m/z 148 and m/z 178. Compared with the standard, compound 46 was unambiguously assigned as trans-N-feruloyltyramine. Similarly, compound 44 was plausibly identified as trans-N-coumaroyltyramine [31]. The MS2 spectrum of compound 44 was shown in ESM Fig. S3A. The major product ions at m/z 162 and m/z 119 were resulted from the cleavage of the C-N bond and C-C bond around the amide moiety. As shown in ESM Fig. S3B, compounds 19, 47, and 50 had the same fragment ions at m/z 94 (C5H4NO) and m/z 66 (C4H4N) in MS2 spectra, which indicated that they shared the same pyrrole skeleton. Compared with the reference [32], compounds 19, 47, and 50 were tentatively deduced as 2-formyl-5-(hydroxymethyl)-1H-pyrrole-1-butanoic acid, 2-formyl-5-(hydroxyl-methyl)-1H-pyrrole-1-hexanoic acid, and 5-formyl-1-methyl-3-[(3-methylphenoxy) methyl]-1H-pyrrole-2-carboxylic acid, respectively. The loss of butyric acid group yielded the product ion at m/z 124. In particular, compounds 19, 47, and 50 were reported in the Dendrobium species for the first time. As presented in the ESM in Fig. S3C, compound 24 produced the [M−H]− ion at m/z 250 and the major fragment ion at m/z 132. Compound 24 was tentatively assigned as N-phenacetyl-l-aspartic acid by comparing with the reference [33]. The cleavage of the amide moiety led to the product ion at m/z 132.0307. Further loss of carboxyl group gave the product ion at m/z 88.0422.
Screening for ligands binding to lipase using the established approach
The ultrafiltration approach was applied to screen lipase binders in D. officinale and the results were shown in Fig. 3. The binding capacities of constituents in the stem, leaf, and flower were shown in Fig. 4. According to ref. [34], the authors set a rule for ligand selection as follows: If the peak area of a ligand in the incubation with active enzyme exceeded the peak area observed in the control incubation by at least twofold, it can be sent for functional assay. By integration of the area of the ligand in our experiments, we found that this rule was also suitable for our screening system. We ranked the binding capacities of 53 compounds and only showed the top six binding constituents with binding capacities above 67 %. The binding capacities of other compounds could also be found in Fig. 4. The major constituents of D. officinale were phenolics, flavonoids, pyrrole derivatives, and unsaturated fatty acids. It seems like that the optimum incubation condition optimized using rutin may be suitable for the binding between flavonoids and lipase. However, the results showed that the top six binding constituents were naringenine (52, 81.05 %), trans-N-coumaroyltyramine (44, 77.84 %), 9,10,13-trihydroxy-(E)-11-octadecenoic acid (53, 77.32 %), trans-N-feruloyltyramine (46, 76.98 %), 2,6-dihydroxybenzoic acid 2-O-β-d-apiofuranosyl-(1 → 2)-β-d-xylopyranoside (13, 71.87 %), and 2,6-dihydroxybenzoic acid 2-O-β-d-apiofuranosyl-(1 → 2)-β-d-glucopyranoside (8, 70.12 %), respectively. Among them, only naringenine belongs to the class of flavonoids. trans-N-Coumaroyltyramine and trans-N-feruloyltyramine were amide derivatives. 9,10,13-Trihydroxy-(E)-11-octadecenoic acid was an unsaturated fatty acid. 2,6-Dihydroxybenzoic acid 2-O-β-d-apiofuranosyl-(1 → 2)-β-d-xylopyranoside (13, 71.87 %) and 2,6-dihydroxybenzoic acid 2-O-β-d-apiofuranosyl-(1 → 2)-β-d-glucopyranoside (8, 70.12 %) were phenolic compounds. The bias of the proposed approach does exist; however, except for flavonoids, other classes of lipase binders were also identified using the proposed approach. The binding capacities of constituents in stem were larger than those in leaf and flower, which indicated that the lipase inhibitory effect of the stem extract might be better. Competitive binding experiments between a high concentration of rutin and naringenine or plant extracts on lipase were performed. As shown in Figs. S4 and S5 in the ESM, the peak areas of all the ligands were increased in the presence of a high concentration of rutin. The results revealed that these ligands in the extracts of D. officinale bound to the active site of the lipase.
Binding capacities of constituents in different extracts (stem, leaf, and flower) of Dendrobium officinale. Binding capacities of constituents to lipase were determined by calculating the ratio (A 0 − A t)/A 0 × 100 %. A 0 and A t represented TIC peak areas of constituents before and after interaction with pancreatic lipase (PL), respectively
With the development of affinity selection approaches, new bioactive compounds were rapidly discovered from natural medicines. In our study, the proposed approach could be employed to directly identify ligands that interacted with the lipase from different extracts of D. officinale. New compounds were also identified, such as compounds 12, 19, 20, 39, 47, 48, 49, and 50. However, due to the lack of standard compounds, the structures and their inhibitory effects were not well elucidated at present. In the future, their structures and inhibitory effects will undoubtedly be elucidated with the endeavor of phytochemistry research.
Lipase inhibitory assay
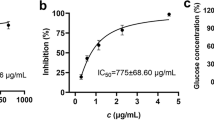
In our study, 13 active ligands were evaluated for their inhibitory activities against lipase using traditional lipase inhibitory assay. Orlistat served as the positive control. The assay results were listed in Table 3. Surprisingly, naringenine, which had the largest binding degree, only exhibited a weak lipase inhibitory activity (IC50 291.89 ± 2.36 μM). trans-N-Feruloyltyramine even showed less than 10 % inhibition at the concentration of 200 μM. Due to the unavailability of 9,10,13- trihydroxy-(E)-11-octadecenoic acid in commercial, two fatty acids, which had been reported in D. officinale, were purchased and tested for their lipase inhibitory activity. Linolenic acid, also called 9,12,15-octadecatrienoic acid, was an unsaturated fatty acid. The IC50 values of linolenic acid and palmitic acid were 32.44 ± 2.19 and 119.86 ± 2.60 μM, respectively. Five flavone C-glycosides and two flavone O-glycosides were purchased and tested for lipase inhibitory effect. Rutin and isoschaftoside showed moderate lipase inhibitory effects with IC50 values of 57.44 ± 3.18 and 63.83 ± 0.89 μM, respectively. Syringoside, which belongs to the phenolic compounds, showed less than 10 % inhibition at the concentration of 200 μM.
Docking studies
The enzyme-binding capacity is not equivalent to inhibitory effects due to the fact that the enzymatic binding sites are not necessary for substrate transformation [17]. Docking studies were performed to investigate the interaction between the active constituents and the active site of the lipase. Both 3D and 2D of the best-docked conformations of lipase-naringenine complexes were shown in Fig. S6 in the ESM. Naringenine was found in the vicinity of Asp-80, Val-260, Leu-265, Ile-79, His-152, Arg-257, Phe-78, Ser-153, and Tyr-115 with a free binding energy of −9.6 kcal mol−1. The hydrogen bond was formed between the hydroxyl group of naringenine and Asp-80* of lipase as well as the hydroxyl group of naringenine and Arg-257* of lipase. Hydrophobic interaction was observed between the C-ring of naringenine and Phe-216 of lipase. The catalytic sites of porcine lipase (Ser-153, Asp-177, and His-264) [35] surround naringenine directly. Thus, the lipase inhibitory activity of naringenine is a result of blocking the catalytic site. The preferred binding sites of rutin, isoschaftoside, and 9,10,13-trihydroxy-(E)-11-octadecenoic acid were shown in Fig. S7 in the ESM. In rutin-lipase adduct, rutin was surrounded by Asp-80* (H-bonding), Arg-257, Phe-78, Ile-79, Val-260, Thr-256, Trp-253, Tyr-115, Ala-179, and Phe-236 (π-π interaction) with a free binding energy of −8.5 kcal mol−1. In isoschaftoside-lipase adduct, isoschaftoside was located in the vicinity of His-264, Val-260, Trp-253, Arg-257, Thr-256, Ile-79, Phe-78, Phe-216, and Ser-153 with a free binding energy of −6.5 kcal mol−1. Noncovalent interactions between phenolic compounds and proteins are hydrophobic in nature and may stabilize with hydrogen binding. Both rutin and isoschaftoside formed π-π interaction (with benzene ring of Phe-216) and one hydrogen bond with the lipase. These noncovalent interactions may alter the enzyme molecular conformation. The conformational mobility of the protein structure can influence the catalytic activity of enzymes. Thus, rutin or isoschaftoside might act as a pancreatic lipase inhibitor due to the protein conformational change with lipase binding to rutin or isoschaftoside. In 9,10,13-trihydroxy-(E)-11-octadecenoic acid-lipase adduct, 9,10,13-trihydroxy-(E)-11-octadecenoic acid was surrounded by Leu-265, Arg-257* (H-bonding), Ala-261, His-264, Asp-80, Phe-78* (H-bonding), Ser-153* (H-bonding), Tyr-115, Phe-216, and Val-260 with a free binding energy of −6.8 kcal mol−1. The binding pattern of 9,10,13-trihydroxy-(E)-11-octadecenoic acid is different with that of phenolic compounds. 9,10,13-Trihydroxy-(E)-11-octadecenoic acid formed four hydrogen bonds with the lipase: one between the carbonyl group of Ser-153 and the hydroxyl group of ligand, one between the hydroxyl group of Ser-153 and the carbonyl group of ligand, and the others between the hydroxyl group of ligand and Phe-78 and Arg-257. The hydrogen bonding with the Ser-153 could block the catalytic site of lipase. Thus, 9,10,13-trihydroxy-(E)-11-octadecenoic acid exerted lipase inhibitory effects by direct blocking of the catalytic site. In summary, naringenine formed more stable complexes with lipase than those of rutin and isoschaftoside and the order is naringenine > rutin > 9,10,13-trihydroxy-(E)-11-octadecenoic acid > isoschaftoside. It was assumed that the interaction of ligand-enzyme would be in charge of inhibiting the enzyme activity.
Conclusions
In the study, we have demonstrated a new approach of ultrafiltration coupled with HPLC-MS for discovering lipase binders in the stem, leaf, and flower of D. officinale. The newly established approach provides an alternative to the methods for screening lipase binders from the medicinal plants and will accelerate the discovery of antiobesity drugs. Particularly, 11 lipase inhibitors were identified, eight of which were reported for the first time. Meanwhile, the different interaction modes between the inhibitors and lipase may be responsible for their varying activities.
References
Birari RB, Bhutani KK (2007) Pancreatic lipase inhibitors from natural sources: unexplored potential. Drug Discov Today 12:879–889
Wu X, He W, Zhang H, Li Y, Liu Z, He Z (2014) Acteoside: a lipase inhibitor from the Chinese tea Ligustrum purpurascens kudingcha. Food Chem 142:306–310
Nakai M, Fukui Y, Asami S, Toyoda-Ono Y, Iwashita T, Shibata H, Mitsunaga T, Hashimoto F, Kiso Y (2005) Inhibitory effects of oolong tea polyphenols on pancreatic lipase in vitro. J Agric Food Chem 53:4593–4598
Tao Y, Zhang Y, Wang Y, Cheng Y (2013) Hollow fiber based affinity selection combined with high performance liquid chromatography-mass spectroscopy for rapid screening lipase inhibitors from lotus leaf. Anal Chim Acta 785:75–81
Ng TB, Liu J, Wong JH, Ye X, Wing Sze SC, Tong Y, Zhang KY (2012) Review of research on Dendrobium, a prized folk medicine. Appl Microbiol Biotechnol 93:1795–1803
Gong CY, Yu ZY, Lu B, Yang L, Sheng YC, Fan YM, Ji LL, Wang ZT (2014) Ethanol extract of Dendrobium chrysotoxum Lindl ameliorates diabetic retinopathy and its mechanism. Vascul Pharmacol 62:134–142
Pan LH, Li XF, Wang MN, Zha XQ, Yang XF, Liu ZJ, Luo YB, Luo JP (2014) Comparison of hypoglycemic and antioxidative effects of polysaccharides from four different Dendrobium species. Int J Biol Macromol 64:420–427
Lee W, Eom DW, Jung Y, Yamabe N, Lee S, Jeon Y, Hwang YR, Lee JH, Kim YK, Kang KS, Kim SN (2012) Dendrobium moniliforme attenuates high-fat diet-induced renal damage in mice through the regulation of lipid-induced oxidative stress. Am J Chin Med 40:1217–1228
Xu J, Li SL, Yue RQ, Ko CH, Hu JM, Liu J, Ho HM, Yi T, Zhao ZZ, Zhou J, Leung PC, Chen HB, Han QB (2014) A novel and rapid HPGPC-based strategy for quality control of saccharide-dominant herbal materials: Dendrobium officinale, a case study. Anal Bioanal Chem 406:6409–6417
Annis DA, Cheng CC, Chuang CC, McCarter JD, Nash HM, Nazef N, Rowe T, Kurzeja RJ, Shipps GW Jr (2009) Inhibitors of the lipid phosphatase SHIP2 discovered by high-throughput affinity selection-mass spectrometry screening of combinatorial libraries. Comb Chem High Throughput Screen 12:760–771
Deng YQ, Shipps GW, Cooper A Jr, English JM, Annis DA, Carr D, Nan Y, Wang T, Zhu HY, Chuang CC, Dayananth P, Hruza AW, Xiao L, Jin WH, Kirschmeier P, Windsor WT, Samatar AA (2014) Discovery of novel, dual mechanism ERK inhibitors by affinity selection screening of an inactive kinase. J Med Chem 57:8817–8826
Zhu H, Liu S, Li X, Song F, Liu Z (2013) Bioactivity fingerprint analysis of cyclooxygenase-2 ligands from radix Aconiti by ultrafiltration-UPLC-MSn. Anal Bioanal Chem 405:7437–7445
Munigunti R, Mulabagal V, Calderón AI (2011) Screening of natural compounds for ligands to PfTrxR by ultrafiltration and LC-MS based binding assay. J Pharm Biomed Anal 55:265–271
Wang J, Liu S, Ma B, Chen L, Song F, Liu Z, Liu CM (2014) Rapid screening and detection of XOD inhibitors from S. tamariscina by ultrafiltration LC-PDA-ESI-MS combined with HPCCC. Anal Bioanal Chem 406:7379–7387
Passicos E, Santarelli X, Coulon D (2004) Regioselective acylation of flavonoids catalyzed by immobilized Candida antarctica lipase under reduced pressure. Biotechnol Lett 26:1073–1076
Yang Z, Zhang Y, Sun L, Wang Y, Gao X, Cheng Y (2012) An ultrafiltration high-performance liquid chromatography coupled with diode array detector and mass spectrometry approach for screening and characterising tyrosinase inhibitors from mulberry leaves. Anal Chim Acta 719:87–95
Tao Y, Chen Z, Zhang Y, Wang Y, Cheng Y (2013) Immobilized magnetic beads based multi-target affinity selection coupled with high performance liquid chromatography-mass spectrometry for screening anti-diabetic compounds from a Chinese medicine “Tang-Zhi-Qing”. J Pharm Biomed Anal 78–79:190–201
Abu-Reidah IM, Arraez-Roman D, Lozano-Sanchez J, Segura-Carretero A, Fernandez-Gutierrez A (2013) Phytochemical characterisation of green beans (Phaseolus vulgaris L.) by using high-performance liquid chromatography coupled with time-of-flight mass spectrometry. Phytochem Anal 24:105–116
Zhao C, Liu Q, Halaweish F, Shao B, Ye Y, Zhao W (2003) Copacamphane, picrotoxane, and alloaromadendrane sesquiterpene glycosides and phenolic glycosides from Dendrobium moniliforme. J Nat Prod 66:1140–1143
Meng Z, Dong H, Wang C, Guo S (2013) Chemical constituents of Dendrobium devonianum. Chin Pharm J 48:855–859
He Y, Dong X, Jia X, Li M, Yuan T, Xu H, Qin L, Han T, Zhang Q (2015) Qualitative and quantitative analysis on chemical constituents from Curculigo orchioides using ultra high performance liquid chromatography coupled with electrospray ionization quadrupole time-of-flight tandem mass spectrometry. J Pharm Biomed Anal 102:236–245
Sahakitpichan P, Chadmuk P, Disadee W, Chimnoi N, Ruchirawat S, Kanchanapoom T (2014) New trans- and cis-p-coumaroyl flavonol tetraglycosides from the leaves of Mitragyna rotundifolia. Phytochem Lett 8:65–68
Li Y, Wang C, Wang F, Dong H, Guo S, Yang J, Xiao P (2010) Phenolic components and flavanones from Dendrobium candidum. Chin Pharm J 45:975–979
Ono H, Kuwahara Y, Nishida R (2004) Hydroxybenzoic acid derivatives in a nonhost rutaceous plant, Orixa japonica, deter both oviposition and larval feeding in a rutaceae-feeding swallowtail butterfly, Papilio xuthus L. J Chem Ecol 30:287–301
Itokawa H, Oshida Y, Ikuta A, Inatomi H, Adachi T (1982) Phenolic plant growth inhibitors from the flowers of Cucurbita pepo. Phytochemistry 21:1935–1937
Zi J, Li S, Liu M, Gan M, Lin S, Song W, Zhang Y, Fan X, Yang Y, Zhang J, Shi J, Di D (2008) Glycosidic constituents of the tubers of Gymnadenia conopsea. J Nat Prod 71:799–805
Xu SJ, Yang L, Zeng X, Zhang M, Wang ZT (2006) Characterization of compounds in the Chinese herbal drug Mu-Dan-Pi by liquid chromatography coupled to electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom 20:3275–3288
Xiong L, Cao Z, Peng C, Li X, Xie X, Zhang T, Zhou Q, Yang L, Guo L (2013) Phenolic glucosides from Dendrobium aurantiacum var. denneanum and their bioactivities. Molecules 18:6153–6160
Zhou GF, Lv GY (2012) Study on eight flavone C-glycosides in Dendrobium officinale leaves and their fragmentation pattern by HPLC-DAD-ESI-MS. Chin Pharm J 47:13–19
Xu J, Zhao WM, Qian ZM, Guan J, Li SP (2010) Fast determination of five components of coumarin, alkaloids and bibenzyls in Dendrobium spp. using pressurized liquid extraction and ultra-performance liquid chromatography. J Sep Sci 33:1580–1586
Zhang J, Guan S, Sun J, Liu T, Chen P, Feng R, Chen X, Wu W, Yang M, Guo DA (2015) Characterization and profiling of phenolic amides from Cortex Lycii by ultra-high performance liquid chromatography coupled with LTQ-Orbitrap mass spectrometry. Anal Bioanal Chem 407:581–595
Zhou Z, Luo J, Pan K, Kong L (2014) A new alkaloid glycoside from the rhizomes of Aristolochia fordiana. Nat Prod Res 28:1065–1069
Gianfagna T, Davies P (1980) N-Benzoylaspartate and N-phenylacetylaspartate from pea seeds. Phytochemistry 19:959–961
Cao H, Yu R, Choi Y, Ma ZZ, Zhang H, Xiang W, Lee DY, Berman BM, Moudgil KD, Fong HH, van Breemen RB (2010) Discovery of cyclooxygenase inhibitors from medicinal plants used to treat inflammation. Pharmacol Res 61:519–524
Hermoso J, Pignol D, Kerfelec B, Crenon I, Chapus C, Fontecilla-Camps JC (1996) Lipase activation by nonionic detergents. The crystal structure of the porcine lipase–colipase–tetraethylene glycol monooctyl ether complex. J Biol Chem 271:18007–18016
Acknowledgments
This study was supported by the Natural Science Foundation for the Youth of Jiangsu Province (No. BK20140963) and the Priority Academic Program Development of Jiangsu Higher Education Institution (PAPD).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 5188 kb)
Rights and permissions
About this article
Cite this article
Tao, Y., Cai, H., Li, W. et al. Ultrafiltration coupled with high-performance liquid chromatography and quadrupole-time-of-flight mass spectrometry for screening lipase binders from different extracts of Dendrobium officinale . Anal Bioanal Chem 407, 6081–6093 (2015). https://doi.org/10.1007/s00216-015-8781-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-015-8781-4