Abstract
Food safety is a major concern for suppliers in the food chain to ensure the safety of their products. The identification procedure requested by norms is tedious, and it often requires systematic controls and qualified staff to perform the necessary analyses. Raman spectroscopy offers new opportunities to rapidly and efficiently ascertain the presence of pathogens in samples. Nevertheless, this technique requires a standardized procedure to be applied in the industrial context. Our study shows that the variability between spectral fingerprints is related to the physiological state of the microbial species and the growth phase of the bacteria plays a crucial role in its identification by Raman spectroscopy. To improve the discrimination between closely related bacterial species, a procedure based on the selection of bacterial spectra in the exponential growth phase was proposed. Different ways to introduce Raman spectroscopy in the ISO 6579:2002 standards are also proposed from the entire process to a shorter protocol. In the latter case, the identification of bacterial colonies after the selective enrichment step was proposed with the advantages of this path in terms of simplicity and rapidity (analysis time is reduced up to 50 h from the 100 h required by the standard). The protocol validated using six food categories from industrial partners have presented a good correlation by confirmation with other laboratory classical methods. In the future, this procedure could be introduced to the control system of the food production chain with a reliable database for various microorganisms encountered in this field.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The detection of bacteria responsible for food contamination is a key step in the processes of the food industry. The identification of pathogens is recommended by international standards, including the International Standard Organization (ISO), and these regulations define the process that should be followed by operators to ensure the safety of food for consumers [1]. Despite their effectiveness, the current identification process standards are tedious and time-consuming. In our case, the ISO 6579:2002 standard requires the detection of one Salmonella cell in 25 g of a food sample. To perform this task, the standard proposes a series of tedious steps (pre-enrichment, enrichment, selective isolation, and incubation; Fig. 1). The first step (pre-enrichment) is performed in buffered peptone water (BPW), which increases the number of Salmonella cells (if present in the sample) so that they are detectable and compete with the microbiological species present in the sample. The enrichment step in Muller-Kauffmann tetrathionate novobiocine broth (MKTTn) selectively promotes the growth of Salmonella and inhibits Gram-positive bacteria. Bacterial isolation with selective media indicates visually the presence of Salmonella colonies. To assign a species name to the isolated bacteria, each suspect colony is incubated on an agar plate for 15 h, and the obtained colonies are identified by molecular, biochemical, or other methods [2]. This process takes 4 to 5 days to confirm or deny the presence of this pathogen in the analyzed sample. Simplifying this operation and reducing the analysis time is essential to identify pathogens before the product is marketed.
Scheme representing the different paths introduced to apply Raman spectroscopy to the 6579:2002 standards. Path A: Raman analysis was performed on colonies isolated from BPW agar plates, path B: Raman analysis of colonies isolated from XLD agar plates during the selective isolation, and path C: Raman analysis of colonies isolated from BPW agar plates after selective enrichment in MKTTn
Noninvasive strategies have attempted to provide microbiologists more rapid and user-friendly methods to identify bacterial contamination, such as surface plasmons resonance [3], nuclear magnetic resonance [4], mass spectrometry [5], or vibrational spectroscopy including infrared and Raman [6–8]. The use of Raman spectroscopy has been proposed for the analysis of bacteria in various fields [9–11]. In fact, it has been used to identify the presence of bacterial infections isolated from clinical samples [12, 13] and to identify biological contaminants in clean room air [14]. Despite this success, the application of Raman spectroscopy in the food industry is novel and relatively uncommon despite the benefits in this field [15, 16]. This technique, not disrupted by water, the main component of food samples, provides a considerable advantage for the analysis of Clostridium in dairy products compared with other vibrational techniques [17]. Raman microspectroscopy can detect the presence of pathogens frequently encountered in food samples; the lower limit of detection with microspectroscopy is a single bacterial cell [18–23]. However, the methods used to isolate microorganisms can affect their identification by Raman spectroscopy. The spectroscopic detection of certain serotypes of Pseudomonas spp. and Escherichia coli in milk differed depending on the method of isolation that was used [24]. Nevertheless, the undeniable proof of concept provided by published studies, the discrimination between bacterial species remains very complex due to the nature of biological samples. In fact, bacteria are living samples that are evolving with time, and the continuous physiological changes in the composition of bacterial cells complicate their identification by spectroscopic methods. Additionally, their spectral fingerprints depend largely on the culture conditions and the procedure followed for their identification. Some studies have demonstrated the impact of physical factors such as culture media, temperature, and incubation time, on the Raman spectra of bacteria. For example, Harz et al. [25] noted that the Raman spectra of E. coli displayed differences in the bands of tryptophan and phenylalanine when the bacteria were incubated at different temperatures (30 and 37 °C). Other studies reported significant variations in the intensity of nucleic acids bands for Salmonella epidermidis cultured at different times (6, 30, 54, and 72 h). Liu et al. [26] demonstrated that the decrease in the intensity of bacterial nucleic acid bands after long periods of incubation is mainly due to degradation of the genetic material in the bacterial cells. Other than physical factors, bacterial colonies are a source of variability between spectra as shown by Maquelin et al. [27]. These fluctuations were attributed to the presence of physiologically different bacteria, according to their locations in the colony. Choo-Smith et al. [28] confirmed that the spectra of colonies from the same bacterial species are different depending on the incubation time. For analysis, authors recommend the use of small colonies (short time of culture).
To simplify the procedure of analysis, our study suggests Raman spectroscopy as an identification method that allows food manufacturers to quickly detect the presence of pathogenic bacteria in their products. An experimental procedure was proposed to improve the discrimination between the spectra of bacterial colonies with respect to food analysis requirements (robustness, reliability, and simplicity). Different paths to use Raman spectroscopy in ISO 6579:2002 standards are also suggested for the discrimination of bacteria and validated on samples issued from food industry.
Materials and methods
Bacterial strains and culture conditions
Three bacterial strains were analyzed in this study: Salmonella enterica typhimurium (CIP 58.58), E. coli K12 MG1655 (ATCC 700926), and Pseudomonas aeruginosa (CIP A22). These strains were stored in cryotubes at −80 °C in buffered peptone water (BPW). Precultures were carried out in flasks (100 ml) containing 10 ml of BPW from cryotubes and then incubated overnight at 30 °C with stirring at 250 rpm (Infors, AG CH-4103, Bottmingen). Precultures were used to inoculate two cultures an optical densities (OD620 nm initial) = 0.1 in two flasks (250 ml), each containing 65 ml of BPW. Growth was followed over time, 100 μl of bacterial culture are added in disposable cuvettes (Brand GmBH, ref. 759015) containing 900 μl of sterile BPW, and the OD was measured by a spectrophotometer UV–vis (Hitachi U 1800) at 620 nm.
Growth media
Buffered peptone water
Buffered peptone water (Biokar, ref. BK018HA) was prepared with 25.5 g in 1 l of deionized water according to the manufacturer’s instructions. For agar plates, 15 g of bacteriological agar type E (Biokar, ref. A1012HA) was added to 1 l of the liquid medium. These media were autoclaved at 120 °C for 20 min. After sterilization, BPW agar was poured into sterile petri dishes that were stored at 4 °C until their use.
Muller-Kauffmann tetrathionate novobiocine enrichment broth
This medium was prepared by adding 89.5 g of the dehydrated complete medium (Merck, ref. 1058780500) to 1 l of deionized water. The mixture was heated at 95 °C for 15 min and homogenized regularly. After cooling to 50 °C, 20 ml of the iodine-iodide solution was added. The iodine-iodide solution was freshly prepared by adding 5 g of potassium iodide (KI) (Labogros, ref. 9016801) and 4 g of solid iodine beads (I2) (Panreac, ref. 131771.1608) to 20 ml of deionized water.
Xylose-lysine-tergitol 4 broth
Xylose-lysine-tergitol 4 (XLT4) agar was prepared by adding 59 g of dehydrated base medium (Merck, ref. 1139190500) and 4.6 ml of tergitol 4 (Biokar, ref. BS03908) to 1 l of deionized water. The medium was heated slowly to boiling with constant stirring until all components were dissolved. After cooling to approximately 50 °C, the solution was poured into sterile petri dishes that were stored at 4 °C for up to one month.
Xylose-lysine-deoxycholate broth (XLD)
XLD agar was prepared following the same procedure used for XLT4. Briefly, 55.2 g of dehydrated medium (Merck, ref. 1052870500) was added to 1 l of deionized water. The mixture was heated and stirred frequently until it had a transparent, red-orange color. After cooling to approximately 50 °C, the solution was poured into sterile petri dishes that were stored at 4 °C for up to 1 month.
Food samples
Six food categories (rice, oat, wheat, maize, chicken, and pork) received from anonymous industrial partners were distributed in bags in 25-g aliquots and then stored under vacuum at 4 °C until their use. These storage conditions guarantee the chemical and the biological stability of samples over time. Samples were prepared as described by Thouand et al. [29]. Briefly, each sample was performed in duplicate; for negative controls, Stomacher bags containing the food sample and the culture medium (BPW). For positive controls, 10 μl of S. typhimurium (OD620nm = 2) were added to the food samples + BPW (around 2.107 cells were added to the sample). The pre-enrichment, enrichment, and selective isolation steps were performed according to the ISO 6579:2002 standards.
ISO 6579:2002
The incubation of samples was performed according to the horizontal method described by ISO 6579:2002, which is used for the detection of Salmonella spp. in food (Fig. 1). For pre-enrichment, 25 g of sample were added to a Stomacher bag containing 275 ml of BPW (total mass: sample + BPW = 300 g); l0 μl of S. enterica typhimurium (OD = 2) were added only to positive controls. After 18 h of incubation at 37 °C, 1 ml of the resulting solution was added independently to two flasks (100 ml), each containing 10 ml of MKTTn broth. These cultures were then incubated at 37 °C for 15 h with stirring at 100 rpm. After the selective enrichment step in MKTTn, the cultures were spread with a sterile loop onto XLD and XLT4 agar and incubated for 24 h at 37 °C. After the selective isolation, two to four isolated colonies on each selective medium were collected using a sterile loop and cultured again on a BPW agar plate for 15 h at 37 °C. Finally, the obtained colonies were analyzed by Raman spectrometer, identified using API strips (BioMérieux, ref. 20100), and an accredited analytical laboratory confirmed the results (Laboratory for Analysis of the Vendée, France).
Raman analysis
The strains were grown in the recommended conditions (temperature and growth media). For the analysis of bacteria in different growth phases, 20 ml of each culture were collected at different ODs: 2, 4, and 3.8. Samples were centrifuged at 10,000 g for 5 min (Heraeus Primo R Biofuge, Germany) and 10 μl of the obtained pellet were deposited directly onto gold slides. In the case of colonies, after incubation, one isolated colony was spread onto gold surfaces using the rounded tip of a sterile Pasteur pipette. In the both case, the deposit was analyzed by Raman spectrometer after 5 min of drying at room temperature. A total of 15 spectra were acquired on each deposit and 4 to 5 colonies were analyzed from each agar plate.
Gold surface preparation
Gold surfaces were prepared according to the procedure used at the Jean Rouxel Institute (University of Nantes, France). Briefly, ordinary glass microscope slides (ISO 8037, ref. RS, France) were cut into rectangular areas (26 × 9 × 1 mm) and heated in freshly prepared Piranha solution (70 % v/v H2SO4 and 30 % v/v H2O2) at 70 °C for 30 min. The surfaces were washed several times with deionized water and ethanol (Labogros, ref. 9006902). After drying, a 30-nm chromium layer (Sigma-Aldrich, ref. 266264) and a 100-nm gold layer (Goodfellow, ref. AU005160/72) were deposited onto surfaces by physical vapor deposition using Alcatel built machine.
Raman microspectroscopy measurements and analysis conditions
Raman spectra were performed using an Xplora TM spectrometer (Horiba Jobin Yvon, Lille, France) driven by the LabSpec software (V5). This device was equipped with multiple gratings (1,800, 1,200, and 600 lines/mm), a camera CCD cooled to −80 °C, and a BX41 Olympus microscope with multiple objectives (LCLanN100xIR/0.85 and LMPLanFLN50x/0.5). Analyses were performed at 785 nm and approximately 30 mW on the sample. The spectral resolution was approximately 4 cm−1, and the spectral range was recorded from 400 to 1,800 cm−1. This region contained all the Raman bands necessary for the discrimination of bacteria. For each spectrum, five acquisitions of 15 s each were used for measurements.
Preprocessing spectra and data analysis
Raman spectra were processed using the Opus software (Bruker optics GmBH, V 6.5). Elastic concave (64° and 10 iterations) was used for baseline corrections in the range of interest (400 to 1,800 cm−1). The Savitzky-Golay algorithm order 9 was used to smooth the spectra. The maximum-minimum bands or vector normalization was used to normalize the spectra.
Hierarchical cluster analysis (HCA) based on Ward’s algorithm (squared Euclidean distance) and principal component analysis (PCA) were used to make comparison between Raman spectra (XLSTAT, Addinsoft v2008). Before classification, the spectra were centered by subtraction of the average of each group [30]. As first principal components explain the majority of variance of the dataset, our argumentation and observation was based on the PC1 and PC2 scores.
MATLAB (v.2013) and “SAISIR package” [31] was used to perform analysis of variance (ANOVA) using the Anova1 function. The correlation coefficient between the spectra was calculated using Opus software (option: spectra comparison). For the selection of good spectra, spectra are compared with the average spectrum of the exponential phase; only spectra with a correlation ≥98.5 % were selected to make comparison.
Results
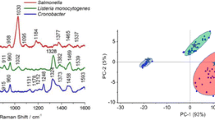
Raman spectroscopy has been used to analyze S. enterica typhimurium from liquid culture or colonies to evaluate its introduction in ISO 6579:2002 standard. This method should be robust, reproducible, and reliable for pathogen identification in the industrial context. Preliminary results show a significant variability between the spectra of bacterial colonies (approximately 40 %), making discrimination between analyzed species impossible (Fig. 2). In fact, the hierarchical cluster analysis performed on the spectra considering the total spectral ranges (400–1,800 cm−1) indicates the absence of separate clusters specific to each analyzed species; spectra of S. enterica typhimurium, E. coli, and P. aeruginosa are frequently classified in the same group. The observed confusion between spectra prevents the direct use of Raman spectroscopy for pathogen identification.
Hierarchical cluster analysis based on Ward’s algorithm (squared Euclidean distance) of Raman spectra of three bacterial species: Escherichia coli (1 to 40), Salmonella enterica typhimurium (1 to 35), and Pseudomonas aeruginosa (1 to 21). Bacterial colonies were cultured on BPW agar plates at 30 °C for 18 h. Raman parameters: λ = 785 nm, laser power = 30 mW, objective: LMPLanFLN50x /0.5, grating = 600 l/mm, and acquisition time = 15 s/5 cycles
Raman analysis of S. enterica typhimurium in different growth phases (liquid culture)
To improve the discrimination between strains, the effect of bacterial age on the spectral fingerprint of S. enterica typhimurium was studied. The influence of the growth phase on the Raman spectra was measured in exponential phase (OD620nm = 2.0, approximately 4 h of culture), stationary phase (OD620nm = 4.0, approximately 15 h), and prolonged stationary phase (OD620nm = 3.8, approximately 24 h of culture).
Raman bands are assigned according to Maquelin et al. [11]; our obtained spectra displayed several Raman band intensity differences (Fig. 3). The most significant difference observed between spectra was attributed to nucleic acids bands (DNA and RNA at 778 and 810 cm−1, respectively). The RNA/DNA ratios calculated from 30 spectra under the test conditions were 0.77 (±0.06), 0.52 (±0.06), and 0.25 (±0.09) for the exponential, stationary, and prolonged stationary phases, respectively. This ratio can be collected from a bacterial cell’s Raman spectrum and used as an age indicator for that bacterial cell. The higher ratio was close to 1 (intensity of RNA band is high), indicating that the analyzed bacteria were in exponential phase. If the ratio is lower, the bacterial cells are older. Other slight band differences were found at 1,350, 1,450, and 1,660 cm−1; these bands were attributed to protein and lipid structures. The discrimination between spectra by principal component analysis showed the presence of three separate groups that were classified according to their growth phase (exponential, stationary, and prolonged stationary; Fig. 4). This classification indicates that the dispersion of spectra is not similar between groups, as the spectra of bacteria in exponential and early stationary phases were homogeneous (the correlation rate is 98 and 97 %, respectively), whereas the spectra of the prolonged stationary phase were more dispersed (the correlation is approximately 90 %).
Raman spectra of Salmonella enterica typhimurium from liquid culture (BPW) performed on different growth phases. A) spectrum at 4 h of culture, B) spectrum at 15 h of culture and C) spectrum at 24 h of culture. Areas colors were assigned based on the difference of intensity between the Raman bands and their shapes. Raman parameters: λ = 785 nm, laser power = 30 mW, objective: LMPLanFLN50x /0.5, Grating = 600 l/mm and acquisition time = 15 seconds/5 cycles
Discrimination between S. enterica typhimurium and E. coli according to their growth phases (liquid culture)
The results presented above were used to discriminate between S. enterica typhimurium and E. coli, two species that are very similar physiologically. The discrimination between spectra obtained from different cultures of different ages confirms that the exponential phase is the best to discriminate between these two species (Fig. 4). In fact, the Raman spectra were separated into two groups. In contrast, bacteria in stationary phase formed two groups with very similar spectra and were frequently classified as the same species, particularly during prolonged stationary phase. Therefore, the exponential phase was chosen as the best stage to discriminate between bacterial species for two reasons: (1) the homogeneity of spectra is very high, and (2) the culture time is approximately 4 h, which reduces the analysis time.
Raman analysis of S. enterica typhimurium and E. coli grown on agar plates of BPW (bacterial colonies)
Despite the successful discrimination between bacteria that have high-intensity RNA bands, Raman analysis of bacteria from liquid culture in exponential phase is not a realistic prospect for application in the food industry. In fact, this culture method is not recommended for industrial application due to the need to cultivate each suspect colony and monitor its growth over time in culture. ISO 6579:2002 standard is able to identify bacteria from microbial colonies; this method of cultivation is very practical, as bacteria in this form have the advantage of being easily handled and stored. Despite these advantages, colonies contain bacterial cells that are physiologically very heterogeneous, fulfilling the requirement of a reliable process to generate good spectra to ensure the best identification.
As previously noted, the exponential phase achieved the best discrimination between bacteria. Thus, the average spectrum of this phase was considered as a reference spectrum. After comparison, a correlation coefficient of 98.5 % with this spectrum was necessary to obtain perfect identification (100 %). Subsequently, this quality criterion was applied to all of the spectra from randomly measured colonies. This procedure aimed to select only spectra of bacterial cells in the colony that are still in exponential phase (high intensity of nucleic acid bands) and eliminate those that had a lower rate with this criterion. The comparison between the spectra of colonies cultured during 18 h and the average spectrum of exponential phase revealed several good spectra. To determine the minimum number of spectra required for optimal identification, 15 spectra were collected from each colony, and 6 colonies in the same culture were analyzed. Out of 90 spectra (15 spectra × 6 colonies), only 29 showed a correlation coefficient ≥98.5 % with the average spectrum of the exponential phase, a success rate of approximately 32 % (±0.75 %). Then, the reproducibility of measurements was evaluated between colonies from different cultures. Ninety spectra were collected from 6 colonies from 6 different cultures, providing 26 spectra with a correlation coefficient ≥98.5 % and a success rate of approximately 28 % (±0.8). These results demonstrate the collection of the same number of spectra selected from colonies grown under the same incubation conditions despite their affiliation to different cultures. This result indicates that the operator must perform at least three spectra from the same bacterial colony to obtain a reliable spectrum for identification. The application of this strategy on the spectra of S. enterica typhimurium and E. coli was effective. Figure 5 shows that the spectra are separated into two distinct groups and that each is specific for one bacterial species. Therefore, the selection of spectra from only young bacteria in a colony improves the classification between strains.
Discrimination between the Raman spectra of Escherichia coli and Salmonella enterica typhimurium colonies after the selection of good spectra from bacteria (correlation ≥98.5 % with the spectrum of the exponential growth phase). (Black square) Spectra of Salmonella enterica typhimurium, (black triangle) spectra of Escherichia coli
Evaluation of the food matrix and the selective media effects on the Raman spectra of S. enterica typhimurium
The standard applied to Salmonella spp. research can be divided into several paths; only path (A) is required to identify isolated colonies by the recommended methods (biochemical, molecular, or otherwise). To simplify the analysis, two other ways was considered to identify bacteria: path (B) directly analyzes bacterial colonies inoculated on an agar plate containing selective media (XLD and XLT4) and path (C) analyzes bacterial colonies inoculated on BPW agar plates after the selective enrichment in MKTTn (Fig. 1).
The quality of the Raman spectrum is critical for the identification process. Evaluating the impact of the food matrix on the spectral fingerprint helps the operator to choose, create, and organize the database necessary for the discrimination. The impact of two-sample matrix (oat and rice) was evaluated on the quality of Raman spectra from S. enterica typhimurium colonies isolated from positive cultures and incubated following the entire process of the standard (path A in Fig. 1). The data were compared with the reference spectra of S. enterica typhimurium inoculated onto BPW agar plates directly from precultures. ANOVA has been used on PC1 and PC2 scores of PCA done on this data to testing the level of similarity from a statistic point of view. The ANOVA results showed a p value equal to 0.0688. This value was higher than 0.05, indicating that all sample spectra were not significantly different at 95 % of confidence level (see Electronic Supplementary Material Fig. S1). Therefore, the food matrix did not influence the final spectra. Next, the impact of the selective growth media (XLD and XLT4) used in the ISO 6579:2002 standard was evaluated on the quality of Salmonella Raman spectra. On this media, Salmonella colonies often have a characteristic black color, while interfering strains are yellow. Raman analysis of black colonies of S. enterica typhimurium deposited on gold surfaces showed that the spectra were disturbed by fluorescence; some bands, from unidentified sources, were very different from the reference Raman spectra obtained from Salmonella colonies grown on BPW agar. Washing the black colonies several times with water or MgSO4 (10−2 M) did not eliminate the molecules causing the spectral changes (see Electronic Supplementary Material Fig. S2). The spectra from young Salmonella colonies (isolated before the appearance of the black color at approximately 10 h of culture) lacked fluorescence, but they were remarkably different from the spectra of Salmonella grown on BPW agar plates (77 % of correlation), mainly in the 400–1,200 cm−1 region. Given the small colony size, many colonies were picked to perform the analysis. The analysis of colonies after 48 h of culture (after disappearance of the black color) showed new bands at 476 and 511 cm−1, which were attributed to the chemical products present in the selective media. These results show that even after the selective process, Salmonella should always be inoculated on BPW agar plates to ensure the quality of the spectra.
Choice of the analysis path and the validation of protocol in industrial food context
The analysis using path A requires 100 h and lots of incubation and isolation steps (Fig. 1). Path B cannot be used to identify colonies grown on selective media because the media interfere with spectra collection, complicating the bacterial identification. To simplify the analysis, samples were inoculated onto BPW agar plates after the selective enrichment step in MKTTn (path C in Fig. 1), which reduced the analysis time up to 50 h. The comparison between the obtained and reference spectra showed an average correlation rate of 98.5 % (result not shown). This procedure was therefore adopted for all remaining samples. Six types of samples were analyzed, including four types of cereal (maize, rice, wheat, and oat) and two types of meat (pork and chicken). Raman spectra of all the isolated species other than S. typhimurium are plotted in Fig. 6a; the visual inspection of these spectra detected different bands of the S. enterica typhimurium fingerprint, mainly in the 400–800 cm−1 region. Principal component analysis of approximately 150 spectra showed the presence of 5 distinct groups (Fig. 6b). The S. enterica typhimurium introduced into samples (positive cultures) were classified in a separate group. Isolated strains were classified into groups according to their similarities. The spectra of Citrobacter freundii (isolated from oat and pork samples) and Citrobacter youngae (isolated from chicken) were classified into two neighboring groups. The group formed from the spectra of Enterobacter sakazakii (isolated from maize) and Enterobacter claocae (isolated from rice and pork samples) were very close. Finally, Klebsiella pneumonia spectra were isolated from wheat and oat samples. Thus, Raman spectroscopy discriminated between species of the same genus at an early stage of the identification process.
a Comparison between Raman spectra of different species isolated from food industry samples via path C. ES: Enterobacter sakazakii isolated from maize samples, EC: Enterobacter claocae isolated from rice and pork samples, CY: Citrobacter youngae isolated from chicken, CF: Citrobacter freundii isolated from oat and pork samples, KP: Klebsiella pneumonia isolated from wheat and oat samples. Raman parameters: λ = 785 nm, laser power = 30 mW, objective: LMPLanFLN50x /0.5, grating = 600 l/mm, and acquisition time = 15 s/5 cycles. b Discrimination between Raman spectra of Salmonella enterica typhimurium (black triangle) and bacterial species isolated from various samples. (Gray square) Enterobacter cloacae isolated from rice and pork samples, (red circle) Klebsiella pneumonia isolated from wheat and oat samples, (blue square) Citrobacter youngae isolated from chicken, (diamond) Citrobacter freundii isolated from oat and pork samples, and (brown square) Enterobacter sakazakii isolated from maize samples. Raman parameters: λ = 785 nm, laser power = 30 mw, and i = 10 s × 5 cycles. The average values from five Raman spectra were used in this figure
Discussion
Raman spectroscopy identifies microorganisms very quickly based on their spectral fingerprints without the use of reagents [32–34]. This identification method is comparable to phenotypic and genotypic methods and its discriminatory power is very high for bacterial species of different families [10, 35, 36]. Regardless of the power of this technique, this study demonstrates that using Raman spectroscopy to discriminate between physiologically closely related bacterial species, such as S. enterica typhimurium and E. coli K12, should follow a standard procedure. Otherwise, the discrimination between these strains is difficult (Fig. 2), reducing the utility of this technique for the reliable identification of bacteria in an industrial context.
The analysis of bacterial suspensions of S. enterica and E. coli according to their growth phases demonstrated that the Raman spectra of these species depend on their physiological state, in accordance with previous studies [26, 37]. The differences are remarkable, especially for the region of nucleic acid bands. Moreover, the results demonstrate the existence of a relationship between the age of a bacterium and its ability to be identified. In fact, it is preferable that the bacteria are in the exponential growth phase for identification. The heterogeneity between spectra from the colonies is related to the presence of bacterial cells in several physiological states. This characteristic complicates the application of this cultivation method for the identification due to the high variability observed between spectra. To reduce this variability, the use of small-sized colonies was proposed because at this stage, bacteria are homogenous [28]. In contrast, a procedure to select the spectra of bacteria was proposed in our study without changing the usual conditions described by ISO 2002:6579. For identification, only the spectra corresponding to the quality criterion with a correlation coefficient ≥98.5 % relative to the spectrum of the exponential growth phase was selected. A minimum of three spectra were needed to be assessed from a bacterial colony to obtain at least one that is suitable for the identification. The use of these selection criteria could successfully discriminate between species that are physiologically closely related, and it shows that the application of a standardized procedure improves the discriminatory power of Raman spectroscopy.
Our analyses were performed on samples from the food industry, while other studies have mostly used laboratory samples that are more easily controlled and manipulated [19, 38]. The same problems and the same difficulty were encountered as in industrial operations for analyzing samples with unknown contaminants. In our conditions, the food matrix did not affect the identification of contaminants if the bacteria were inoculated on the same growth media. Spectra of S. enterica typhimurium were grouped together despite their different origins (reference spectra and those from bacteria isolated by the enrichment process). While our study showed that the quality of Raman spectra depended on the culture media, the problem of detection related to the isolation method mentioned by Meisel et al. [24] was resolved with a standardized method using BPW agar plates. Following the entire process of the standard (path A), the colonies of S. enterica typhimurium were identified; in this case, Raman spectroscopy is comparable to other identification techniques proposed by the ISO standard. Nevertheless, the method is very rapid (50 s/spectrum) and did not require any extraction of the cellular materials as for polymerase chain reaction (PCR). Spectra from colonies grown on agar plates containing selective media showed that path B could not be used during identification, as many Raman bands were generated by the selective media. The analysis of colonies at different growth stages on selective media did not provide interesting results. The good discrimination provided by path C (bacteria inoculated from MKTTn broth media) confirmed that this approach is a potential alternative for industrial applications. This approach significantly reduced the analysis time by 50 %; it also reduced the number of isolation and culture steps, which simplified the analysis procedure. Path C has other interesting advantages; following this path, the easy process provided by Raman spectroscopy may be possible in the identification of the most cultivable bacteria after the MKTTn enrichment step. If the standard process had only been used, pathogens such as E. sakazakii and other dangerous species would not have been identified in the industrial samples.
Conclusion
Raman spectroscopy offers manufacturers an interesting tool to systematically monitor their production due to its speed and ability to identify pathogens. Despite the advantages of this technique, some of the pitfalls associated with species variability remain to be overcome. This study proposes an experimental approach to improve the discrimination between bacterial colonies with the goal of introducing this technique in the ISO (6579:2002) standard. Our study showed that the exponential phase is the best for identifying bacterial species. A procedure was proposed to select spectra from young bacterial cells by comparing the spectra performed on colonies with the average spectrum of the exponential phase. After selection, the spectra of S. typhimurium and E. coli were identified at 100 %. Results show that the food matrix did not affect the discrimination between bacteria cultivated by the standard process (path A). Despite its efficiency, the standard method requires a series of culture and isolation steps, which are often too time consuming and tedious to be used routinely. Given that bacteria on selective media cannot be analyzed directly, the colonies inoculated on BPW agar after the selective enrichment step (path C) were analyzed. The advantages of this path include simplicity and reduced time (analysis time is reduced up to 50 h from the 100 h required by the standard). This path enabled the identification of bacterial strains in only three steps. In contrast, the standard identification method requires at least five culture steps to confirm the presence of one bacterial species (Salmonella spp.). Thanks to Raman spectroscopy, it is possible to make the identification of most cultivable bacteria after the MKKTn enrichment; the mesh size is then narrowed and much more pathogenic bacteria can be identified in a limited time and steps. Despite the advantages of the proposed method, this procedure should be validated with other types of samples; this validation will be addressed in future studies.
References
McMeekin T A (ed) (2003) Detecting pathogens in food. Woodhead Publishing Series in Food Science, Technology and Nutrition No.84
ISO 6579:2002, Horizontal method for the detection of Salmonella spp. Microbiology of food and animal feeding stuffs
Spadavecchia J, Manera MG, Quaranta F, Siciliano P, Rella R (2005) Surface plasmon resonance imaging of DNA based biosensors for potential applications in food analysis. Biosens Bioelectron 21(6):894–900
Kern T, Giffard M, Hediger S, Amoroso A, Giustini C, Bui NK, Joris B, Bougault C, Vollmer W, Simorre P (2010) Dynamics characterization of fully hydrated bacterial cell walls by solid-state NMR: evidence for cooperative binding of metal ions. J Am Chem Soc 132(31):10911–10919
Claydon MA, Davey SN, Edwards Jones V, Gordon DB (1996) The rapid identification of intact microorganisms using mass spectrometry. Nat Biotechnol 14(11):1584–1586
Mariey L, Signolle JP, Amiel C, Travert J (2001) Discrimination, classification, identification of microorganisms using FTIR spectroscopy and chemometrics. Vib Spectrosc 26(2):151–159
Naumann D, Helm D, Labischinski H (1991) Microbiological characterizations by FTIR spectroscopy. Nature 351(6321):81–82
Kudelski A (2008) Analytical applications of Raman spectroscopy. Talanta 76(1):1–8
Choo–Smith LP, Maquelin K, Endtz P, Bruining HA, Puppels GJ (1999) A novel method for rapid identification of micro-organisms using confocal Raman microspectroscopy. Spectrosc Biol Mol 8:537–540
Kirschner C, Maquelin K, Pina P, Thi NAN, Choo-Smith LP, Sockalingum GD, Sandt C, Ami D, Orsini F, Doglia SM, Allouch P, Mainfait M, Puppels GJ, Naumann D (2001) Classification and identification of enterococci: a comparative phenotypic, genotypic, and vibrational spectroscopic study. J Clin Microbiol 39(5):1763–1770
Maquelin K, Kirschner C, Choo-Smith LP, van den Braak N, Endtz HP, Naumann D, Puppels GJ (2002) Identification of medically relevant microorganisms by vibrational spectroscopy. J Microbiol Methods 51(3):255–271
Maquelin K, Kirschner C, Choo-Smith LP, Ngo-Thi NA, van Vreeswijk T, Stammler M, Endtz HP, Bruining HA, Naumann D, Puppels GJ (2003) Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J Clin Microbiol 41(1):324–329
Jarvis RM, Goodacre R (2004) Ultra-violet resonance Raman spectroscopy for the rapid discrimination of urinary tract infection bacteria. Fems Microbiol Lett 232(2):127–132
Rosch P, Harz M, Schmitt M, Peschke KD, Ronneberger O, Burkhardt H, Motzkus HW, Lankers M, Hofer S, Thiele H, Popp J (2005) Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: application to clean-room-relevant biological contaminations. Appl Environ Microbiol 71(3):1626–1637
Hassing S, Jernshøj KD and Christensen L S (2012) In: Kostas Kapiris (ed) Raman spectroscopy: a non-destructive and on-site tool for control of food quality. InTech
Das RS, Agrawal YK (2011) Raman spectroscopy: recent advancements, techniques and applications. Vib Spectrosc 57:163–176
Fehrmann A, Franz M, Hoffmann A, Rudzik L, Wust E (1995) Dairy product analysis: Identification of microorganisms by mid-infrared spectroscopy and determination of constituents by Raman spectroscopy. J AOAC Int 78(6):1537–1542
Mizrach A, Schmilovitch Z, Korotic R, Irudayaraj J, Shapira R (2007) Yeast detection in apple juice using Raman spectroscopy and chemometric methods. Am Soc Agric Biol Eng 50(6):2143–2149
Fan C, Hu ZQ, Mustapha A, Lin MS (2011) Rapid detection of food and waterborne bacteria using surface-enhanced Raman spectroscopy coupled with silver nanosubstrates. Appl Microbiol Biotechnol 92(5):1053–1061
Meisel S, Stockel S, Elschner M, Melzer F, Rosch P, Popp J (2012) Raman spectroscopy as a potential tool for detection of Brucella spp. in milk. Appl Environ Microbiol 78(16):5575–5583
Oust A, Møretrø T, Naterstad K, Sockalingum K, Adt I, Manfait M, Kohler A (2006) Fourier transform infrared and raman spectroscopy for characterization of Listeria monocytogenes strains. Appl Environ Microbiol 72(1):228
Sengupta A, Mujacic M, Davis EJ (2006) Detection of bacteria by surface enhanced Raman spectroscopy. Anal Bioanal Chem 386(5):1379–1386
Meisel S, Stockel S, Rosch P, Popp J (2013) Identification of meat-associated pathogens via Raman microspectroscopy. Food Microbiol 38:36–43
Meisel S, Stockel S, Elschner M, Rosch P, Popp J (2011) Assessment of two isolation techniques for bacteria in milk towards their compatibility with Raman spectroscopy. Analyst 136(23):4997–5005
Harz A, Rosch P, Popp J (2009) Vibrational spectroscopy—a powerful tool for the rapid identification of microbial cells at the single-cell level. Cytometry Part A 75A(2):104–113
Liu TT, Lin YH, Hung CS, Liu TJ, Chen Y, Huang YC, Tsai TH, Wang HH, Wang DW, Wang JK, Wang YL, Lin CH (2009) A high speed detection platform based on surface-enhanced Raman scattering for monitoring antibiotic-induced chemical changes in bacteria cell wall. PLoS ONE 4(5)
Maquelin K, Choo-Smith LP, van Vreeswijk T, Endtz HP, Smith B, Bennett R, Bruining HA, Puppels GJ (2000) Raman spectroscopic method for identification of clinically relevant microorganisms growing on solid culture medium. Anal Chem 72(1):12–19
Choo-Smith LP, Maquelin K, van Vreeswijk T, Bruining HA, Puppels GJ, Ngo-Thi NA, Kirschner C, Naumann D, Ami D, Villa AM, Orsini F, Doglia SM, Lamfarraj H, Sockalingum GD, Mainfait M, Allouch P, Endtz HP (2001) Investigating microbial (micro)colony heterogeneity by vibrational spectroscopy. Appl Environ Microbiol 67(4):1461–1469
Thouand G, Vachon P, Liu S, Dayre M, Griffiths MW (2008) Optimization and validation of a simple method using P22:luxAB bacteriophage for rapid detection of Salmonella enterica serotypes A, B, and D in poultry samples. J Food Prot 71(2):380–385
Cordella C B Y (2012) PCA: The Basic Building Block of Chemometrics, Analytical Chemistry. In: Ira S. Krull (ed) Available from: http://www.intechopen.com/books/analytical-chemistry/pca-the-basic-building-block-of-chemometrics
Cordella C B Y, Bertrand D (2013) SAISIR®: a new general chemometric toolbox. Trends Anal Chem 54:75--82
Greve J, Puppels GJ (1993) Raman microspectroscopy of single whole cells. Biomolecular Spectroscopy. Wiley, Chichester, pp 231–265
Naumann D (1998) Infrared and NIR Raman spectroscopy in medical microbiology. SPIE, Washington, USA, pp 245–257
Huang YS, Karashima T, Yamamoto M, Ogura T, Hamaguchi H (2004) Raman spectroscopic signature of life in a living yeast cell. J Raman Spectrosc 35(7):525–526
Tripathi A, Jabbour RE, Treado PJ, Nelson MP, Jensen JL, Sneyder AP (2007) Detection and identification of a water mixture of E. coli cells and B. subtilis spores with Raman chemical imaging microscopy-art. no. 65540J. Chemical and Biological Sensing VIII. A. W. Fountain. Bellingham. Spie-Int Soc Opt Eng 6554:J5540–J5540
Jarvis RM, Brooker A, Goodacre R (2006) Surface-enhanced Raman scattering for the rapid discrimination of bacteria. Faraday Discuss 132:281–292
Harz M, Rosch P, Peschke KD, Ronneberger O, Burkhardt H, Popp J (2005) Micro-Raman spectroscopic identification of bacterial cells of the genus Staphylococcus and dependence on their cultivation conditions. Analyst 130(11):1543–1550
Yang H, Irudayaraj J (2003) Rapid detection of foodborne microorganisms on food surface using Fourier transform Raman spectroscopy. J Mol Struct 646(1–3):35–43
Acknowledgments
The authors thank the Conseil General de la Vendée, the town of la Roche sur Yon, and the region of Pays de la Loire for the financial assistance. Authors thank also Pr. Ganesh Sockalingum from the University of Reims Champagne-Ardenne and Dr. Guy Louarn from the Nantes Materials Institute for their help to achieve this work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 312 kb)
Rights and permissions
About this article
Cite this article
Assaf, A., Cordella, C.B.Y. & Thouand, G. Raman spectroscopy applied to the horizontal methods ISO 6579:2002 to identify Salmonella spp. in the food industry. Anal Bioanal Chem 406, 4899–4910 (2014). https://doi.org/10.1007/s00216-014-7909-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-014-7909-2