Abstract
The combination of ultrahigh-resolution mass spectrometry imaging (UHRMSI) and ultrahigh-performance liquid chromatography coupled with tandem mass spectrometry (UHPLC/MS/MS) was used for the identification and the spatial localization of atorvastatin (AT) and its metabolites in rat tissues. Ultrahigh-resolution and high mass accuracy measurements on a matrix-assisted laser desorption/ionization (MALDI)-Orbitrap mass spectrometer allowed better detection of desired analytes in the background of matrix and endogenous compounds. Tandem mass spectra were also used to confirm the identification of detected metabolites in complex matrices. The optimization of sample preparation before imaging experiments included the tissue cryogenic sectioning (thickness 20 μm), the transfer to stainless steel or glass slide, and the selection of suitable matrix and its homogenous deposition on the tissue slice. Thirteen matrices typically used for small molecule analysis, e.g., 2,5-dihydroxybenzoic acid (DHB), 1,5-diaminonaphthalene (DAN), 9-aminoacridine (AA), etc., were investigated for the studied drug and its metabolite detection efficiency in both polarity modes. Particular matrices were scored based on the strength of extracted ion current (EIC), relative ratio of AT molecular adducts, and fragment ions. The matrix deposition on the tissue for the most suitable matrices was done by sublimation to obtain the small crystal size and to avoid local variations in the ionization efficiency. UHPLC/MS profiling of drug metabolites in adjacent tissue slices with the previously optimized extraction was performed in parallel to mass spectrometry imaging (MSI) measurements to obtain more detailed information on metabolites in addition to the spatial information from MSI. The quantitation of atorvastatin in rat liver, serum, and feces was also performed.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
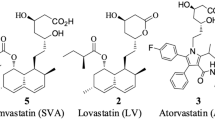
Atorvastatin (Fig. 1), as an inhibitor of 3-hydroxy-3-methylglutaryl reductase, belongs to the group of statins used in hyperlipidemia therapy [1] and is the best-selling drug in history. Most published articles are related to its determination in biological samples [2, 3], the characterization of its metabolism [4, 5], and the description of its oxidative degradation products [6, 7]. Atorvastatin analytical characterization and pharmaceutical and medical effects are described as well [1, 2]. The choice of suitable analytical technique for the sample characterization in the area of medical and pharmaceutical research is an important task. The coupling of separation techniques and mass spectrometry (MS) is usually used for the analysis of body fluids [8], but this technique is unable to localize the spatial distribution of particular compounds in studied tissues and organs. The visualization of biological tissues is usually done by optical microscopy after various histological stainings with acidic or basic stains, immunohistochemistry, or fluorescence microscopy. However, the imaged species are observed indirectly without the molecular specificity. These techniques can only be used in a targeted manner for known compounds, but only a limited number of target compounds can be visualized from a given sample at the same time [9]. MS imaging (MSI) allows the label-free detection and the mapping of wide range of biological compounds in one analysis based on the direct measurement of m/z values. Several ionization techniques can be used for MSI, e.g., matrix-assisted laser desorption/ionization (MALDI) [9–11], desorption electrospray ionization [12], secondary ion mass spectrometry [13, 14], and other desorption ionization techniques. Most widespread MSI scanning method is MALDI with a prominent role in the analysis of synthetic polymers [15] and biopolymers [16], such as proteins, peptides, oligonucleotides, and oligosaccharides since its introduction by Hillenkamp and Karas [17]. Nowadays, the MALDI is also considered in the area of small molecule analysis [10, 18–20]. The successful application of this technique is, however, hampered by low molecular weight matrix-derived interference signals and poor reproducibility of signal intensity arising from the difficulty of uniform matrix deposition. Several strategies for the improvement of MALDI-MSI performance for small molecules were developed, e.g., the use of high molecular weight matrices [19], solvent-free matrix deposition [21], cationization agents [19], ion-less matrices [22], matrix-free approaches [23], or tandem mass spectrometry (MS/MS) strategies [24]. Another possibility to filter matrix-derived signal is the use of ultrahigh-resolution mass spectrometry (UHRMS) [11, 25–27], such as Orbitrap or ion cyclotron resonance mass analyzers. In addition to matrix interferences, signals of endobiotic or better endogenous compounds (mainly lipids) with m/z values close to ions of desired drug or its metabolites can be present in mass spectra of biological tissues. Their insufficient separation can provide false-positive results and distorts pharmaceutical studies.
In our work, ultrahigh-resolution MALDI tandem mass spectrometer linear trap quadropole (LTQ) Orbitrap XL was used to localize atorvastatin and its metabolites in selected rat tissues. The application of ultrahigh-performance liquid chromatography (UHPLC)/electrospray ionization (ESI)-MS/MS of adjacent tissue slides was performed in parallel to verify the presence of metabolites and quantify atorvastatin (AT) in studied rat samples. The model for evaluation of individual matrices for small molecule analysis was developed and applied for the analysis of AT and its metabolites.
Materials and methods
Chemicals and reagents
Atorvastatin (3R,5R)-7-[2-(4-fluorophenyl)-3-phenyl-4-(phenylcarbamoyl)-5-propan-2-ylpyrrol-1-yl]-3,5-dihydroxyheptanoic acid) calcium trihydrate was purchased from the European Directorate for the Quality of Medicines & HealthCare Council of Europe (Strasbourg, France). Atorvastatin-d5 was purchased from Toronto Research Chemicals (Ontario, Canada). Acetonitrile, methanol, acetic acid, ammonium acetate, sodium formate, and all studied matrices (2,5-dihydroxybenzoic acid (DHB) and its sodium salt (sodium 2,5-dihydroxybenzoate (NaDHB)), 1,5-diaminonaphthalene (DAN), 2-mercaptobenzothiazole (MBT), 9-aminoacridine (AA), 3-aminoquinoline (AQ), N,N,N′,N′-tetramethyl-1,8-naphthalenediamine (DMAN), 2,6-dihydroxyacetophenone (DHA), 2,4,6-trihydroxyacetophenone (THA), 2-aminopyrazine (AP), 3-aminoacetophenone (AAP), α-cyano-4-hydroxycinnamic acid (CHCA), and butylamine salt of α-cyano-4-hydroxycinnamic acid (BuCHCA)) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Deionized water was prepared with Demiwa 5-roi purification system (Watek, Ledeč nad Sázavou, Czech Republic).
In vivo experiment design
Animals
Males of laboratory rats (Wistar Han II, average weight 255 ± 12 g) were purchased from the breeding facility Velaz (Koleč, Czech Republic). Atorvastatin was administered orally using the gastric gavage in the dose of 100 mg/kg of body weight (diluted in 0.5 % methylcellulose). The physiological saline solution was orally administered in case of control group (average weight 235 ± 9 g). Animals were placed into metabolic cages for the collection of urine and feces. Rats were sacrificed in halothane inhalation anesthesia 3.5 h after the drug administration, and plasma, urine, tissue samples (brain, heart, kidney, and liver), and the content of intestine (feces) were collected. All samples were stored at −80 °C before the sample preparation. The study was approved by the Institutional Review Board of the Animal Care Committee. Animals were held and treated in accordance with the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (Council of Europe 1986). Rat tissues were sectioned at 20-μm thickness using Leica CM1950 cryostat microtome (Leica Microsystems, Wetzlar, Germany) before MALDI-MSI experiments. Adjacent tissue slides of 60 μm were obtained and subsequently subjected to UHPLC/MS analysis.
Sublimation matrix deposition [28]
Each tissue slide for the imaging experiment was immediately thaw-mounted onto stainless or microscopic glass plate and placed on the bottom side of sublimation condenser. The matrix deposition was performed in a modified sublimation apparatus (Chemglass Life Science, Vineland, NJ) with a constant vacuum of 0.03 mbar. The stable vacuum was obtained using the rotary pump Edwards E2M28 (Edwards, Crawley, UK) supplemented with the solenoid valve DN15 (Vacuubrand, Wertheim, Germany) and controlled by the vacuum controller CVC 3000 (Vacuubrand). The sublimation time and temperature obtained in oil bath differed for particular used matrices (details in Table 1). Our experimental setup for the sublimation deposition is shown in the Electronic Supplementary Material (Fig. S1).
Extraction of AT and metabolites
Tissue slides of 60-μm thickness were transferred to tempered vials. The weight of tissue slice was between 6 and 12 mg depending on the particular organ or tissue section. The extraction of AT and its metabolites from rat tissues was done by the addition of 2 ml mixture of methanol–water (9:1, v/v). Of the same solution mixture, 4 ml was added to 202 mg of rat feces and 4 ml of pure methanol was used in case of other rat samples involving 1 ml of serum and 0.5 ml of urine, respectively. The internal standard AT-d5 was dissolved in methanol and added to all samples before the extraction procedure. All samples were further homogenized and centrifuged at 3,000×g for 5 min. Supernatants were evaporated to dryness using the nitrogen stream. Dry samples were quantitatively dissolved in 300-μl mixture of acetonitrile–water (50:50, v/v) and injected into the UHPLC/MS system. Samples of rat feces were diluted 1,000× before the analysis to provide an adequate signal within the linear range of calibration curve.
MALDI mass spectrometry
Mass spectra for profiling and imaging experiments were measured using ultrahigh-resolution MALDI mass spectrometer LTQ Orbitrap XL (Thermo Fisher Scientific, Waltham, MA, USA) equipped with nitrogen UV laser (337 nm, 60 Hz) with a beam diameter of about 80 μm × 100 μm. The LTQ Orbitrap instrument was operated in both positive-ion and negative-ion mode over a normal mass range (m/z 100–1,000). Tuning parameters were optimized individually for used matrices. The number of laser shots and power was determined based on tests performed with automatic gain control turned-on on a small area of testing tissue slide (target value was set to 5.105).
Selection of suitable matrices
The suitability of matrices was compared on standards and extract from tissue samples to achieve the strongest signal of AT in positive- and negative-ion mode. Matrices were dissolved in methanol or acetonitrile–water mixture (9:1, v/v) to provide the concentration of 20 mg/ml and mixed in various analyte/matrix molar ratios with the AT standard (1:10, 1:50, 1:100, 1:500, 1:1,000) or the tissue extract. Dried droplet crystallization was used for the sample deposition on the target plate. The deposited amount of samples was 0.7 μl. The survey crystal positioning system was set for the random choice of shot position by automatic crystal recognition. For one measurement, 32 laser shots (4 microscans/scan, 8 laser shots per microscan) at 16 different positions were accumulated to achieve a reproducible signal. Each sample (spotted matrix and AT molar ratio mixture) was prepared three times and measured in both MALDI-MS polarity modes.
UHRMSI experiments
All experiments were performed with the automatic gain control turned off to achieve the identical number of laser shots for each pixel. The step size of the sample was set to 150–200 μm for the full scan mass spectra measurement and 300 μm for MS/MS. Four microscans were summed for each mass spectrum (each pixel). The mass resolution was set to R = 100,000 (full width at half maximum definition, at m/z 400) for full scan mass spectra and 30,000 for MS/MS experiments. Mass spectra were internally calibrated using the lock mass feature of the instrument using known matrix ions (m/z 273.0394 [2 × MDHB − 2 × H2O+H]+ for DHB and m/z 315.1615 [2 × MDAN−H]− for DAN). The isolation width Δm/z 1, normalized collision energy 25 %, activation Q value 0.250, activation time 30 ms, and helium as the collision gas were used for collision-induced dissociation (CID) experiments in LTQ linear ion trap. For higher energy collisional induced dissociation (HCD), 35 % normalized collision energy was applied. Mass spectrometric images were generated using a tissue imaging visualization software ImageQuest 1.0.1 (Thermo Fisher Scientific).
UHPLC/MS/MS conditions
Ultrahigh-performance liquid chromatography-tandem mass spectrometry (UHPLC/MS/MS) chromatograms of samples were measured using ESI on a hybrid quadrupole-time-of-flight mass analyzer (micrOTOF-Q, Bruker Daltonics, Germany). UHPLC was performed on an Agilent 1290 Infinity liquid chromatograph (Agilent Technologies, Santa Clara, CA, USA) using Zorbax Eclipse C18 column 150 × 2.1 mm, 1.8 μm (Agilent Technologies), temperature 25 °C, flow rate 0.4 mL/min, and injection volume 1 μL. The mobile phase consisted of 0.5 mmol/L ammonium acetate adjusted to pH 4.0 (A) and acetonitrile (B). The linear gradient was as follows: 0 min–50 % B, 6 min–56 % B, 9 min–95 % B, and finally washing and reconditioning of the column. The quadrupole-time-of-flight (QTOF) mass spectrometer was used with the following setting of tuning parameters: capillary voltage 4.5 kV, drying temperature 220 °C, and flow rate and pressure of nitrogen were 8 L/min and 1.3 bar, respectively. The external calibration was performed with sodium formate clusters before individual measurements. ESI mass spectra were recorded in the range of m/z 50–1,000 in positive-ion and negative-ion modes. The isolation width Δm/z 4 and the collision energy 20–30 eV using argon as the collision gas were used for MS/MS experiments.
UHPLC/MS quantitation experiments
The stock solution of atorvastatin 0.27 mmol/L was prepared by dissolving the accurately weighed reference in methanol. The stock solution was further diluted 1:100, 1:300, 1:500, 1:700, 1:1,000, 1:2,000, 1:3,000, and 1:5,000 by mobile phase used at the initial step of gradient elution—acetonitrile, ammonium acetate buffer 0.5 mM, pH 4.0 (50:50)—to get individual points of calibration curve in the range 54 nmol/L–2.7 μmol/L. The internal standard AT-d5 was further added to all standard solutions (0.3 μmol/L). The calibration curve for AT (Fig. S2, Electronic Supplementary Material) was conducted in triplicate at eight concentration levels (Table S1, Electronic Supplementary Material) and constructed by plotting the ratio between the integrated areas of analyte peaks vs. the IS peak.
Results and discussion
The scheme of our drug metabolism experiment is shown in Fig. 1. Selected tissues, serum, urine, and feces of rats euthanized 3.5 h after the oral administration of AT suspension (100 mg/kg of body weight) together with appropriate blank samples were obtained for this study. The sample preparation included the tissue cryogenic sectioning, extraction, and filtration. MALDI-MS measurement with the aim to find the best matrix for the subsequent MALDI-MSI in both polarity modes was performed. UHPLC/ESI-MS was applied for the analysis of extract to reveal present AT metabolites in rat biotransformation samples. Adjacent tissue slides were analyzed for each rat organ to obtain accurate quantitative information for the correlation with the spatial distribution obtained by MSI. The optimization of UHPLC/ESI-MS separation conditions and the matrix selection for MALDI was done with AT standard and the extract of rat liver tissue, where the most of AT metabolites were expected. The mass resolution higher than 100,000 as well as sub-parts per million mass accuracy of Orbitrap mass analyzer enabled to distinguish AT-related ions from the matrix and endogenous compound ions with similar m/z values, e.g., lipids. In addition, the fragmentation using two different approaches, CID and HCD, was performed to identify metabolites.
UHPLC/MS of AT metabolites in selected rat tissues, body fluids, and feces
Separation conditions optimized in our previous article [5] were applied for qualitative and quantitative UHPLC/MS analyses of all studied rat biotransformation samples. The combination of optimized reversed-phase UHPLC separation with subsequent high mass accuracy measurements in full scan and tandem mass spectra using QTOF analyzer was sufficient for the identification of all compounds. The presence of AT metabolites identified in rat in vivo experiment (see Table 2) was in accordance with previously performed in vitro study, i.e., β-oxidation (M2, M3), aromatic hydroxylation on phenylaminocarbonyl part (M1 and M3), AT lactone (M4), and glycol formation (M5). β-Oxidation was the prevailing metabolic pathway. The quantitation of initial compound was performed using the addition of deuterated internal standard (AT-d5) into all calibration solutions and rat tissue samples before the extraction procedure. The calibration curve is shown in the Electronic Supplementary Material (Fig. S2). Concentrations of AT in rat liver, plasma, urine, and feces are listed in Table S2 (Electronic Supplementary Material). The limit of detection (LOD), 30 nmol/L, was determined based on S/N ratio by the injection of smallest concentration of standard AT to provide S/N = 5. The high concentration of AT in feces showed that the drug is eliminated mainly through the intestine in accordance with previously published data [4] and MALDI-ultrahigh-resolution mass spectrometry imaging (UHRMSI) experiments (see “MALDI-UHRMSI experiments” section).
Selection of suitable matrices
Thirteen matrices for the small molecule analysis, i.e., AA, AAP, AP, AQ, BuCHCA, CHCA, DAN, DHA, DHB, DMAN, MBT, NaDHB, and THA, were tested in both polarity modes with drug standards to select the best candidates for MALDI-MSI experiments. The optimal signal was obtained for atorvastatin/matrix molar ratio 1:100 for all matrices. The signal of all important ions related to AT, i.e., m/z 559 [M+H]+, m/z 581 [M+Na]+, m/z 597 [M+K]+, and their product ions at m/z 466 and m/z 440 for the positive-ion mode or m/z 557 [M−H]− together with product ions at m/z 453, m/z 397, and m/z 278 for the negative-ion mode were considered for the data evaluation. Structures of fragment ions or neutral losses associated with their formation were described in our previous work [5]. Table 3 shows results obtained for all tested matrices and includes average extracted ion currents of all above-mentioned AT-related ions with their M+1 and M+2 isotopes (extracted ion current (EIC)AT), their percentage ratio of the total ion current (EICAT × 100 / TIC), the fragmentation degree (FD, the ratio between EIC of AT product ions and EICAT multiplied by 100), and particular standard deviations from three different measurements (see experimental part). Finally, the score illustrating the matrix suitability for MALDI-MS measurements of AT was calculated according to the following equation: Score = (EICAT)2 × (100 − FD) / TIC. In general, the higher signal and percentage ratio of AT-related ions result in higher score and therefore better matrix suitability. On the contrary, the higher fragmentation yields the lower score. All score values were divided by the highest score value for each polarity mode, multiplied by 10,000, and the logarithm of all values was calculated to obtain four logarithmic sectors for better visualization of differences among tested matrices (see Table 3). Figure 2 created based on these modified score values shows two-dimensional map of matrix suitability for AT ionization and illustrates that matrices are suitable either for positive-ion or negative-ion MS analysis. The highest value, 4, of this modified score represents the best, 3 means good, 2 is weak, and 1 means bad matrix suitability for AT analysis. No tested matrix provided an excellent signal of AT ions in both polarity modes. When the higher laser energy is applied, AT also provides a low signal without the matrix application, but the excess of fragmentation including unusual AT product ions is observed. Most suitable matrices with regard to AT and its metabolite signal enhancement were DHB and CHCA, but relatively good score showed also MBT, BuCHCA, NaDHB, and THA for positive-ion MALDI-MS (Table 3). The higher fragmentation of protonated molecules yielding characteristic product ions at m/z 440 and 466 was observed in case of two hot matrices, CHCA or MBT. The extensive fragmentation can lead to significant decrease of intensity of protonated molecules, and for this reason, DHB was preferred for further MSI experiments. The application of DAN provided the best results in the negative-ion MALDI mode. The good signal of AT was also provided for AA, AQ, and DMAN. Particular matrices were also tested on rat liver extract. Results were in accordance with experiments with AT standard. Some matrices, such as DMAN, THA, DHA, and AQ, are not stable enough in vacuum for longer time, and their application can be considered only in short-time experiments. These matrices start to evaporate in the ion source during the time-consuming UHRMSI experiments with Orbitrap mass analyzer (the pressure in the ion source was 0.01 Torr). In general, AT and its metabolites can be measured using both polarity modes (see Table 3). The higher sensitivity was obtained for DHB as matrix in positive-ion mode, and this matrix is also safer for the manipulation compared to DAN as a potential carcinogen.
Two-dimensional map illustrating matrix suitability for MALDI-MS analysis of AT in both polarity modes. Horizontal lines show log (104 × score / scoremax) values (see Table 3) and the matrix suitability (best, good, weak, or bad) for the positive-ion MALDI, while vertical lines show the same for the negative-ion MALDI
MALDI-UHRMSI experiments
Several subsequent slices for each tissue section were measured to verify the repeatability. All MS images represented EICs of m/z values were reconstructed with mass tolerance Δm/z 0.005. The matrix coating was performed by the sublimation. Particular parameters of sublimation (temperature, pressure, and time) were optimized in order to achieve homogenous but not too thick layer of matrix. The sublimation of five different matrices that provided the best signal with AT standard and rat liver extract was performed on liver tissue slices to verify their suitability. Particular sublimation parameters for these tested matrices are shown in Table 1. Results were in agreement with matrix testing on the standard, and the application of DHB for MALDI positive-ion and DAN for MALDI negative-ion mode was evaluated as the best matrices for AT and its metabolite localization in rat tissues. Both matrices were also tested in opposite polarities, but markedly decreased sensitivity was observed in case of DAN (positive-ion mode) and no AT-related ions were found for DHB in negative-ion mode (Table 1). The benefit of used matrices is their suitability for MS imaging in lipidomics [29]. Lipids can be visualized simultaneously with studied hypolipidemic drug and its metabolites within one run. The application of identical matrices as for lipid analysis is also in agreement with the hydrophobic character of AT. The visualization of particular ions has been performed using the rainbow color scheme (Fig. 3), where the peak intensity is color coded: blue and green colors represent a lower intensity, while red and orange colors are used for the higher intensity. Intensity values at minimum and maximum are written above particular images. MALDI-UHRMSI showed a homogenous distribution of AT and its metabolites in rat liver. Examples of rat liver images involving EIC of protonated or deprotonated molecules of AT and β-oxidated AT [5] are shown in Fig. 3A. The concentration of AT in rat liver evaluated from parallel UHPLC/MS quantitation analysis of adjacent tissue slides was about 20 mg/kg (see Table S2, Electronic Supplementary Material). Among other metabolites, hydroxylated atorvastatin, β-oxidated AT, AT glycol, and lactone were detected. The homogenous distribution of AT and its metabolites was also found in rat heart. On the contrary, no AT and its metabolites were found in rat brain. Two different sections were measured for MSI of rat kidney. The excess of renal veins and arteries was present in the first measured section, while the second section included strictly differentiated kidney cortex and medulla parts (see Fig. 3B, C). The distribution study in rat kidney showed the absence of AT, but β-oxidated metabolites were found mainly in kidney cortex or in the area of veins and arteries. Negligible concentration of AT metabolites in kidney medulla and urine confirmed that the major route of AT extraction is through the intestine (high concentration of AT was found in feces, see Table S2 in the Electronic Supplementary Material). The drug is eliminated primarily in bile after hepatic and extrahepatic metabolism. In comparison to AT, drugs with better water solubility, e.g., imatinib studied by Spengler group [11], are concentrated mainly in kidney medulla. The presence of all identified metabolites in MALDI-MS images was in accordance with parallel UHPLC/MS measurement of adjacent tissue slices.
Optical and MALDI-UHRMS images. A EICs of protonated and deprotonated molecules of AT and β-oxidated metabolite M2 in rat liver tissue slice (positive-ion mode with DHB matrix, negative-ion mode with DAN matrix). B Optical and MS images (EICs of [M2+H]+ and [M3+H]+) of rat kidney tissue slice (kidney section 1). C Optical and MS images (EICs of [M2+H]+ and its product ion at m/z 380) of rat kidney tissue slice (kidney section 2)
Importance of ultrahigh-resolution and tandem mass spectra measurement in MSI
Ultrahigh-resolution and mass accuracy of Orbitrap analyzer enabled to reveal AT and its metabolites in the complex mixture of ions belonging to matrix and endogenic compounds. Figure 4A shows the average mass spectrum for rat liver images recorded in the positive-ion mode. The zoomed region around m/z 559 shows 12 peaks in the mass window of 0.27 m/z. Despite the high number of other ions, AT-protonated molecule at m/z 559.2609 is clearly distinguished from other ions, even in the case of the ion at m/z 559.2692 with very close m/z value (minimum mass resolution to distinguish these two peaks is 67,000). The similar situation is evident in case of protonated molecule of AT metabolite M3 (β-oxidation and hydroxylation of AT) illustrated in zoomed mass range around m/z 515. The power of ultrahigh-resolution Orbitrap analyzer is demonstrated on the extracted ion current of m/z 515.2344 in the rat kidney measurement. Two MS images of this ion were reconstructed with different mass tolerance of Δm/z 0.005 and Δm/z 0.5 (Fig. 4B). The comparison demonstrated that false-positive results would be obtained in case of the application of mass analyzer with insufficient mass resolution. In general, MS images in positive-ion mode (DHB matrix) contain more complex spectra compared to negative-ion mode (DAN), but the application of ultrahigh-resolution as well as high mass accuracy is necessary to avoid false-positive results in both polarity modes. All peaks corresponding to AT metabolites contained no interference ions and provided narrow peaks corresponding to applied resolution (R = 100,000). However, widened peaks, originated from two ion superpositions, were observed for some other ions as evident in Fig. 4A. The drawback of ultrahigh-resolution Orbitrap MS imaging is that the scan speed markedly decreases compared to low-resolution measurements. The scan rate is at maximum 1 Hz (R = 100,000) or even less in case of high averaging used to obtain reproducible spectra. Due to this fact, only 200-μm lateral resolution was used because it was sufficient to localize AT and its metabolites in studied tissues. Tandem mass spectra of protonated molecules of AT and all present metabolites were acquired to verify compounds’ origin. Product ions can be even present in full scan mass images with respect to used matrix and ionization parameters (laser energy and type of laser). The identical distribution of precursor and product ions confirmed the metabolite origin, which is shown on the example of β-oxidated AT metabolite at m/z 499 and its product ion at m/z 380 in rat kidney section (Fig. 3C).
Conclusions
In this work, we have performed the parallel application of UHPLC/MS and MALDI-UHRMSI to identify, quantify, and localize AT and its metabolites in various rat organs. UHRMSI experiments showed an excellent correlation with parallel UHPLC/MS measurements of adjacent tissue slices. Among 13 tested matrices, the sublimation deposition of DHB matrix for positive-ion MS imaging and DAN matrix for negative-ion MS imaging provided the best results with respect to score values calculated based on AT-extracted ion currents, the percentage ratio of AT-related ions, and their fragmentation degree. In general, the application of these matrices is useful in lipid distribution studies and lipid-related drugs, such as AT. The homogenous distribution of AT and its metabolite, i.e., β-oxidation, aromatic hydroxylation on phenylaminocarbonyl part, AT lactone, and glycol formation, was found in rat liver. AT concentration was determined 20 mg/kg in rat liver using UHPLC/MS measurement of adjacent tissue slices. MALDI-UHRMSI of rat kidney showed that AT metabolites are concentrated mainly in kidney cortex and around renal veins and arteries. Negligible amount of AT metabolites was found in kidney medulla. The high concentration of AT and the number of AT metabolites were found in rat feces, which confirmed that AT metabolites are transported in blood, and the major elimination route is through the bile after the hepatic metabolism. The experiment also showed the importance of ultrahigh-resolution mass analyzer for MSI of small molecules. Interferences in close m/z region originated from matrix and endogenic compounds and the visualization of these interferences instead of or together with desired ions can provide false-positive results. The minimum mass resolution required to distinguish AT-related compounds from interferences was calculated 67,000 for positive-ion mode and 12,000 in case of negative-ion mode based on detailed spectra interpretation in our experiment, which shows that the negative-ion mode is more suitable in case of lower resolution analyzer application. High mass accuracy, resolution, tandem mass spectra measurement, or possible use of non-conductive MALDI plate can be mentioned among benefits of MALDI-Orbitrap-MS configuration. On the other hand, the data acquisition time is the most significant limitation in case of Orbitrap application. UHRMSI is recommended for applications, where the benefit of ultrahigh-resolution mass spectra outperforms the limitation of longer acquisition time, i.e., drug metabolite studies.
Abbreviations
- AA:
-
9-Aminoacridine
- AAP:
-
3-Aminoacetophenone
- AP:
-
2-Aminopyrazine
- AQ:
-
3-Aminoquinoline
- AT:
-
Atorvastatin
- BuCHCA:
-
Butylamine salt of α-cyano-4-hydroxycinnamic acid
- CHCA:
-
α-Cyano-4-hydroxycinnamic acid
- CID:
-
Collision-induced dissociation
- DAN:
-
1,5-Diaminonaphthalene
- DHA:
-
2,6-Dihydroxyacetophenone
- DHB:
-
2,5-Dihydroxybenzoic acid
- DMAN:
-
N,N,N′,N′-tetramethyl-1,8-naphthalenediamine
- EIC:
-
Extracted ion current
- ESI:
-
Electrospray ionization
- FD:
-
Fragmentation degree
- HCD:
-
Higher energy collisional induced dissociation
- HPLC:
-
High-performance liquid chromatography
- LOD:
-
Limit of detection
- MALDI:
-
Matrix-assisted laser desorption/ionization
- MBT:
-
2-Mercaptobenzothiazole
- MS:
-
Mass spectrometry
- MSI:
-
Mass spectrometry imaging
- MS/MS:
-
Tandem mass spectrometry
- NaDHB:
-
Sodium 2,5-dihydroxybenzoate
- QTOF:
-
Quadrupole-time-of-flight
- THA:
-
2,4,6-Trihydroxyacetophenone
- UHPLC:
-
Ultrahigh-performance liquid chromatography
- UHPLC/MS/MS:
-
Ultrahigh-performance liquid chromatography-tandem mass spectrometry
- UHRMS:
-
Ultrahigh-resolution mass spectrometry
- UHRMSI:
-
Ultrahigh-resolution mass spectrometry imaging
References
Curran MP (2010) Amlodipine/atorvastatin: a review of its use in the treatment of hypertension and dyslipidaemia and the prevention of cardiovascular disease. Drugs 70:191–213
Nováková L, Satinský D, Solich P (2008) HPLC methods for the determination of simvastatin and atorvastatin. Trac Trends Anal Chem 27:352–367
Hermann M, Christensen H, Reubsaet JLE (2005) Determination of atorvastatin and metabolites in human plasma with solid-phase extraction followed by LC-tandem MS. Anal Bioanal Chem 382:1242–1249
Black AE, Sinz MW, Hayes RN, Woolf T (1998) Metabolism and excretion studies in mouse after single and multiple oral doses of the 3-hydroxy-3-methylglutaryl-CoA reductase inhibitor atorvastatin. Drug Metab Dispos 26:755–763
Jirásko R, Mikysek T, Chagovets V, Vokřál I, Holčapek M (2013) Structural characterization of electrochemically and in vitro biologically generated oxidation products of atorvastatin using UHPLC/MS/MS. Anal Bioanal Chem 405:7181–7193
Kracun M, Kocijan A, Bastarda A, Grahek R, Plavec J, Kocjan D (2009) Isolation and structure determination of oxidative degradation products of atorvastatin. J Pharm Biomed 50:729–736
Shah RP, Kumar V, Singh S (2008) Liquid chromatography/mass spectrometric studies on atorvastatin and its stress degradation products. Rapid Commun Mass Spectrom 22:613–622
Holčapek M, Kolářová L, Nobilis M (2008) High-performance liquid chromatography-tandem mass spectrometry in the identification and determination of phase I and phase II drug metabolites. Anal Bioanal Chem 391:59–78
Norris JL, Caprioli RM (2013) Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem Rev 113:2309–2342
Prideaux B, Stoeckli M (2012) Mass spectrometry imaging for drug distribution studies. J Proteome 75:4999–5013
Römpp A, Spengler B (2013) Mass spectrometry imaging with high resolution in mass and space. Histochem Cell Biol 139:759–783
Wu CP, Dill AL, Eberlin LS, Cooks RG, Ifa DR (2013) Mass spectrometry imaging under ambient conditions. Mass Spectrom Rev 32:218–243
Fletcher JS, Lockyer NP, Vickerman JC (2011) Developments in molecular SIMS depth profiling and 3D imaging of biological systems using polyatomic primary ions. Mass Spectrom Rev 30:142–174
Passarelli MK, Winograd N (2011) Lipid imaging with time-of-flight secondary ion mass spectrometry (ToF-SIMS). Biochim Biophys Acta Mol Cell Biol Lipids 1811:976–990
Weidner SM, Falkenhagen J (2009) Imaging mass spectrometry for examining localization of polymeric composition in matrix-assisted laser desorption/ionization samples. Rapid Commun Mass Spectrom 23:653–660
Seeley EH, Caprioli RM (2008) Molecular imaging of proteins in tissues by mass spectrometry. Proc Natl Acad Sci U S A 105:18126–18131
Karas M, Hillenkamp F (1988) Laser desorption ionization of proteins with molecular masses exceeding 10000 daltons. Anal Chem 60:2299–2301
Svatoš A (2010) Mass spectrometric imaging of small molecules. Trends Biotechnol 28:425–434
van Kampen JJA, Burgers PC, de Groot R, Gruters RA, Luider TM (2011) Biomedical application of maldi mass spectrometry for small-molecule analysis. Mass Spectrom Rev 30:101–120
Law KP, Larkin JR (2011) Recent advances in SALDI-MS techniques and their chemical and bioanalytical applications. Anal Bioanal Chem 399:2597–2622
Kawasaki H, Ozawa T, Hisatomi H, Arakawa R (2012) Platinum vapor deposition surface-assisted laser desorption/ionization for imaging mass spectrometry of small molecules. Rapid Commun Mass Spectrom 26:1849–1858
Shroff R, Rulíšek L, Doubský J, Svatoš A (2009) Acid–base-driven matrix-assisted mass spectrometry for targeted metabolomics. Proc Natl Acad Sci U S A 106:10092–10096
Woo HK, Northen TR, Yanes O, Siuzdak G (2008) Nanostructure-initiator mass spectrometry: a protocol for preparing and applying NIMS surfaces for high-sensitivity mass analysis. Nat Protoc 3:1341–1349
Pirman DA, Reich RF, Kiss A, Heeren RMA, Yost RA (2013) Quantitative MALDI tandem mass spectrometric imaging of cocaine from brain tissue with a deuterated internal standard. Anal Chem 85:1081–1089
Cornett DS, Frappier SL, Caprioli RM (2008) MALDI-FTICR imaging mass spectrometry of drugs and metabolites in tissue. Anal Chem 80:5648–5653
Jun JH, Song ZH, Liu ZJ, Nikolau BJ, Yeung ES, Lee YJ (2010) High-spatial and high-mass resolution imaging of surface metabolites of Arabidopsis thaliana by laser desorption-ionization mass spectrometry using colloidal silver. Anal Chem 82:3255–3265
Jungmann JH, Heeren RMA (2012) Emerging technologies in mass spectrometry imaging. J Proteome 75:5077–5092
Hankin JA, Barkley RM, Murphy RC (2007) Sublimation as a method of matrix application for mass spectrometric imaging. J Am Soc Mass Spectrom 18:1646–1652
Thomas A, Charbonneau JL, Fournaise E, Chaurand P (2012) Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1,5-diaminonaphthalene deposition. Anal Chem 84:2048–2054
Acknowledgments
This project was supported by the Czech Science Foundation (Grant No. P206/12/P065). M.H. acknowledges the support of the ERC CZ Project No. LL1302 sponsored by the Ministry of Education, Youth and Sports of the Czech Republic. M.K. acknowledges the support of research project MH CZ-DRO (UHHK, 00179906).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 758 kb)
Rights and permissions
About this article
Cite this article
Jirásko, R., Holčapek, M., Kuneš, M. et al. Distribution study of atorvastatin and its metabolites in rat tissues using combined information from UHPLC/MS and MALDI-Orbitrap-MS imaging. Anal Bioanal Chem 406, 4601–4610 (2014). https://doi.org/10.1007/s00216-014-7880-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-014-7880-y