Abstract
The spatial distribution of an anticancer drug and its intended target within a tumor plays a major role on determining how effective the drug can be at tackling the tumor. This study provides data regarding the lateral distribution of sunitinib, an oral antiangiogenic receptor tyrosine kinase inhibitor using an in vitro animal model as well as an in vitro experimental model that involved deposition of a solution of sunitinib onto tissue sections. All tumor sections were analyzed by matrix-assisted laser desorption/ionization mass spectrometry imaging and compared with subsequent histology staining. Six tumors at four different time points after commencement of in vivo sunitinib treatment were examined to observe the patterns of drug uptake. The levels of sunitinib present in in vivo treated tumor sections increased continuously until day 7, but a decrease was observed at day 10. Furthermore, the in vitro experimental model was adjustable to produce a drug level similar to that obtained in the in vivo model experiments. The distribution of sunitinib in tissue sections treated in vitro appeared to agree with the histological structure of tumors, suggesting that this approach may be useful for testing drug update.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is one of the most common causes of cancer-related death in the Western world [1, 2] with a strong environmental association [3]. While the 5-year survival rate for metastatic colorectal cancer is approximately 10 % [1], chemotherapy alone can produce a median survival of about 20 months [4–6]. Fluorouracil has been commonly used to treat CRC for a long time, but new drugs appeared recently, including cetuximab, bevacizumab, irinotecan, oxaliplatin, and capecitabine. The inhibition of angiogenesis is one of the most promising therapies for CRC as the inhibition of vascular endothelial factor (VEGF) can significantly improve survival in patients treated with the humanized monoclonal antibody, bevacizumab [7, 8]. Further studies have revealed that the coordinated inhibition of platelet-derived growth factors (PDGF) is essential in modulating angiogenesis and inducing production of VEGF, leading to reduced tumor vascularization and improved tumor regression [9, 10].
Sunitinib is an oral antiangiogenic receptor tyrosine kinase inhibitor (previously known as SU11248, Pfizer®) [11, 12]. It can block the intracellular signaling and kinase-dependent functions of VEGF and PDGF receptors. Inactivation of these signaling pathways is thought to reduce angiogenesis and tumor growth [11]. Sunitinib is commonly used or tested in the treatment of gastrointestinal stromal tumor [13], pancreatic neuroendocrine tumors [14], meningioma [15], renal cell carcinoma [11], metastatic breast cancer [16], and non-small cell lung cancer [17]. In preclinical CRC models, sunitinib demonstrated a single-agent antitumor activity at well-tolerated doses. However, a phase II trial of sunitinib was discontinued due to its efficacy [18], underlining the importance of the ability to predict efficacy during drug development.
A key event in the drug development process is the linking of specific disease mechanisms to targets, which is interconnected with the target-drug binding properties and directly related to the pharmacokinetic and the pharmacodynamic properties of any given drug compound [19]. It is important that an in vitro experimental model closely correlates with the corresponding in vivo model in order to be able to prove its applicability. The spatial localization of the drug is dictated by the binding affinity to its target molecules in tissue, most commonly protein receptors [19]. Matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI) is a technology that allows for the simultaneous mapping of peptides, proteins, or molecules in thin-tissue sections with a lateral resolution of up to 30–50 μm [20]. MALDI-MSI also has the added advantage of not being dependent on supplementary labels (i.e., on the use of drug compounds with modified chemical structure). Despite its lower resolving power compared to autoradiography, in tissue studies, MALDI-MSI has the unique advantage of delivering the absolute mass identity of intact compounds, their molecular fragments, and metabolites (thus information that are not available in other imaging technology studies) [21].
The first study utilizing MALDI-MS for the detection of a drug compound directly from tissue has profiled human ovarian tumor sections treated with paclitaxel and also rat liver tissue spiked with spiperone [22]. Later on, the technique was further developed to map the localization of orally administered antitumor drug substances in rat brain and in mouse tumor tissue by using MALDI-MS for sampling the tissue sections in an array of positions with a predefined step size [23]. Subsequent studies have focused on the localization of active compounds and its metabolites in animal models and in different biological tissues [24, 25]. The MALDI-MSI technology has been applied for monitoring drug distribution kinetics in animal models as summarized by Prideaux and Stoeckli [26]. As the quantitative aspect of the technique has been explored, various normalization methods [27, 28] and quantitation approaches were introduced [29–34].
The drug distribution in tissue is an important pharmacological property, identifying the accumulation sites and providing valuable information about whether the drug may be affective or toxic [35]. Because in vivo animal models for displaying drug distribution are time-consuming to develop and may require many experimental animals to be involved, we introduced an in vitro approach that employs drug deposition onto the tissue sections [21]. In this study, our goal was to verify the in vitro method in comparison with matched in vivo experiments using sunitinib in a colorectal mouse model. We have found that sunitinib localization correlated closely in the in vitro and in vivo models that may promote the use of the in vitro model for drug development and preclinical studies in the future.
Materials and methods
In vivo mouse model
Tumors were obtained from 10-week-old female Balb/C mice, which were inoculated subcutaneously with 2 × 106 cells from the mouse colon-26 (C-26) adenocarcinoma cell line from Cell Line Service (CLS, Mason Research Institute), cultured in RPMI 1640 with 10 % fetal bovine serum (FBS) and 1 % penicillin/streptomycin (all from Sigma-Aldrich, Steinheim, Germany) at 37 °C, 5 % CO2 in a humidified atmosphere, as described previously [36]. The treatment began 2 weeks after tumor cell injection and was performed once daily at a dose of 100 mg/kg per mouse with a feeding tube for 10 consecutive days. Sunitinib (LC Laboratories, Woburn, MA, USA) at >99 % purity (CAS. No. 557795-19-4) was suspended in 2 % carboxymethyl cellulose with 2 mg/mL methyl-4-hydroxybenzoate (both from Sigma-Aldrich). Control mice received only the suspending medium. Animals were sacrificed at different time points but always 2 h after the final drug treatment. Tumors were surgically removed, snap frozen by submerging the tissues into liquid nitrogen-cooled isopentane, as described previously [37], and stored at −80 °C until sectioning. All animal model protocols were carried out in accordance with the Guidelines for Animal Experiments and were approved for the Department of Experimental Pharmacology in the National Institute of Oncology, Budapest, Hungary (permission number 22.1/722/3/2010).
Tissue sectioning
Tumors were removed from the −80 °C freezer and placed in the −20 °C freezer for an hour before cutting at −20 °C in a cryostat (Leica CM1950 Cryostat, Nussloch, Germany). To avoid polymer interference in mass spectra originating from optimal cutting temperature media, the tumors were mounted on the block using distilled water. The tumors were then trimmed at 50 μm until a full surface was visible, after which 10-μm sections were cut onto glass slides (Thermo Superfrost, Thermo Scientific, Germany) and stored at ambient temperature and pressure until further use.
In vitro experimental model
In the in vitro model (immersion overlay), sunitinib was deposited on the tissue surface as described earlier [21]. In brief, a tissue paper was placed on the bottom section of the moisture chamber and moistened using distilled water, while the tumor section on the slide was placed in the top section of the moisture chamber. Tissue sections then were covered with 125 μL of 1 μg/mL (2.5 μM) sunitinib solution in 50 % methanol (MeOH) that was pipetted onto the tissue surface and incubated for 1 h at room temperature. Following incubation, the in vitro treated tissue surface was washed by pipetting 3 mL of 50 % methanol and 3 mL of distilled water across the surface of the tissue. To confirm that non-specific binding was removed, a tissue section was washed by dipping the slide into 50 % MeOH and agitating the slide in methanol for 5 min followed by a 20-s wash using deionized water. The sections were allowed to dry prior to deposition of the matrix.
Sample preparation
The matrix was made from α-cyano-4-hydroxycinnamic acid (CHCA, Sigma-Aldrich, Steinheim, Germany) at a concentration of 7.5 mg/mL in 50 % acetonitrile at hypergrade for LC-MS (Merck, Darmstadt, Germany) and 0.1 % trifluoroacetic acid (TFA, Sigma-Aldrich, Steinheim, Germany) and was made up fresh every week. The airbrush which was used to spray the matrix was kept in a fume hood for safety. The airbrush was run off compressed nitrogen. One milliliter of CHCA matrix solution was added to an Aztek A470 airbrush (Testor Corp. Rockfort, IL) and sprayed in a short-burst fashion onto each tissue section. The tissue sections were held 10–12 cm from the airbrush nozzle, and a 3–5-s gap in between each spray was allowed for the matrix to dry and crystallize on the surface [38]. After the matrix was applied, the slide had a yellow frosted appearance and the tissue section was covered with the matrix.
MALDI mass spectrometry imaging analysis
The MALDI-MS acquisition was performed in a MALDI LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) using a method that employed 10-10 laser shots at 10 μJ for two scan events. Automatic gain control was switched off. The tissue surface was sampled in meandering mode at a 100-μm distance between positions. First, full scans in profile mode in a mass range from m/z 200 to 500 were generated in the Orbitrap at a resolution of 60,000 (at m/z 400) in positive polarity. The second scan event producing collision-induced dissociation (CID) fragmentation MS/MS spectra in a mass range from m/z 105 to 500 was performed in the linear ion trap in positive polarity.
Data visualization
The raw files were opened using ImageQuest™ software (Thermo Fisher Scientific, San José, CA), and the precursor or fragment ion masses of sunitinib were extracted showing their localization within the tissue sections. Screen shots were taken of the distribution of sunitinib precursor ion (m/z 399.218) normalized on the total ion count (TIC) and of its product ion (m/z 326.1) at the maximum image resolution allowed. For estimation of sunitinib concentrations in tissue sections, the normalized precursor ion maps of m/z 399.218 were used. The average signal intensity of five non-overlapping regions of interest (ROIs) (sized 1 × 1 mm of each containing 100 positions) randomly distributed over the analyzed surface was determined in each tissue section after 1, 4, 7, and 10 days of treatment using the rectangular averaging function of ImageQuest™.
Histological staining
The slides were dipped in and out of 70 % ethanol until they turned clear. After removal of the matrix, the slides were immersed in 4 % phosphate-buffered formaldehyde (Histolab, Gothenburg, Sweden) for 30 min, which was followed by a distilled water wash for 5 s. The slides were then immersed in Meyer’s hematoxylin (Histolab, Gothenburg, Sweden) for 3 min, followed by a blueing in running water for 10 min. Staining in eosin (Histolab, Gothenburg, Sweden) for 1 min was followed by a wash with deionized water. Then the slides were dipped in 70 % ethanol (Histolab AB, Gothenburg, Sweden) for 30 s, in 90 % ethanol for 1 min, and in 100 % ethanol for 1 min. Following, the slides were put in four different xylene (VWR, Stockholm, Sweden) baths for 1 min in each bath and finally were cover slipped by applying Pertex® (Histolab AB, Gothenburg, Sweden) to a cover slip.
Cover-slipped hematoxylin and eosin (H&E)-stained slides were loaded into the slide scanner (Mirax Midi Slide Scanner, Zeiss, Germany) to take a non-detailed image for the selection of regions of interest to be scanned at high resolution. The files were opened in Aperio ImageScope Viewer v12.1 (Leica Biosystems Imaging Inc., Vista, CA), where a photo of the detailed H&E scan could be taken and saved as a .tiff image file.
Results and discussion
Sunitinib localization in treated tumors—in vivo model
Animal models of cancer cell lines are valuable tools in understanding disease progression and also in drug therapy developments. We have used a mouse colorectal model that was induced by injecting C-26 adenocarcinoma cells subcutaneously. Following 2 weeks of tumor growth, three mice were treated with a single dose of per os sunitinib (100 mg/kg). The animals were sacrificed after 2 h, and their tumors were removed surgically and snap frozen. These treated tumors were selected to investigate sunitinib distribution in tissue sections, expecting that the single dose of drug treatment had a minimal effect on tumor histology. All tumors appeared with similar histological characteristics displaying a homogeneous distribution of cancer cells within the tissue sections as shown in the H&E panels of Fig. 1. Three adjacent tissue sections from the middle region of each tumor were subjected to MALDI-MS imaging using CHCA as the matrix.
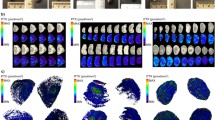
a–c Three technical replicates of a mouse colorectal adenocarcinoma (C-26) treated with a single dose (100 mg/kg) of sunitinib displayed a homogeneous distribution of tumor cells (H&E), yet the drug was unevenly localized. The lateral distribution of sunitinib in these adjacent tissue sections showed that more drug molecules were present in the peripheral area than in the center as the precursor mass (m/z 399.218) intensities normalized on TIC exposed. The extracted product ion of sunitinib (m/z 326.1) presented a very similar localization
The ionization of sunitinib was characterized in pure solution (dissolving 1 mg sunitinib in 50 % MeOH) spotted, and mixed with 3 mg/mL CHCA on a MALDI target plate. Precursor and product ions of sunitinib were recognized at m/z 399.218 (accurate mass by Orbitrap) and m/z 326.1 and 283.1 (obtained by ion trap), respectively. The distribution of drug signals was found to be similar in three tissue sections of a tumor treated with a single dose of sunitinib. Typically, the highest levels of sunitinib were observed in the peripheral areas of the tumor as the extracted ion chromatograms (XIC) of precursor mass (m/z 399.218) normalized on TIC showed in Fig. 1 (middle panels). The identification of sunitinib in tissue section was verified by a second scan event in each pixel generating tandem mass spectra. The XIC of the most intensive product ion (m/z 326.1), as shown in the right panels of Fig. 1, displayed excellent colocalization with the precursor mass. The levels of drug in the C-26 adenocarcinoma clearly indicated that the drug could be absorbed in the tissue section.
We have investigated whether this specific distribution of sunitinib in treated tumor sections might be a result of technical artifacts related to matrix deposition. An untreated tumor section was prepared applying the same protocol, spraying 7.5 mg/mL CHCA onto the tissue surface (Fig. 2(A)). As expected, no sunitinib signal (detected as precursor mass at m/z 399.218) was detected over the tissue section as shown in Fig. 2(B). Furthermore, the matrix signal, clearly observed as a Na adduct [M + Na]+ at m/z 212.032, was distributed rather homogeneously on the tissue surface with significantly lower intensities compared to the sampled area on the glass slide only (Fig. 2(C)). However, the peripheral area of the tissue displayed no specific crystallization properties or higher matrix signals, indicating that sunitinib distribution was specific and not due to technical artifacts.
Manual spaying of the matrix over a tissue section of an untreated mouse colorectal adenocarcinoma (C-26) typically provided homogeneous crystallization over the tissue and glass surfaces (A). The untreated tissue displayed no signals of sunitinib in this control experiment (B) but rather homogenous matrix intensities as the Na adduct of CHCA (m/z 212.032) demonstrated (C). Uniform crystallization was observed on both tissue and glass surface using transmission microscopy (ROI1 and ROI2, respectively). However, the signal intensities of CHCA were significantly lower and less homogenously distributed on the tissue (ROI 1 ) than on the glass surface (ROI 2 )
We have also investigated the crystallization optically in two ROIs using transmission microscopy with a fluorescent filter at 50-fold magnification as shown in Fig. 2. The size of matrix crystals was smaller, and the layer was more homogeneous in ROI2 than in ROI1. The mapped signals of the matrix (Na adduct) in the ROIs revealed that the crystallization was somewhat less heterogeneous and lower signal levels were detected in the tissue section (ROI1) than that from the glass surface (ROI2).
The comparison of two additional tumors identically treated with a single dose of sunitinib exposed similar peripheral distribution in one sample with lower signal intensities (Fig. 3a) compared with the tumor presented in Fig. 1. However, the third tumor showed sunitinib localization somewhat more homogeneously and also the average signal intensity was the highest among the three biological replicates (Fig. 3b). The variation of the sunitinib levels may be due to differences in the individual drug uptake rate after receiving a single dose.
Two additional mouse colorectal adenocarcinomas (C-26) treated with a single dose (100 mg/kg) presented some biological variability as the precursor mass (m/z 399.218) intensities normalized on TIC indicated (a). The tumor cells were homogeneously distributed, but their density might be higher in the second tumor (b) that also supported the higher signal intensities of sunitinib
Temporal distribution of sunitinib in treated tumors—in vivo model
Obviously, in vivo experiments are needed if the temporal distribution of the drug is a subject of investigation. Continued treatment with sunitinib has demonstrated that the drug could accumulate in the entire tumor as high levels of the active compound was detected more homogeneously in the center of tissue sections already after 4 days of administration (data not shown). The measured levels of sunitinib were estimated by averaging the signal intensities of the normalized precursor ion (m/z 399.218) in five smaller-sized DOIs in each tissue section and compared in tumors treated for different periods of time as shown in Fig. 4. A continuous increase of sunitinib signal was obtained between days 1 and 7, while the last time point at day 10 showed a significantly lower intratumoral drug level. The increase in levels may be caused by the drug entering the mouse tumor faster than the tumor cells can metabolize and excrete the drug, which will then reach saturation at a time point between day 7 and day 10. A similar effect was observed previously on studying saturation of drug binding sites [39].
The average signal intensities of sunitinib as calculated from small ROIs within each tissue section (500 analyzed positions) after 1, 4, 7, and 10 days of treatment. The estimated levels of sunitinib in treated mouse colorectal adenocarcinomas (C-26) showed a fast accumulation of the drug until day 7 that was, however, found to be significantly lower at day 10
Sunitinib localization in untreated tumors—in vitro model
The in vitro model was adjusted to detect similar levels of sunitinib to those of the in vivo method. Sections from untreated tumors were used to investigate drug distribution following the overlay of sunitinib solution for 1 h and washing with MeOH and water. The concentration of sunitinib and optimal incubation time was determined with different concentrations of sunitinib solutions applied onto the surface of different tissue sections from an untreated tumor at concentrations of 1000, 100, 10, 1, and 0.1 μg/mL in 50 % MeOH. The concentration of 1 μg/mL (2.5 μM) was found to be most suitable for detection with similar signal intensities to the in vivo levels. Triplicates of tissue sections from each tumor were then covered with 1 μg/mL sunitinib according to the experimental immersion overlay protocol and applied throughout the entire study in the in vitro model.
In comparison of the in vivo and in vitro models, similar levels of sunitinib with a homogenous distribution were observed in two tumors treated in vitro (Fig. 5). Although both tumors were treated identically with the same volume and concentration of drug and were analyzed using the same method, the drug signals were higher in the lower area of the second tumor (Fig. 5b). Nevertheless, the immersion overlay apparently provided satisfactory results in terms of detectable signal intensities, as well as the drug adequately covered and distributed in the tissue sections after washing. The fact that the drug was deposited onto thin-tissue sections of the tumor allowed the compound to directly access the cytoplasm without passing through the cell membrane. Additionally, the high concentration of organic modifier (50 % MeOH) in the sunitinib solution might alter the conformation of many proteins and thus enabling new interaction sites and increasing non-specificity.
Comparing the in vivo and the immersion overlay in vitro models, a similarity between the distributions of sunitinib was found. The immersion overlay model has the strongest resemblance to the treated tissue sections at later stage when sunitinib is fully diffused into the tumor. However, subtle biological differences in the tumors may contribute to the specific localization and the detected signal intensities of the drug. Individual variations may also be explained by a quenching factor present in a tumor that is not present in the other, which is amplifying or muting the signal from MALDI-MSI. It is generally accepted that ion suppression can be caused by a variety of factors, including salts, ion-pairing agents, exogenous compounds, metabolites, or proteins [40] as well as by the tissue type [41]. Increased intratumoral ion suppression may be caused by one or a combination of these factors, such as dehydration leading to increased salt levels in the tissue. Levels of drug in the tumor may be also affected by some of the drug’s side effects such as diarrhea [42]. Furthermore, it is possible that the mice may have not been able to absorb the entire given dose, resulting in reduced levels of detected drug in the tumors.
By this series of experiment, we intended to investigate if our in vitro model could recapitulate the in vivo model when the major question was whether the active compound could enter the tumor and reach its target receptors in tumor cells. Based on the similarities in sunitinib distribution in the in vivo treated tumor samples and in the in vitro model, we conclude that the latter experimental setting has the potential to be used to investigate or predict whether a drug can interfere with a tissue surface expressing specific target proteins. A combination of drug solution may also be investigated hence to provide fast predictive information how compounds could be combined to improve treatment efficiency.
Conclusions
The comparison of sunitinib distributions in in vivo animal and in vitro experimental models revealed that a single-dose treatment was sufficient to localize the drug in tumors as measured by MALDI-MSI. While technical reproducibility was good, more variance was found in the comparison of tumors identically treated with sunitinib. An increasing amount of drug could be detected through continued treatment. Based on our data presented here, we can assume that biological factors in the tumor growth are influencing the levels of sunitinib being detected during the course of treatment. Applying the experimental model on tumor sections from untreated animals could accurately reproduce the levels of drug obtained in the in vivo treated tumors. This suggests that this simple experimental approach may be successfully applied to investigate drug distributions in tissue sections. The possibility of being able to test multiple drugs on the same sample would open up the option of using this new experimental model for testing how drugs penetrate and bind to the tumor most effectively in a tumor biopsy, which in turn may open up the prospects of a more personalized therapy.
References
Siegel R, Desantis C, Jemal A (2014) Colorectal cancer statistics, 2014. CA: Cancer J Clin 64(2):104–117. doi:10.3322/caac.21220
Malvezzi M, Bertuccio P, Levi F, La Vecchia C, Negri E (2014) European cancer mortality predictions for the year 2014. Ann Oncol: Off J Eur Soc Med Oncol / ESMO. doi:10.1093/annonc/mdu138
Durko L, Malecka-Panas E (2014) Lifestyle modifications and colorectal cancer. Curr Color Cancer Rep 1(10):45–54
Douillard JY, Sobrero A, Carnaghi C, Comella P, Diaz-Rubio E, Santoro A, Van Cutsem E (2003) Metastatic colorectal cancer: integrating irinotecan into combination and sequential chemotherapy. Ann Oncol: Off J Eur Soc Med Oncol / ESMO 14(Suppl 2):ii7–ii12
Goldberg RM, Sargent DJ, Morton RF, Fuchs CS, Ramanathan RK, Williamson SK, Findlay BP, Pitot HC, Alberts S (2006) Randomized controlled trial of reduced-dose bolus fluorouracil plus leucovorin and irinotecan or infused fluorouracil plus leucovorin and oxaliplatin in patients with previously untreated metastatic colorectal cancer: a North American Intergroup Trial. J Clin Oncol : Off J Am Soc Clin Oncol 24(21):3347–3353. doi:10.1200/jco.2006.06.1317
Tournigand C, Andre T, Achille E, Lledo G, Flesh M, Mery-Mignard D, Quinaux E, Couteau C, Buyse M, Ganem G, Landi B, Colin P, Louvet C, de Gramont A (2004) FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: a randomized GERCOR study. J Clin Oncol : Off J Am Soc Clin Oncol 22(2):229–237. doi:10.1200/jco.2004.05.113
Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S, Holmgren E, Ferrara N, Fyfe G, Rogers B, Ross R, Kabbinavar F (2004) Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med 350(23):2335–2342. doi:10.1056/NEJMoa032691
Yamada Y, Takahari D, Matsumoto H, Baba H, Nakamura M, Yoshida K, Yoshida M, Iwamoto S, Shimada K, Komatsu Y, Sasaki Y, Satoh T, Takahashi K, Mishima H, Muro K, Watanabe M, Sakata Y, Morita S, Shimada Y, Sugihara K (2013) Leucovorin, fluorouracil, and oxaliplatin plus bevacizumab versus S-1 and oxaliplatin plus bevacizumab in patients with metastatic colorectal cancer (SOFT): an open-label, non-inferiority, randomised phase 3 trial. lancet Oncol 14(13):1278–1286. doi:10.1016/s1470-2045(13)70490-x
Bergers G, Song S, Meyer-Morse N, Bergsland E, Hanahan D (2003) Benefits of targeting both pericytes and endothelial cells in the tumor vasculature with kinase inhibitors. J Clin Invest 111(9):1287–1295. doi:10.1172/jci17929
Erber R, Thurnher A, Katsen AD, Groth G, Kerger H, Hammes HP, Menger MD, Ullrich A, Vajkoczy P (2004) Combined inhibition of VEGF and PDGF signaling enforces tumor vessel regression by interfering with pericyte-mediated endothelial cell survival mechanisms. FASEB J: Off Publ Fed Am Soc Exp Biol 18(2):338–340. doi:10.1096/fj.03-0271fje
Motzer RJ, Rini BI, Bukowski RM, Curti BD, George DJ, Hudes GR, Redman BG, Ka M, Merchan JR, Wilding G, Ginsberg MS, Bacik J, Kim ST, Baum CM, Michaelson MD (2006) Sunitinib in patients with metastatic renal cell carcinoma. JAMA : J Am Med Assoc 295(21):2516–2524
George S, Blay JY, Casali PG, Le Cesne A, Stephenson P, Deprimo SE, Harmon CS, Law CN, Morgan JA, Ray-Coquard I, Tassell V, Cohen DP, Demetri GD (2009) Clinical evaluation of continuous daily dosing of sunitinib malate in patients with advanced gastrointestinal stromal tumour after imatinib failure. Eur J Cancer (Oxford, Engl : 1990) 45(11):1959–1968. doi:10.1016/j.ejca.2009.02.011
George S, Blay JY, Casali PG, Le Cesne A, Stephenson P, Deprimo SE, Harmon CS, Law CNJ, Ja M, Ray-Coquard I, Tassell V, Cohen DP, Demetri GD (2009) Clinical evaluation of continuous daily dosing of sunitinib malate in patients with advanced gastrointestinal stromal tumour after imatinib failure. Eur J Cancer 45(11):1959–1968
Kulke MH, Bendell J, Kvols L, Picus J, Pommier R, Yao J (2011) Evolving diagnostic and treatment strategies for pancreatic neuroendocrine tumors. J Hematol Oncol 4:29. doi:10.1186/1756-8722-4-29
Norden AD, Drappatz J, Wen PY (2007) Targeted drug therapy for meningiomas. Neurosurg Focus 23(4):E12. doi:10.3171/foc-07/10/e12
Houk BE, Bello CL, Poland B, Rosen LS, Demetri GD, Motzer RJ (2010) Relationship between exposure to sunitinib and efficacy and tolerability endpoints in patients with cancer: results of a pharmacokinetic/pharmacodynamic meta-analysis. Cancer Chemother Pharmacol 66(2):357–371
Novello S, Scagliotti GV, Rosell R, Ma S, Brahmer J, Atkins J, Pallares C, Burgess R, Tye L, Selaru P, Wang E, Chao R, Govindan R (2009) Phase II study of continuous daily sunitinib dosing in patients with previously treated advanced non-small cell lung cancer. Br J Cancer 101(9):1543–1548
Saltz LB, Rosen LS, Marshall JL, Belt RJ, Hurwitz HI, Eckhardt SG, Bergsland EK, Haller DG, Lockhart AC, Rocha Lima CM, Huang X, DePrimo SE, Chow-Maneval E, Chao RC, Lenz HJ (2007) Phase II trial of sunitinib in patients with metastatic colorectal cancer after failure of standard therapy. J Clin Oncol: Off J Am Soc Clin Oncol 25(30):4793–4799. doi:10.1200/jco.2007.12.8637
Moss DM, Siccardi M (2014) Optimising nanomedicine pharmacokinetics using PBPK modelling. Br J Pharmacol. doi:10.1111/bph.12604
Chaurand P, Schwartz SA, Reyzer ML, Caprioli RM (2005) Imaging mass spectrometry: principles and potentials. Toxicol Pathol 33(1):92–101
Végvári Á, Fehniger TE, Rezeli M, Döme B, Jansson B, Welinder C, Marko-Varga G (2013) Experimental models to study drug distributions in tissue using MALDI mass spectrometry imaging. J Proteome Res 12(12):5626–5633
Troendle FJ, Reddick CD, Yost RA (1999) Detection of pharmaceutical compounds in tissue by matrix-assisted laser desorption/ionization and laser desorption/chemical ionization tandem mass spectrometry with a quadrupole ion trap. J Am Soc Mass Spectrom 10(12):1315–1321
Reyzer ML, Hsieh YS, Ng K, Korfmacher WA, Caprioli RM (2003) Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom 38(10):1081–1092. doi:10.1002/jms.525
Rohner TC, Staab D, Stoeckli M (2005) MALDI mass spectrometric imaging of biological tissue sections. Mech Ageing Dev 126(1):177–185. doi:10.1016/j.mad.2004.09.032
Hsieh Y, Chen J, Korfmacher WA (2007) Mapping pharmaceuticals in tissues using MALDI imaging mass spectrometry. J Pharmacol Toxicol Methods 55(2):193–200
Prideaux B, Stoeckli M (2012) Mass spectrometry imaging for drug distribution studies. J Proteome 75(16):4999–5013. doi:10.1016/j.jprot.2012.07.028
Källback P, Shariatgorji M, Nilsson A, Andrén PE (2012) Novel mass spectrometry imaging software assisting labeled normalization and quantitation of drugs and neuropeptides directly in tissue sections. J Proteome 75(16):4941–4951. doi:10.1016/j.jprot.2012.07.034
Deininger SO, Cornett DS, Paape R, Becker M, Pineau C, Rauser S, Walch A, Wolski E (2011) Normalization in MALDI-TOF imaging datasets of proteins: practical considerations. Anal Bioanal Chem 401(1):167–181. doi:10.1007/s00216-011-4929-z
Pirman DA, Yost RA (2011) Quantitative tandem mass spectrometric imaging of endogenous acetyl-L-carnitine from piglet brain tissue using an internal standard. Anal Chem 83(22):8575–8581. doi:10.1021/ac201949b
Takai N, Tanaka Y, Inazawa K, Saji H (2012) Quantitative analysis of pharmaceutical drug distribution in multiple organs by imaging mass spectrometry. Rapid Commun Mass Spectrom: RCM 26(13):1549–1556. doi:10.1002/rcm.6256
Hamm G, Bonnel D, Legouffe R, Pamelard F, Delbos JM, Bouzom F, Stauber J (2012) Quantitative mass spectrometry imaging of propranolol and olanzapine using tissue extinction calculation as normalization factor. J Proteomics 75(16):4952–4961. doi:10.1016/j.jprot.2012.07.035
Groseclose MR, Castellino S (2013) A mimetic tissue model for the quantification of drug distributions by MALDI imaging mass spectrometry. Anal Chem 85(21):10099–10106. doi:10.1021/ac400892z
Pirman DA, Reich RF, Kiss A, Heeren RM, Yost RA (2013) Quantitative MALDI tandem mass spectrometric imaging of cocaine from brain tissue with a deuterated internal standard. Anal Chem 85(2):1081–1089. doi:10.1021/ac302960j
Takai N, Tanaka Y, Saji H (2014) Quantification of small molecule drugs in biological tissue sections by imaging mass spectrometry using surrogate tissue-based calibration standards. Mass Spectrom (Tokyo, Japan) 3(1):A0025. doi:10.5702/massspectrometry.A0025
Schinkel AH, Wagenaar E, van Deemter L, Mol CA, Borst P (1995) Absence of the mdr1a P-Glycoprotein in mice affects tissue distribution and pharmacokinetics of dexamethasone, digoxin, and cyclosporin A. J Clin Invest 96(4):1698–1705. doi:10.1172/jci118214
Manea M, Leurs U, Orban E, Baranyai Z, Ohlschlager P, Marquardt A, Schulcz A, Tejeda M, Kapuvari B, Tovari J, Mezo G (2011) Enhanced enzymatic stability and antitumor activity of daunorubicin-GnRH-III bioconjugates modified in position 4. Bioconjug Chem 22(7):1320–1329. doi:10.1021/bc100547p
Dome B, Paku S, Somlai B, Timar J (2002) Vascularization of cutaneous melanoma involves vessel co-option and has clinical significance. J Pathol 197(3):355–362. doi:10.1002/path.1124
Fehniger TE, Végvári Á, Rezeli M, Prikk K, Ross P, Dahlbäck M, Edula G, Sepper R, Marko-Varga G (2011) Direct demonstration of tissue uptake of an inhaled drug: proof-of-principle study using matrix-assisted laser desorption ionization mass spectrometry imaging. Anal Chem 83(21):8329–8336
Kuh H, Jang S, Wientjes M (1999) Determinants of Paclitaxel penetration and accumulation in human solid tumor. J Pharmacol Exp Ther 290(2):871–880
Annesley TM (2003) Ion suppression in mass spectrometry. Clin Chem 49(7):1041–1044
Fujimura Y, Miura D (2014) MALDI mass spectrometry imaging for visualizing in situ metabolism of endogenous metabolites and dietary phytochemicals. Metabolites 4(2):319–346
Schwandt A, Wood LS, Rini B, Dreicer R (2009) Management of side effects associated with sunitinib therapy for patients with renal cell carcinoma. Oncol Targets Ther 2:51–61
Acknowledgments
AV is grateful for funding support Innovate Support 2011–03926 from CREATE Health. BD was supported by KTIA AIK 12-1-2013-0041, TÁMOP 424A/1-11-1-2012-0001, OTKA K109626, OTKA K108465, EUREKA_HU_12-1-2012-0057, ÖNB Jubiläumsfondsprojekt Nr. 14043, and the Vienna Fund for Innovative Interdisciplinary Cancer Research. JT was supported by OTKA K84173 and INNO 08-3-2009-0248 (2010).
Conflict of interest
The authors declare no potential conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Published in the topical collection Mass Spectrometry Imaging with guest editors Andreas Römpp and Uwe Karst.
Rights and permissions
About this article
Cite this article
Connell, J.J., Sugihara, Y., Török, S. et al. Localization of sunitinib in in vivo animal and in vitro experimental models by MALDI mass spectrometry imaging. Anal Bioanal Chem 407, 2245–2253 (2015). https://doi.org/10.1007/s00216-014-8350-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-014-8350-2