Abstract
Accurate detection of certain chemical vapours is important, as these may be diagnostic for the presence of weapons, drugs of misuse or disease. In order to achieve this, chemical sensors could be deployed remotely. However, the readout from such sensors is a multivariate pattern, and this needs to be interpreted robustly using powerful supervised learning methods. Therefore, in this study, we compared the classification accuracy of four pattern recognition algorithms which include linear discriminant analysis (LDA), partial least squares-discriminant analysis (PLS-DA), random forests (RF) and support vector machines (SVM) which employed four different kernels. For this purpose, we have used electronic nose (e-nose) sensor data (Wedge et al., Sensors Actuators B Chem 143:365–372, 2009). In order to allow direct comparison between our four different algorithms, we employed two model validation procedures based on either 10-fold cross-validation or bootstrapping. The results show that LDA (91.56 % accuracy) and SVM with a polynomial kernel (91.66 % accuracy) were very effective at analysing these e-nose data. These two models gave superior prediction accuracy, sensitivity and specificity in comparison to the other techniques employed. With respect to the e-nose sensor data studied here, our findings recommend that SVM with a polynomial kernel should be favoured as a classification method over the other statistical models that we assessed. SVM with non-linear kernels have the advantage that they can be used for classifying non-linear as well as linear mapping from analytical data space to multi-group classifications and would thus be a suitable algorithm for the analysis of most e-nose sensor data.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
According to the “no free lunch theorems” in search and optimization [1], the application of an algorithm to different types of data may result in diverse outputs, and in general, no single algorithm is optimal for solving all problems. Each set of chemical data therefore requires the choice of an optimal (or a near-optimal) algorithm for that particular data set [1]. Hence, optimization in data analysis is essential, especially in the analysis of data from electronic noses (e-noses) where for real-time analysis, an algorithm must be able to classify data rapidly. This realisation that chemical sensing on the fly in real time is a distinct possibility has led to the increased popularity of e-noses in recent years [2].
Due to the complexity of the output from chemical sensor pattern recognition is an essential part of the analysis and has been used previously for characterisation of data from e-noses [3]. For example, several studies have employed chemometrics such as discriminant analysis which is a supervised statistical tool for studying the association between a set of chemical descriptors (inputs) and categorical response (the output(s) or target(s)) and is commonly used as a dimensionality reduction technique and for classification [4]. Linear discriminant analysis (LDA) [4] has been used successfully for the analysis of the data from chemical sensors applied to volatile analytes [5], the analysis of data from e-nose sensors used for the detection of explosives and flammable vapours (toluene, acetone and ethanol) [6], and for discrimination between patients with asthma from healthy controls [7]. Partial least squares-discriminant analysis (PLS-DA) [8] has been used previously in combination with e-noses for the classification of wines [9], in the diagnosis of lung cancer by breath analysis [10] and for the diagnosis of urinary tract cancers [11].
Although LDA and PLS-DA are very powerful classifiers, they generally only permit linear mapping from inputs to outputs [12], and therefore there is a desire to employ more sophisticated machine learning algorithms. Random forests (RF) are a technique used for classification and for the estimation of variable importance based on multiple decision trees [13] and have been used previously in e-nose data analysis for food quality control [14]. Another popular machine learning technique is based on support vector machines (SVM). These are kernel-based classification methods that determine the optimum boundaries (support vectors) that accurately separate classes with the maximum margin between them [15]. These methods have also been effectively used in the classification of e-nose data [16], for the measurement of vapour mixtures by using metal oxide gas sensors [17], in the detection of lung cancer [18], for the assessment of lymph nodes in the course of breast cancer diagnostics [19] and in olfactory signal recognition [20].
Wedge and colleagues [21] employed genetic programming (GP) for vapour classification and reported an average sensitivity and specificity of 0.91 and 0.96 for acetone, 0.86 and 0.88 for dimethyl methylphosphonate (DMMP), 0.79 and 0.87 for methanol, and 0.79 and 0.83 for propanol [21]. However, GP requires a high of expertise in programming, as the relevant cross-over and mutation rates need to be selected and the models constrained in order to reduce bloat, and thus cannot be widely used by non-experts [22, 23]. Therefore, in the present study, we have programmed four different chemometric approaches using the same data as a tool to compare more accessible chemometrics methods for the analysis of e-nose data. These included two discriminant analysis approaches (viz. LDA and PLS-DA), random forests, as well as four SVM which were employed with different kernel-based functions [24]. In order to allow objective comparisons, we used k-fold cross-validation [25] and bootstrapping with replacement [26], and for the test sets only, we compute classification prediction accuracy, sensitivity and specificity from all methods. Prediction accuracy (not to be confused with precision) corresponds to the proportion of the total number of correct predictions as calculated from confusion matrix (for further details, please see Table S1 and its descriptive statistics as shown in the Electronic Supplementary Material (ESM)).
Materials and methods
Data
In a previous study by Wedge et al., an e-nose sensor composed of arrays of organic field-effect transistors (OFETs) had been developed for vapour sensing [21]. As a result of difficulties in the fabrication of sensing materials that are essential for universal sensing [27], the authors focus only on four chemicals: acetone, DMMP, methanol and propan-1-ol. Additionally, these authors indicated that significant enhancement of sensitivity and specificity was accomplished by coating multiple transistors with different semiconducting polymers. Therefore, four OFETs based upon amorphous polytriarylamines (PTAA) were used, and these comprised three terminals (source, drain and gate) based on organic semiconducting polymers in their conductive channel. These were chosen to deliver materials with different electron-donating properties and flexibilities and hence chemical-sensing abilities [21].
Upon exposure to the different vapours to the OFETs, data were collected for 4 s. The main characteristics of these OFETs are the “threshold voltage,” reflecting the lowest gate voltage essential to produce an accumulation layer, where the value above indicates that the OFETs is “on” whereas the values below the threshold means that the sensor is “off.” The main data collection characteristics that have been measured from these four OFETs are the following: (1) off current shows the bulk conductivity of the polymer; (2) on resistance shows conductivity in the presence of field effect; and finally, (3) mobility calculated from a series of on current values acquired at altered stepped voltages. This resulted in 12 measurements per sample [21], and these were used for chemometrics analysis in the current study.
As explained above three measurements—on resistance, off current and mobility—were aggregated from each of four transistors coated with different semiconducting polymers based upon amorphous polytriarylamines. The four sensors are abbreviated to J49, JM116, OMe and PTAA and are described in full detail in a previous publication. In summary, the complete data matrix consists of 127 observations and 12 variables (4 sensors × 3 measurements) [21].
Modelling process
The R 2.15.0 software environment (http://cran.r-project.org/) was used for data analysis. This environment contains a variety of packages useful for data analysis and is a free open-source program [28]. Data analysis techniques were evaluated in terms of their ability to discriminate between the four different classes (viz. acetone, DMMP, methanol and propanol), and full details of the statistics we used are provided in the Electronic Supplementary Material. All R scripts are freely available from the authors on request.
A flow chart showing the overall data analysis is shown in Fig. 1. Additionally, data pre-processing (auto-scaling) was performed to place all the variables on a single comparable scale. This process was used as it reduces the influence of large values that may overtly dominate the chemometric analysis [29].
Flow chart showing data preparation steps prior to classification with supervised learning. This classification process also included two different validation methods, and these are detailed within this flow chart. The superscript letter a denotes the seven models that were used for supervised learning which include LDA, PLS-DA, RF and SVM with four different kernel functions
Data preparation
The SVM, RF and PLS-DA algorithms are governed by certain parameters which for SVM depend on the kernel type used for training and prediction [24]; for RF, it will be the number of variables included in each tree and the number of trees in the forest [13]; and, finally, for PLS-DA, the number of non-zero components used in classification [30]. Due to the no free lunch theorems [1] discussed above, it is inadvisable to use the default parameters, as these are not guaranteed to be optimal. In order to assess the most optimal method, we used cross-validation methods. Whilst methods such as leave-one-out (LOO) cross-validation have been used previously to determine the optimal parameters for SVM [31], this may be more biased as only a single sample is removed before the model is constructed with the rest of the samples, and this often provides an overoptimistic impression of model accuracy as the class distribution is kept constant and hence unreliable predictions [32]. Jain et al. reported that statistical confidence intervals constructed for models using bootstrapping are more robust compared to LOO cross-validation [33]. In addition, if one wants to use an experimental design to set up the calibration, bootstrapping is necessary to avoid the position where a single validation split of the training and test sets may inadvertently not reflect the overall data structure and thus may significantly influence the validation process. Therefore, in this study, we estimated these parameters for each of the methods by using bootstrapping which tests several random class distributions as proposed by Efron and Tibshirani [32], and as this gives more representative estimates of the “average” model, it is thus likely to be less biased [12, 32, 33].
The optimisation of the most appropriate parameters for SVM and RF was based on model accuracy from these bootstraps calculated using grid search. This involved (1) setting up the range for each parameter in a two-step grid search as proposed by Xu et al. [34] where, in the first stage, the analyses were conducted in large ranges and distances between parameters values to group possible targets and then in a second stage the analyses were performed on narrower search intervals (by way of example for a polynomial kernel, this varies from 0 to ∞ (or for practical reasons at least a reasonable and sensible number that avoids redundant computation; see below) for the cost parameter [35]); (2) creating a table with a coarse search of each parameter; for the above example, this would be 0–10 in steps of 0.1 to narrow the search area, where for instance, gamma with large values may lead to overfitting as shown by Ben-Hur and Weston [36]; (3) measuring the classification accuracy for the coarse search using 100 bootstraps to evaluate each combination of parameters and approximate a suitable, narrower range of parameters; and finally (4), analysing this reduced range of parameters in more depth by shrinking the step size. The results of grid search for SVM can be found in the Electronic Supplementary Material (Table S2).
Discriminant analysis
In this study, two discriminant analysis approaches have been used, namely LDA and PLS-DA which try to find linear combinations of features that aim to separate two or more groups of samples [4, 30]. Although the aim of these supervised methods is classification, certain differences can be observed as described by Brereton and Lloyd [30]. For PLS-DA, when two groups are to be classified a single output (Y vector) is used and PLS1 is used to predict whether a sample belongs to group A (coded “1”) or group B (coded “0” or “−1”). By contrast, when multiple groups are analysed, the same number of multiple outputs are used and PLS2 is applied. It is strange that in many analytical areas, people still use the PLS scores plots for interpretation rather than the values on the Y vector. This happens despite the fact that (amongst others) Westerhuis and colleagues suggested that PLS-DA scores plots should not be used for interpretation of class differences, as it present an overoptimistic understanding of the separation between two or more classes, and they show that without suitable validation, similar results can be accomplished when random data are classified [37]. For LDA, in this paper, we use discriminant function analysis (DFA; also known as canonical variate analysis (CVA) [4]), which simultaneously rotates and scales input space to reduce within-group variance and maximise between-group variance [4]. However, unlike PLS, LDA is highly sensitive to collinearity and can be only used with the data where the number of features is smaller than the number of observations [38]. This can, of course, be overcome using PCA, and care needs to be taken to select the most appropriate number of PCs to feed into LDA [4, 30, 39]. The following R packages were employed for discriminant analysis:
-
(1)
MASS “Support Functions and Datasets for Venables and Ripley’s MASS” version 7.3-19 [40] was used for LDA.
-
(2)
CARET “Classification and Regression Training” version 5.15-023 [41] was used for PLS-DA.
Random forests
In RF, a collection of trees is generated and the ensemble is used for prediction [13, 42]. This approach relies on constructing a series of tree-based “learners” which use a subset of the input space and so can also be used for variable selection, or to understand the important features that are used for prediction. As the subset selection can be random, it is important to allow judicious input selection. See ESM Fig. S1 for a diagrammatic representation of RF and ESM, and Fig. S2 illustrates an example of RF used for variable selection [13, 42]. For RF, we employed “Breiman and Cutler’s random forests for classification and regression” version 4.6-6 [42].
Support vector machines
SVM is a machine learning approach that can deal with linear and non-linear classification problems [15, 24]. SVM are computed to only separate two classes, and therefore for multi-classification tasks, several SVM are used where one particular class is compared to all. The performance of SVM relies upon the kernel selection and on the parameters that each of these uses during calibration. In this study, four types of kernel were used: (1) linear—the simplest and most commonly used kernel function used to map data into a space where the classes are linearly separable; (2) polynomial—for non-linearly separable classes; (3) radial—based on class-conditional Gaussian probability distribution which maps data into a different space where linear separation can occur; and finally (4), sigmoid—usually used when the structure of the data is unknown [15, 24]. SVM analysis used four different kernels (vide infra), and these were implemented using e1071 “Misc Functions of the Department of Statistics (e1071), TU Wien” version 1.6 [43].
Model validation
To develop a good classifier and to obtain an appropriate estimation error, it is essential to use as much of the data as possible for training and testing and to ensure that the data are correctly organised for analysis. Hence, in order to compare accurately the classification ability of all four techniques, it is crucial to assess these procedures with robust validation. In this process, a separate preliminary data set is analysed by the algorithm before it is applied into common usage, i.e. out in the field. The basic concept of validation is to assess the performance of the algorithm on these preliminary data so that confidence can be had on the ability of the sensor chemometrics approach on real-world “unseen” data. For model validation, the idea is to take a certain number of samples that are not used in training (that is to say the calibration phase of the analysis), and samples that are not used in this phase are used to validate the algorithm [25].
In this study, model validation methods such as k-fold cross-validation (where k = 10) [32] and bootstrapping with replacement (1000 bootstraps) were used to generate multiple training and test data sets [26]. In this study, we applied 1000 bootstrap replicates to approximate estimations of accuracy. In initial experiments (data not shown), we used fewer (e.g. 100 bootstraps) and excessive numbers of bootstrap iterations (10,000 and 100,000) and found that the shape of the correct classification results (CCR) for the 1000 iterations approximated those of the larger iteration numbers well, whilst with 100 iterations within the bootstrap the CCR distributions were not very smooth. These two model validation processes are fully detailed in the Electronic Supplementary Material. After classification, the test sets were used to estimate prediction accuracy for each of the models. The statistical terms for this are also fully defined in the Electronic Supplementary Material.
Results and discussion
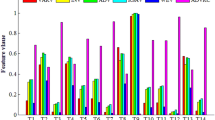
As discussed above, in order to classify the four chemical vapours under study, based on the response of the four sensors, we programmed and applied a variety of supervised learning algorithms and compared their performance in terms of prediction accuracy and time taken for calculations. Furthermore, some of these classification methods can be used for variable selection, rather than for prediction only as shown in our recent study on the Gram-positive bacteria Bacillus [44]. In this study, different variable selection methods such as stepwise forward variable selection for LDA, variable importance for projection (VIP) coefficient for PLS-DA, mean decrease in Gini, and accuracy for RF and finally recursive feature elimination (a method related to stepwise backward variable selection) for SVM have been used to reduce dimensionality and select relevant variables. Hence, the number of variables can be reduced to only include those that are important, thereby decreasing computation time. For instance, in RF analysis, Fig. S2 in the ESM shows variable importance ordered according to their importance by mean decrease in accuracy and mean decrease in Gini index. Moreover, loadings plots from LDA and PLS-DA (see ESM Fig. S5b) can be used to estimate which variables are important and thus influence class separation. Therefore, if one wants to analyse high-dimensional e-nose data, it is recommended that one of the above-mentioned approaches be applied in order to reduce dimensionality. Further explanation of these variable selection methods can be found in the Electronic Supplementary Material.
Figure 2a shows the LDA scores plot of the first two LDs for the classification of both the training and test sets. In this model, an equal split of the samples was used and the LDA model was formed with the training data set pairs only; these pairs are the chemical data and the corresponding class (four in total—one for each of the different vapours). Subsequently, the test set samples were projected into this LDA scores place and plotted. The separation between the four classes is quite clear. Although there is some overlap between the propanol samples and the methanol and DMMP samples, propanol is clearly separated from the other samples in the LD3 (data not shown). The validation observed is very good, as the test samples (open symbols) are coincident with their respective training data sets (closed symbols). The LDA loadings plot (Fig. 2b) was used to identify the variables with the greatest influence on discrimination. This plot clearly indicates that the most important inputs in this classification method are on resistance J49, on resistance JM116, on resistance OMe and off current J49. These variables were also identified by the other chemometric techniques as being important for class discrimination (see ESM Figs. S2 and S5).
Linear discriminant analysis (LDA). a The scores plot for classification (training set, filled symbols) and prediction (test set, open symbols). For ease of visualisation of class prediction, this example is based on equal division of samples between the test and training sets. b Corresponding LDA loadings plot
To measure the prediction accuracy (including standard deviation, sensitivity and specificity), for each chemometric technique, 10-fold cross-validation and bootstrapping with replacement were employed. For these studies, 100 runs were performed for 10-fold cross-validation and 1000 repetitions for bootstrapping with replacement. As it is important to enable a direct comparison between the four different classification methods, we ensured that the same data set splits were employed. The classification models used on both methods were validated with the test samples, and the statistical metrics on the ability of each of the models to classify the vapours correctly were calculated and these are summarised in Tables 1 and 2. These include the computation time, prediction accuracy and its standard deviation (sd), and sensitivity and specificity for each vapour. The standard deviation for each method used both for sensitivity and specificity varied between 0.01 and 0.03 for cross-validation sampling and 0.01–0.08 for bootstrapping with replacement (data not shown). It can be seen that all methods show similar classification ability in terms of prediction accuracy, although these were always slightly higher for the 10-fold cross-validation model compared with the more rigorous bootstrap analysis.
Table 1 shows the comparison of different classifier methods based on 10-fold cross-validation. In general, all four methods used here were better than the GP (see ESM Table S3), although we do note that the GP will not have used the same validation splits that are used here and did not include bootstrapping with replacement. For all methods, the accuracy was between 81.74 and 91.66 %, the latter from the SVM with polynomial kernel. This may be related to the classification rule which is determined by only a small number of training set samples called support vectors (SVs), which lie close to the decision boundary and the specific kernel employed [24]. Additionally, the SVM with polynomial kernel approach demonstrated more consistency across each of the vapours than LDA (the next most accurate method) and generally resulted in a higher specificity and sensitivity in detecting propanol. This may be related to the non-linear mapping provided by this particular type of kernel function used in the SVM. By contrast, it can be seen that LDA outperforms SVM (polynomial) with respect to sensitivity for acetone, DMMP and methanol, because these classes are well-separated (Fig. 2a); therefore, a linear boundary appears to be sufficient. Propanol is a little harder to predict, and it seems that SVM with a polynomial function is better at classifying this vapour compared to the other methods.
A closer inspection of the confusion matrices (see ESM Fig. S3–S3) reveals that the misclassified propanol samples are in general more likely to be assigned to the methanol class (and vice versa), which may be expected, as they are both alcohols and therefore the –OH group interacts with the chemical sensor in a similar fashion.
PLS-DA gave better results than those obtained using GP (see ESM Table S3) with the exception of methanol sensitivity. Compared with SVM (polynomial) and LDA, PLS-DA resulted in slightly poorer prediction accuracy but with similar analysis speed. However, when PCA has been applied prior to LDA following the same pre-processing procedure as for PLS-DA, similar results have been achieved (data not shown). This only confirms the arguments of Brereton and Lloyd [30] that, in many cases, simpler algorithms may provide better results and there is no need to use more complicated techniques.
Random forests improved upon the results obtained using GP in all areas with the exception of propanol sensitivity and acetone specificity. However, the analysis times were much longer (>10 times) in comparison to the other methods, and this is due to the fact that 500 trees were computed within each RF. The lower prediction accuracy for RF compared to SVM and LDA may be due to the fact that RF performs better with a larger number (typically 1000s) of input variables than with a smaller number of variables (12 in this study), but there is no direct evidence for this.
Table 2 shows a comparison of the classification ability of the same four classification methods based on bootstrapping for model validation. We believe that bootstrapping is a more robust validation procedure, as it involves more configurations of the training and test set splits (1000 in our case) and therefore the statistics on these 1000 test set are closer to a random selection of training test set and thus reflect the underlying classification performance better.
The sensitivity and specificity of the models were all above 0.73, except for SVM with a sigmoid kernel which resulted in a sensitivity of 0.64 for propanol. Yet again, SVM with polynomial kernel and LDA display the highest prediction accuracy of 86.39 and 88.60 %, respectively. Nonetheless, the difference between both is much larger than that in the previous analysis based on 10-fold cross-validation, where it differed only by the first decimal place. Moreover, LDA seems to be more consistent across all of the chemicals than SVM with polynomial kernel, whereas for 10-fold cross-validation, it was vice versa. For the other methods such as PLS-DA and RF, we observe a small decrease in accuracy for all vapours, whereas for SVM, all kernels gave a noticeable decrease in model performance with the exception of the sigmoidal function. It is important to highlight the fact that bootstrapping has a slightly higher standard deviation in comparison to 10-fold cross-validation where the difference along all methods is ∼0.04, and this is because more training and test set splits were used.
Table 3 summarises the comparison of the four classification approaches from this study based on both general characteristics and specific findings from our work. In the first part of the table, we present some general characteristics, which include the following:
-
(1)
“Visualization” refers to pictorial interpretation and understanding of the results. For the LDA and PLS-DA models, three pluses were given, as both methods display visible separation in terms of scores plots which contain the information about the observations (samples) and loadings plot which provides information about key variables. RF has received two pluses, as this method is difficult to visualise if we have a large number of variables due to the fact that its output is an assembly of multiple decision trees [13]. Lastly, SVM is given the lowest score, as the separation is rather demanding in terms of visualization and interpretation due to the projection of sample locations into high-dimensional space. This is certainly the case when non-linear kernels are used, although for linear kernels, visualization interpretation is achievable.
-
(2)
“Variable selection” refers to whether the method contains enough information about the identification and ranking of potentially relevant/important variables. Therefore, LDA, PLS-DA and RF received the highest results, as all provide an opportunity to extract relevant information from the model in terms of variable importance. By contrast, SVM cannot be used on its own for feature selection, unless it is combined with recursive feature elimination [45].
-
(3)
“Separation performance” of the models has been scored according to the ability of each of the supervised learning algorithms to solve linear and non-linear problems; therefore, LDA and PLS-DA scored two pluses, as both techniques rely on linear mapping from inputs to outputs. RF scored +++, as the procedure can be implemented both for linear and non-linear problems. Finally, SVM receives the same score (+++) as RF, as SVM apply straightforward linear separation to the data after projecting the input data to a high-dimensional feature space wherein classes are now linearly separable. This ability is especially useful when one has to deal with e-nose data that include overlapping classes as shown by Pardo and Sberveglievi [16].
-
(4)
“Impact of collinearity” in this study indicates the strength of the model towards collinearity in the data set. Collinearity refers to the event in which two or more predictor variables in the analysed data set are highly correlated, and therefore it is difficult to estimate which of them influence the prediction accuracy. According to the above characteristic, LDA was given one plus, as the method is not the best approach when one needs to analyse the highly collinear data set. However, this obstacle as mentioned in the “Materials and methods” section can be circumvented by application of PCA or stepwise forward variable selection. These statistical techniques reduce the problems of collinearity but cannot entirely terminate them. PLS-DA, SVM and RF received the highest scores (+++), as all are resistant to collinearity of the data. Therefore, these approaches should be favoured when analysing highly collinear e-nose data set.
As for the second set of statements in Table 3, we review specific findings/perceptions from this study. As a result, we conclude the following:
-
(1)
Visualization is where we evaluated the characteristics based on visualisation and ease interpretation. Here, our findings reflect general characteristics that have been described above.
-
(2)
“Speed” summarises the time that was needed for the calculations. Here, LDA and PLS-DA receive the same score (+++), as both are computationally fast (<5 s; see Tables 1 and 2 for details). RF has received one plus, as the time taken for calculations was the longest, and this is because of the large number of trees (500) that need to be grown in each forest. Finally, SVM scored ++, as the method depends on different kernels and these affect computational intensity (Tables 1 and 2).
-
(3)
“Predictive power” scores are based on the prediction accuracy, and these reflect the findings discussed in the paper and summarised in Tables 1 and 2. Whilst all score well as LDA generally performed better, we have given it the highest score.
-
(4)
Finally, a “parameter selection” characteristic is used to indicate how easy or difficult it is to tune the internal parameters within each model. Therefore, LDA is the best approach for our data, as no parameters have to be adjusted as all X data are used. The reason that PLS-DA scores two pluses is due to the fact that we have to optimize the number of latent variables (LVs), and these LVs are optimised during each bootstrap iteration. Whilst RF does include several parameters that need to be optimised, it has been reported by Liaw and Wiener [42] that the default RF parameters are generally sufficient, and whilst we did alter these parameters, they did not improve the prediction accuracy (data not shown). Finally, SVM were given the lowest score, as these methods require considerable optimization of several parameters as performed within this studied.
Conclusions
We have compared four chemometrics methods with two common model validation approaches—10-fold cross-validation and bootstrapping with replacement—for the classification of data acquired from e-nose sensors. LDA, PLS-DA, RF and the four SVM all generated very good predictions for the four different vapours and, in general, outperformed the GP. LDA and SVM with a polynomial kernel were found to give the best overall model performance, whilst the SVM with a sigmoidal kernel gave the worst prediction accuracy results.
It has previously been shown that parameter selection for SVM has a big influence on prediction accuracy [24, 43, 46]. We have confirmed this and have shown that careful parameter selection for the four SVM (see ESM Table S2) resulted in a noticeable improvement in the prediction accuracy.
There is of course a subjective trade-off between getting a model that works and assessing model performance. The latter is usually more computationally expensive but gives one credibility in the data generated and the chemometric predictions based on such data. In this study design, we employed a sufficient number of training sets (especially for bootstrapping) in order to gain confidence in the performance of the models tested. Moreover, this enabled direct and objective comparison between the four different algorithms used for supervised learning.
Taking both validation methods into account for these e-nose data, we conclude that LDA and SVM with polynomial kernel provides the best overall results and can be considered as a good choice when analysing these types of data. In addition, the results show that the methods are satisfactorily stable and the time taken for measurements is relatively low. However, we do note, as discussed in [30, 39], that caution needs to be taken when applying LDA to highly collinear data and that it may be appropriate to remove this collinearity using PCA prior to LDA. In addition, as demonstrated in this study, if one wants to analyse the highly collinear or non-linear e-nose sensor data, our findings recommend that SVM with a polynomial kernel should be favoured as a classification method over the other statistical models that we tested.
References
Wolpert DH, Macready WG (1997) No free lunch theorems for optimization. IEEE Trans Evol Comput 1:67–82
Rock F, Barsan N, Weimar U (2008) Electronic nose: current status and future trends. Chem Rev 108:705–725
Scott SM, James D, Ali Z (2006) Data analysis for electronic nose systems. Microchim Acta 156:183–207
Manly BFJ (1986) Multivariate statistical methods: a primer. Chapman and Hall
Jurs PC, Bakken GA, McClelland HE (2000) Computational methods for the analysis of chemical sensor array data from volatile analytes. Chem Rev 100:2649–2678
Dobrokhotov V, Oakes L, Sowell D, Larin A, Hall J, Kengne A, Bakharev P, Corti G, Cantrell T, Prakash T, Williams J, McIlroy DN (2012) Toward the nanospring-based artificial olfactory system for trace-detection of flammable and explosive vapors. Sensors Actuators B Chem 168:138–148
Dragonieri S, Schot R, Mertens BJA, Le Cessie S, Gauw SA, Spanevello A, Resta O, Willard NP, Vink TJ, Rabe KF, Bel EH, Sterk PJ (2007) An electronic nose in the discrimination of patients with asthma and controls. J Allergy Clin Immunol 120:856–862
Wold S, Sjostrom M, Eriksson L (2001) PLS-regression: a basic tool of chemometrics. Chemometr Intell Lab 58:109–130
Cynkar W, Dambergs R, Smith P, Cozzolino D (2010) Classification of Tempranillo wines according to geographic origin: combination of mass spectrometry based electronic nose and chemometrics. Anal Chim Acta 660:227–231
Di Natale C, Macagnano A, Martinelli E, Paolesse R, D’Arcangelo G, Roscioni C, Finazzi-Agro A, D’Amico A (2003) Lung cancer identification by the analysis of breath by means of an array of non-selective gas sensors. Biosens Bioelectron 18:1209–1218
Bernabei M, Pennazza G, Santortico M, Corsi C, Roscioni C, Paolesse R, Di Natale C, D’Amico A (2008) A preliminary study on the possibility to diagnose urinary tract cancers by an electronic nose. Sens Actuators B-Chem 131:1–4
Brereton RG (2009) Chemometrics for pattern recognition. Wiley, Chichester
Breiman L (2001) Random forests. Mach Learn 45:5–32
Pardo M, Sberveglieri G (2008) Random forests and nearest shrunken centroids for the classification of sensor array data. Sens Actuators B-Chem 131:93–99
Vapnik VN (1999) An overview of statistical learning theory. IEEE Trans Neural Netw 10:988–999
Pardo M, Sberveglieri G (2005) Classification of electronic nose data with support vector machines. Sensors Actuators B Chem 107:730–737
Gualdron O, Brezmes J, Llobet E, Amari A, Vilanova X, Bouchikhi B, Correig X (2007) Variable selection for support vector machine based multisensor systems. Sensors Actuators B Chem 122:259–268
Machado RF, Laskowski D, Deffenderfer O, Burch T, Zheng S, Mazzone PJ, Mekhail T, Jennings C, Stoller JK, Pyle J, Duncan J, Dweik RA, Erzurum SC (2005) Detection of lung cancer by sensor array analyses of exhaled breath. Am J Respir Crit Care Med 171:1286–1291
Sattlecker M, Bessant C, Smith J, Stone N (2010) Investigation of support vector machines and Raman spectroscopy for lymph node diagnostics. Analyst 135:895–901
Distante C, Ancona N, Siciliano P (2003) Support vector machines for olfactory signals recognition. Sensors Actuators B Chem 88:30–39
Wedge DC, Das A, Dost R, Kettle J, Madec MB, Morrison JJ, Grell M, Kell DB, Richardson TH, Yeates S, Turner ML (2009) Real-time vapour sensing using an OFET-based electronic nose and genetic programming. Sensors Actuators B Chem 143:365–372
Gilbert RJ, Goodacre R, Woodward AM, Kell DB (1997) Genetic programming: a novel method for the quantitative analysis of pyrolysis mass spectral data. Anal Chem 69:4381–4389
Koza JR (1992) Genetic programming: on the programming of computers by means of natural selection. MIT Press, Cambridge
Burges CJC (1998) A tutorial on support vector machines for pattern recognition. Data Min Knowl Discov 2:121–167
Kohavi R (1995). A study of cross-validation and bootstrap for accuracy estimation and model selection. In: Proceedings of the 14th international joint conference on artificial intelligence, Montreal. Morgan Kaufmann, p 7
Efron B (1979) 1977 Rietz lecture-bootstrap methods: another look at the jackknife. Ann Stat 7:1–26
Pearce TC, Manuel SM (2003) Chemical sensor array optimization: geometric and information theoretic approaches. In: T.C. P, S. SS, T NH, W GJ (eds) Handbook of machine olfaction—electronic nose technology. Wiley, Weinheim
Team RDC (2008) R: A language and environment for statistical computing. R Foundation for Statistical Computing. http://www.R-project.org.
Brereton RG (2006) Consequences of sample size, variable selection, and model validation and optimisation, for predicting classification ability from analytical data. Trac-Trend Anal Chem 25:1103–1111
Brereton RG, Lloyd GR (2014) Partial least squares discriminant analysis: taking the magic away. J Chemometrics 28:213–225
Dixon SJ, Brereton RG (2009) Comparison of performance of five common classifiers represented as boundary methods: Euclidean distance to centroids, linear discriminant analysis, quadratic discriminant analysis, learning vector quantization and support vector machines, as dependent on data structure. Chemometr Intell Lab 95:1–17
Efron B, Tibshirani R (1997) Improvements on cross-validation: the 632 + bootstrap method. JASA 92:548–560
Jain AK, Dubes RC, Chen CC (1987) Bootstrap techniques for error estimation. IEEE Trans Pattern Anal Mach Intell 9:628–633
Xu Y, Zomer S, Brereton RG (2006) Support vector machines: a recent method for classification in chemometrics. Crit Rev Anal Chem 36:177–188
Gunn SR (1998) Support vector machines for classification and regression. Technical Report. http://ce.sharif.ir/courses/85-86/2/ce725/resources/root/LECTURES/SVM.pdf.
Ben-Hur A, Weston J (2010) A user’s guide to support vector machines. Technical report. http://pyml.sourceforge.net/doc/howto.pdf. 609
Westerhuis JA, Hoefsloot HCJ, Smit S, Vis DJ, Smilde AK, van Velzen EJJ, van Duijnhoven JPM, van Dorsten FA (2008) Assessment of PLSDA cross validation. Metabolomics 4:81–89
Goodacre R, Timmins EM, Burton R, Kaderbhai N, Woodward AM, Kell DB, Rooney PJ (1998) Rapid identification of urinary tract infection bacteria using hyperspectral whole-organism fingerprinting and artificial neural networks. Microbiology 144:1157–1170
Goodacre R, Broadhurst D, Smilde AK, Kristal BS, Baker JD, Beger R, Bessant C, Connor S, Calmani G, Craig A, Ebbels T, Kell DB, Manetti C, Newton J, Paternostro G, Somorjai R, Sjostrom M, Trygg J, Wulfert F (2007) Proposed minimum reporting standards for data analysis in metabolomics. Metabolomics 3:231–241
Venables WN, Ripley BD (2002) Modern applied statistics with S. Springer, New York
Kuhn M (2008) Building predictive models in R using the caret package. J Stat Softw 28:1–26
Liaw A, Wiener M (2002) Classification and regression by randomforest. R News 2:18–22
Karatzoglou A, Meyer D, Hornik K (2006) Support vector machines in R. J Stat Softw 15:1–28
Gromski PS, Xu Y, Correa E, Ellis DI, Turner ML, Goodacre R (2014) A comparative investigation of modern feature selection and classification approaches for the analysis of mass spectrometry data. Anal Chim Acta 829:1–8
Guyon I, Weston J, Barnhill S, Vapnik V (2002) Gene selection for cancer classification using support vector machines. Mach Learn 46:389–422
Chapelle O, Vapnik V, Bousquet O, Mukherjee S (2002) Choosing multiple parameters for support vector machines. Mach Learn 46:131–159
Acknowledgments
The authors would like to thank to PhastID (grant agreement no. 258238) which is a European project supported within the Seventh Framework Programme for Research and Technological Development for funding and for the studentship for PSG. Additionally, the authors would like to thank the reviewers for their useful comments and suggestions which have helped us improve our manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 9475 kb)
Rights and permissions
About this article
Cite this article
Gromski, P.S., Correa, E., Vaughan, A.A. et al. A comparison of different chemometrics approaches for the robust classification of electronic nose data. Anal Bioanal Chem 406, 7581–7590 (2014). https://doi.org/10.1007/s00216-014-8216-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-014-8216-7