Abstract
Most oral cancers are oral squamous cell carcinoma (OSCC). The anatomical features of OSCC have been histochemically evaluated with hematoxylin and eosin. However, the border between the cancer and stromal regions is unclear and large portions of the cancer and stromal regions are resected in surgery. To reduce the resected area and maintain oral function, a new method of diagnosis is needed. In this study, we tried to clearly distinguish the border on the basis of biomolecule distributions visualized by imaging mass spectrometry (IMS). In the IMS dataset, eleven signals were significantly different in intensity (p < 0.01) between the cancer and stromal regions. Two signals at m/z 770.5 and m/z 846.6 were distributed in each region, and a clear border was revealed. Tandem mass spectrometric (MS/MS) analysis identified these signals as phosphatidylcholine (PC) (16:0/16:1) at m/z 770.5 in the cancer region and PC (18:1/20:4) at m/z 846.6 in the stromal region. Moreover, the distribution of PC species containing arachidonic acid in the stromal region suggests that lymphocytes accumulated in response to the inflammation caused by cancer invasion. In conclusion, the cancer and stromal regions of OSCCs were clearly distinguished by use of these PC species and IMS analysis, and this molecular identification can provide important information to elucidate the mechanism of cancer invasion.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most cases of head and neck cancer are oral cancers of the tongue, floor of the mouth, gingiva, or salivary glands. Oral cancers are histologically classified as squamous cell carcinomas (SCCs), adenoid cystic carcinoma, or mucoepidermoid. Oral SCCs (OSCCs) account for more than 80 % of all head and neck cancers and are fatal without surgical resection. The extent of cancer is evaluated by a combination invasive biopsies and differentiation on the basis of several criteria, for example the Jakobsson classification [1], the Anneroth classification [2], and the Yamamoto–Kohama classification [3]. Moreover, the surgeon’s experience determines the margin area resected in the stromal region. The stromal region includes the connective tissue area around the cancer lesion, unlike normal tissue. It is known that lymphocyte immune cells are found in the stromal region. The resected tissue is histochemically analyzed and the completeness of resection is judged. However, there is a dangerous possibility of cancer cells remaining because it is very difficult to resect all the cancer in the narrow oral space. If cancer cells remain in the oral region after surgery, infiltration and metastasis will again result in a serious problem. In general, in cancer tissue intercellular adhesion is weakened, and the motor ability of the cancer cells is enhanced. The cancer cells invade the basal membrane and the membrane disintegrates. The cancer cells are carried in the interstitial connective tissue and metastasize through blood vessels toward other organs. Indeed, the remaining cancer cells induce recrudescence [4]. Finally, OSCCs result in death as a result of infiltration and metastasis. Thus, all cancer cells should be completely resected with a sufficient margin. On the other hand, the margin often prevents the maintenance of oral function. Therefore, criteria for precise surgical resection are needed.
In previous studies, immunological analysis has been applied to OSCC. Cancer growth factors, for example PCNA [5] and Ki-67 [6], have been immunohistologically evaluated to elucidate the extent of tumor invasion in comparison with conventional classification of OSCCs. These proteins have been determined to be biomarkers because of their correlation with the classification. However, these proteins are distributed in normal cells, and it is difficult to determine the minimum margin to be resected in surgery by using the distribution of PCNA and Ki-67. Lipids are also important biological materials, with proteins, nucleic acids, and sugars. Lipids are involved in cell proliferation [7] and differentiation, metabolism control, immunity, and inflammation [8]. The location and metabolism of lipids are believed to be related to the growth of cancer, the invasion of cancer, and the mechanism of metastasis [9, 10]. Brasitus et al. analyzed lipid extract from cells and tissues by thin-layer chromatography and gas chromatography and revealed changes of lipid composition which depended on the development of colon cancer [11]. However, the lipid extract is not suitable for determining the border between the cancer and stromal regions, and thus an imaging technique is needed.
Imaging mass spectrometry (IMS) enables us to visualize the distribution of many biomolecules, without any labeling, in a single analysis, and to identify the biomolecules on a tissue section [12]. The technique is usually applied in the form of secondary mass spectrometry (SIMS) and matrix-assisted laser-desorption ionization (MALDI). SIMS has good resolution for imaging and the resolution is <1 μm. However, the analytes are limited to biomolecules with low molecular weight because those of high molecular weight are fragmented. In contrast, MALDI [13] can ionize biomolecules of high molecular weight, for example proteins [14], lipids [15, 16], nucleic acids [17], and sugars [18], on a tissue section. IMS using MALDI-time-of-flight (TOF) is especially useful for lipid analysis [19, 20] because the technique can separately visualize even the differences between the fatty acid components of lipids [16, 20].
The technique has been applied to several pathology samples, including cancerous tissue, and has revealed the altered phospholipid composition in the diseased region compared with the normal region [21]. Shimma et al. used IMS to examine tissue sections from colon cancer liver metastasis and found a cancer-specific increase of sphingomyelin (SM) [22]. Ishikawa et al. found that phosphatidylcholine (PC), oleic acid, and SM were increased in the region of thyroid cancer, and proposed that these distributional differences were related to the biological behavior of cancer, for example invasion and metastasis [23]. Thus, IMS is a useful technique for finding and identifying specific molecules (especially lipids) in such pathology samples as cancer tissues. However, there has been no report of comparison of the signals in the cancer and stromal regions by use of IMS.
In this study, we attempted to find phospholipids specifically distributed in the cancer or stroma of OSCC tissue. Two consecutive tissue sections including cancer and stroma were prepared from OSCC tissue; one section was then analyzed by IMS and the other was stained with hematoxylin and eosin (HE). The mass range in IMS analysis was focused on phospholipids, and specific peaks in each cancer region and stromal region were analyzed by statistical analysis, by use of computer software. Molecules distributed differently between the cancer and stromal regions were visualized, and we determined whether each region was clearly distinguished. To identify these molecules, we conducted tandem mass spectrometric (MS/MS) analysis.
Materials and methods
Clinical samples
Gingival OSCC biopsy tissue blocks were obtained from five patients before surgery, in accordance with protocols approved by the Hamamatsu University School of Medicine. None of the patients had received radiation, chemotherapy, or immunotherapy treatment. Pathological examination was used to confirm these portions of tumor and the histology of invasive SCC. After they were obtained, the tissues blocks were immediately frozen in liquid nitrogen and stored at −80 °C to maintain tissue morphology and minimize molecular degradation until MALDI-IMS analysis.
Chemicals
Methanol, potassium acetate, and ultra-pure water were purchased from Wako Chemicals (Osaka, Japan). 2,5-Dihydroxybenzoic acid (DHB) was purchased from Bruker Daltonics (Bremen, Germany). Standard peptides for calibration of the TOF analyzer were purchased from Sigma–Aldrich (St Louis, MO, USA). All of the chemicals used in this study were of the highest purity available.
Imaging mass spectrometry (IMS)
Tissue section preparation
Tissues blocks were sectioned at −20 °C, by use of a cryostat (CM 1950; Leica, Wetzler, Germany), to a thickness of 8 μm in the direction of the long axis, as described elsewhere [24, 25]. To prevent degradation of mass spectra in IMS analysis, OSCC blocks were not embedded in OCT polymer during this procedure [25]. The frozen sections were thaw-mounted on indium–tin oxide (ITO)-coated glass slides (Bruker Daltonics) and stored at −20 °C until used for IMS analysis.
Spray coating of the matrix solution
A DHB solution (50 mg mL−1 DHB and 10 mmol L−1 potassium acetate in 70 % methanol, 1 mL) was used as the matrix [26]. The potassium salt can help to ionize molecules in the form [M + K]+, without [M + H]+ and [M + Na]+, in positive-ion mode, making analysis of the mass spectrum fairly easy. The matrix solution was sprayed on to the sample sections by use of a 0.2-mm nozzle caliber airbrush (Procon Boy FWA Platinum; Mr Hobby, Tokyo, Japan).
IMS conditions
IMS was performed by use of a MALDI–TOF–TOF-type instrument (Ultraflex II TOF/TOF; Bruker Daltonics). This instrument was equipped with a 355-nm Nd:YAG laser. The ionized molecules were detected in the positive reflectron mode by using an external calibration method. Mono DHB ([M + H]+, m/z 155.03) and human angiotensin II ([M + H]+, m/z 1,046.54) were used for calibration. Mass spectra were acquired in the mass range m/z 400–1,200. The laser energy and detector gain were optimized to maximize the sensitivity of signal detection in the IMS analysis. Raster scans on tissue surfaces were performed automatically by use of flexControl and flexImaging 2.1 software (Bruker Daltonics). The number of laser irradiations was 200 shots in each spot. The distance between data points was 100 μm. Image reconstruction was performed by use of flexImaging 2.1 software.
IMS data analysis
The regions of interest (ROIs) in the cancer and stromal regions were defined by reference to the HE staining in adjacent sections. The IMS data format of the flexImaging 2.1 software was converted into Analyze 7.5 file format to statistically analyze the IMS data by use of SIMtool software (in-house software; Shimadzu, Kyoto, Japan). With the exception of isotope peaks, all peaks were picked up from ROIs that were defined as “cancer region” or “stromal region”, and these distributions were visualized on a tissue section. These signal intensities between the cancer and stromal regions were statistically assessed by use of Welch’s t-test, and p-values <0.01 were regarded as significant. Finally, the peaks with significant differences were applied to four other datasets and visualized as ion images.
MS/MS analysis
Because multiple fatty acid isobars are possible, the MS/MS analysis was performed on tissue sections in the positive-ion mode by use of the QSTAR Elite System (Applied Biosystems/MDS Sciex, Foster City, CA, USA), a hybrid quadrupole–TOF mass spectrometer equipped with an orthogonal MALDI source and a pulsed Nd:YAG laser. The laser energy and collision energy were optimized to maximize the sensitivity of signal detection in the MS/MS analysis. Biomolecules were identified by referring to the results of a Metabolite MS Search (http://www.hmdb.ca/spectra/ms/search)
Results
HE staining of OSCC specimens from case 1
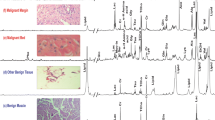
Frozen specimens of OSCC were obtained from gingiva on which no preoperative therapy had been performed. To analyze the morphological features in detail, we used HE staining. The HE-stained section from case 1 is shown in Fig. 1. In Fig. 1a, the epidermal layer is on the upper side and the lamina propria is on the bottom. The cancer was found on the left side, and the histopathological diagnosis of this OSCC was well-differentiated. An enlarged view of the typical cancer region is shown in Fig. 1b. Here, the cancer nests (i.e., clusters of carcinoma cells) are remarkable. Invasion of the cancer from an epidermal layer to the lamina propria can be observed. Cancer cells were observed in connective tissue areas around the cancer nests and were referred to as the “cancer stroma”. An enlarged view of a cancer-free stromal region is shown in Fig. 1c (the right side in Fig. 1a). The stromal region includes the connective tissue area around the cancer lesion, unlike normal tissue. Lymphocyte immune cells accumulated in the stromal regions of OSCC tissue from case 1. By use of HE staining the cancer and stromal regions were almost distinguished. However, a more precise border was needed to reduce the resection area. Therefore, we next applied the IMS technique to an adjacent tissue section.
HE-stained tissue of oral squamous cell carcinoma (OSCC) of the maxillary gingiva from case 1. a An overall view from a continuous frozen section. The cancer region is located on the left and the stromal region is located on the right. Invasion from an epidermal layer to the lamina propria is observed in the cancer region. The border between the cancer region and the stromal region is indistinct, and cancer cells and connective tissue are mixed. b Cancer nests (arrow) and cancer stroma are observed in the cancer region. Cancer nests were clusters of carcinoma cells, and the cancer stroma consisted of connective tissue areas around the cancer nests. c The stromal region shows the neoplasm, indicating connective tissue areas around the cancer region
Comparison of the mass spectra in each region and ion images from case 1
After acquiring the IMS dataset, the average mass spectra from each region were created in the mass range m/z 400–1,200 (inset in Fig. 2a, b). First, with the exception of isotope peaks, the top 80 peaks were acquired in the mass range m/z 400–1,200 by use of SIMtool software. In the m/z ranges 400–700 and 850–1,200, small differences in signal intensity were observed for 65 peaks in each region. In contrast, differences in signal intensity were observed for 15 peaks in the m/z 700–850 range. It is known that this mass range corresponds to phospholipid species in positive-ion mode. The 15 signals, which were m/z 720.1, 741.5, 748.1, 756.5, 770.5, 772.5, 782.6, 794.5, 796.5, 798.5, 800.5, 804.5, 820.5, 824.6, and 846.6, are visualized as ion images in Fig. 3. Different types of distribution were visualized by IMS. The ion images at m/z 720.1, 770.5, and 794.5 were clearly distributed in the cancer region, including cancer cells, whereas those at m/z 804.5, 820.5, and 846.6 were distributed in the stromal region including lymphocytes and plasma cells. Other signals at m/z 741.5, 748.1, 756.5, 772.5, 782.6, 796.5, 798.5, 800.5, and 824.6 were distributed throughout the whole area without significant differences in intensity among regions.
Comparison of mass spectra from the cancer and stromal regions from case 1. a The average mass spectrum from the cancer region was obtained in the range m/z 400–1,200 (inset). The mass spectrum was extracted in the range m/z 700–850. b In the same way, the average mass spectrum from the stromal region was obtained in the ranges m/z 400–1,200 (inset) and m/z 700–850. The characteristic peaks were compared between the cancer and stromal regions. Each labeled number shows a peak of characteristic signal intensity in a cancer region. The relative intensity was based on a peak with maximum intensity in each region. All peaks were observed in common in each mass spectrum, but some of the peaks had different intensities
Ion images of the top 15 signals from case 1. The threshold of the color scale was adjusted in each ion image to show the clear distribution. The ions m/z 720.1, 770.5, and 794.5 were clearly distributed in the cancer region. In contrast, the ions m/z 804.5, 820.5, and 846.6 were distributed in the stromal region. Other signals were distributed over the whole region
Statistical analysis between cancer and stromal regions from case 1
To clarify significant differences between the cancer and stromal regions from case 1, we used the Welch’s t-test for the 15 peaks in the mass range m/z 700–850. As shown in Table 1, 11 peaks were significantly different (p < 0.01) between the regions; average signal intensity, standard error, and p-value for each m/z value are shown. Six molecules in particular, i.e., those associated with the signals m/z 720.1, 770.5, 794.5, 804.5, 820.5, and 846.6, were of significantly different intensity in the cancer and stromal regions; these are shown by using averaged signal intensities as a bar graph in Fig. 4. The signals at m/z 720.1, 770.5, and 794.5 increased in the cancer region. The signals at m/z 804.5, 820.5, and 846.6 increased in stromal region.
Statistical analysis between cancer and stromal regions from case 1. Significances of differences in signal intensities in the cancer and stromal regions were determined by use of Welch’s t-test. The error bar represents the standard error. Each of the signals at m/z 720.1, 770.5 and 794.5 were increased in cancer regions. Each of the signals at m/z 804.5, 820.5, and 846.6 were increased in stromal regions. For all of the m/z signals compared here, p-values less than 0.01 were obtained; statistical significance is indicated by an asterisk
Distributions of cancer region-positive and stromal region-positive signals from cases 1–5
We performed IMS analysis on more four cases, cases 2–5, which were well-differentiated and for which accumulation of lymphocytes was observed, much as in case 1, and used the six m/z values that were significantly different between the cancer and stromal regions in case 1. These signals are visualized as ion images in Fig. 5. The ion images reconstructed by use of signals at m/z 720.1, 770.5, and 794.5 were cancer region-positive. In contrast, the ion images reconstructed by use of the signals at m/z 804.5, 820.5, and 846.6 were stromal region-positive. The merged images of m/z 770.5 and 846.6 revealed the distribution of the cancer and stromal regions with no overlap and the borders are more clearly visualized than those of the HE-stained section. The p-value for m/z 820.5 was better than that for m/z 846.6, as shown in Table 1. However, the ion distribution was limited to a part of the stromal region. Therefore, we selected the signal at m/z 846.6 as a representative stroma-positive signal.
Ion images with statistically significant differences between the cancer and stromal regions from cases 1–5. We applied six peaks with significantly different intensity to the IMS datasets from cases 2–5 and reconstructed the ion images. The histopathological diagnosis of all cases was well differentiated. Ion images captured cancer-specific distributions at m/z 720.1, 770.5, and 794.5. Ion images captured stroma-specific distributions at m/z 804.5, 820.5, and 846.6. The merged image from the signal intensities at m/z 770.5 and 846.6 revealed the complementary distributions. Scale bar = 1 mm
Molecule identification
MS/MS analysis of tissue sections was performed to identify the structures of the biomolecules evaluated by the statistical analysis. A molecule corresponding to m/z 770.5 was assigned to [PC + K]+ because neutral losses of 59 Da (m/z 711.4) and 183 Da (m/z 587.4) from a precursor ion in the MS/MS spectrum, indicative of a trimethylamine and a choline head group, respectively, were observed (Fig. 6a) [27, 28]. Moreover, the signal at m/z 455.1 indicated a neutral loss of 256 Da corresponding to palmitic acid (16:0) from the signal at m/z 711.4. The Metabolite MS Search identified a candidate, [PC (diacyl-32:1) + K]+. Therefore, this molecule was identified as [PC (diacyl-16:0/16:1) + K]+. In the same way, a molecule corresponding to m/z 846.6 was assigned as [M + K]+ ion of PC (diacyl-18:1/20:4), because neutral losses of 59 Da and 183 Da in the MS/MS spectrum and a neutral loss of 282 Da corresponding to oleic acid (18:1) were also observed (Fig. 6b) [27, 28].
Molecular identification by MS/MS analysis of tissue sections. Precursor ions were: m/z 770.5 (a) and m/z 846.6 (b). A molecule corresponding to m/z 770.5 was assigned by neutral loss as [PC (16:0/16:1) + K]+. In the same manner, a molecule corresponding to m/z 846.6 was assigned as [PC (18:1/20:4) + K]+
Discussion
This study is the first IMS analysis performed on OSCC tissue, and the first study to compare signals in the cancer and stromal regions. IMS has also been applied to other cancer samples. Morita et al. reported that histone H4 (m/z 1,325.6) was found specifically in undifferentiated gastric cancer tissue, after they compared normal tissue and gastric cancer tissues with different degrees of differentiation by IMS [29]. Shimma et al. also reported that SM (d18:1/16:0) at m/z 725.4 was specifically distributed in a cancer region by comparing the mass spectrum obtained from colon cancer liver metastasis tissue [22]. Thus, IMS has been proved useful for identification of specific molecules, for example peptides and phospholipids, in cancer regions. In the classification of OSCCs, the criteria for cancer resection in surgery must be modified and reduction of the resection area is needed because resection of a large area sometimes results in a loss of oral function. However, a diagnosis made using HE staining of biopsy tissue does not clearly distinguish between the cancer and stromal region (Fig. 1). Therefore, there has been a need to identify the biomolecules to determine the border between the cancer and stromal regions. Phospholipids can be analyzed by IMS without complex treatment. This technique could simplify diagnosis and distinguish the regions by use of two phospholipids. All IMS and MS/MS experiments were performed using optimized settings, and the reproducibility of the data was evaluated by use of mass spectra acquired from adjacent sections.
The IMS analyses revealed statistically significant differences in intensity (p < 0.01) for 11 peaks (Table 1) and the ion images characterized the stromal region or cancer region. Two signals at m/z 741 and m/z 798 were detected in cancer regions in other research by Shimma [22] and Ishikawa [23]. According to the ion image at m/z 741.5 in the study by Ishikawa et al., the distribution was a little different from that of other cancer-positive signals, for example m/z 796.5 and m/z 798.5. They mentioned that the signal at m/z 741.5 was more intense in both the cancer region and the stromal region. The signal at m/z 725.4 was also identified as SM (d18:1/16:0) with a different ion-adduct form. The ion image in the study by Shimma et al. also revealed greater intensity in the stromal region than in the cancer region. These results are identical with ours (Fig. 3). Even if a signal is evaluated as significantly different, signal which are extremely different should be used to distinguish the border between the cancer and stromal regions. Therefore, the signals at m/z 720.1, m/z 770.5, and m/z 794.5 for the cancer region and m/z 804.5, m/z 820.5, and m/z 846.6 for the stromal region were applied to all IMS datasets in this study (Fig. 5). These signals had good p-values in the statistical analysis (Table 1 and Fig. 4). The merged ion images of m/z 770.5 and m/z 846.6 revealed a clear border between the cancer and stromal regions without any of the overlap observed in the HE staining. In the near future, there is a possibility that these signals could be used as a diagnostic method to determine the resection area before surgery.
In this study, we successfully identified the signals at m/z 770.5 and m/z 846.6 as two PC species, i.e., PC (diacyl-16:0/16:1) in the cancer region and PC (diacyl-18:1/20:4) in the stromal region, from OSCC tissue sections (Fig. 6). Ishikawa et al. identified phospholipid species, for example PC (diacyl-16:0/18:1), PC (diacyl-16:1/18:1), and SM (d18:1/16:0), that were increased in the thyroid cancer region compared with the normal region [23]. In general, malignant cellular proliferation is stimulated by cell-growth factors, for example PC species, that are increased in the components of the cell membrane, with the increase in cell density. However, PC (diacyl-16:0/16:1) and PC (diacyl-18:1/20:4) have not been identified in the cancer and stromal regions. This suggests that each individual cancer cell has a different profile.
Here we focused on the fatty acid bound to PC species in OSCC cancer regions. The fatty acid compositions are also very interesting from the perspective of cancer invasion mechanisms. Lv et al. reported that the saturated fatty acids palmitic acid (C16:0) and stearic acid (C18:0) and the unsaturated fatty acid linoleic acid (C18:2) were increased in the serum of patients with breast cancer, and they proposed these fatty acids as new biomarkers of breast cancer [30]. Such an increase of fatty acids might reflect the synthesis of PC species including 16:0, 18:0, and 18:2. In fact, our results demonstrated that PC (diacyl-16:0/16:1) was increased in the cancer region of OSCCs.
In the HE-stained OSCC tissue section, many of the lymphocyte cells in the stromal region clearly had nuclei with different shapes compared with other cell types (Fig. 1c). It is well-known that lymphocyte cells accumulate in regions of inflammation [31], and histochemical analysis has revealed accumulation of immune lymphocytes in the stromal region of cervical cancer [32]. Hanada et al. reported that arachidonic acid (AA, 20:4) bound to PC species (AA-PCs) was detected in the inflammation region in a rat model of spinal cord injury. These results suggest that the PC (18:1/20:4) distributed in the OSCC stromal regions could have reflected an increase in cell membrane components induced by an accumulation of lymphocyte cells. Moreover, our results revealed an increase of signals at m/z 804.5 and m/z 820.5 in the stromal region. Previous reports showed that the signals at m/z 804.5 and m/z 820.5 were identified as PC (diacyl-16:0/20:4) [33] and PC (diacyl-16:1/20:4) [27, 34], respectively. Thus, three kinds of AA-PCs were increased in the stromal region, and there is a possibility that AA is important in tumor invasion in the stromal region.
AA is a precursor of prostaglandin (PG) and is converted by cyclooxygenase (COX) in the AA cascade. COX-1 is involved in the maintenance of homeostasis and in the constant supply of prostaglandin E2 (PGE2) [35]. COX-2 and the de-novo synthesis of the enzyme microsomal prostaglandin E synthase-1 (mPGES-1) are induced against the growth of tumor tissue, and this process also causes an increase of PGE2. PGE2 produced by COX-2 activation in the AA cascade in a stromal region is important in oncogenesis, initial tumorigenesis, and cancer cell differentiation in various histologic types, including OSCC [35–37]. Therefore, AAPCs identified as PC (diacyl-18:1/20:4) in a stromal region could be a source of lipid mediators of inflammation against cancer invasion.
In this study, we showed that PC (diacyl-16:0/16:1) was distributed in the cancer region. Such distributions in cancer regions have been reported by other researchers using IMS. Morita et al. reported the distribution of PC (diacyl-16:0/16:1) in hepatocellular carcinoma (HCC) [38]. Ide et al. revealed the distribution of PC (diacyl-32:1) in breast cancers but did not identify the fatty acid composition [39]. We speculate that the fatty acid composition could be diacyl-l6:0/16:1. Thus, according to results from OSCC, HCC, and breast cancer, PC species containing 16:0 are commonly distributed in the cancer region. On the other hand, Ishikawa et al. could not detect the signal at m/z 770.5 corresponding to PC (diacyl-16:0/16:1) in thyroid papillary cancers [23]. Therefore, each cancer should be carefully analyzed because of the different fatty acid components of PC species in the cancer region. Moreover, investigation of fatty acid synthesis by a fatty acid synthase and PC synthesis by lysophosphatidylcholine acyltransferase could elucidate the mechanism of proliferation, migration, and invasion in the cancer and stromal regions.
Conclusion
IMS analysis of OSCC tissue sections from one patient revealed eleven signals of significantly different intensities in the cancer and stromal regions. Two signals were specifically distributed in the cancer or stromal regions of the tissue sections from five patients. Therefore these signals, which were identified as those of PC (16:0/16:1) and PC (18:1/20:4) in the MS/MS analyses, could be used to distinguish the border between the cancer and stromal regions in OSCC.
References
Jakobsson PA, Eneroth CM, Killander D et al (1973) Histologic classification and grading of malignancy in carcinoma of the larynx. Acta Radiol Ther Phys Biol 12(1):1–8
Anneroth G, Batsakis J, Luna M (1987) Review of the literature and a recommended system of malignancy grading in oral squamous cell carcinomas. Scand J Dent Res 95(3):229–249
Yamamoto E, Miyakawa A, Kohama G (1984) Mode of invasion and lymph node metastasis in squamous cell carcinoma of the oral cavity. Head Neck Surg 6(5):938–947
Nagata T, Schmelzeisen R, Mattern D et al (2005) Application of fuzzy inference to European patients to predict cervical lymph node metastasis in carcinoma of the tongue. Int J Oral Maxillofac Surg 34(2):138–142
Myoung H, Kim MJ, Lee JH et al (2006) Correlation of proliferative markers (Ki-67 and PCNA) with survival and lymph node metastasis in oral squamous cell carcinoma: a clinical and histopathological analysis of 113 patients. Int J Oral Maxillofac Surg 35(11):1005–1010
Kurokawa H, Zhang M, Matsumoto S et al (2005) The relationship of the histologic grade at the deep invasive front and the expression of Ki-67 antigen and p53 protein in oral squamous cell carcinoma. J Oral Pathol Med 34(10):602–607
Gschwind A, Prenzel N, Ullrich A (2002) Lysophosphatidic acid-induced squamous cell carcinoma cell proliferation and motility involves epidermal growth factor receptor signal transactivation. Cancer Res 62(21):6329–6336
Coussens LM, Werb Z (2002) Inflammation and cancer. Nature 420(6917):860–867
Guo Y, Wang X, Zhang X et al (2011) Ethanol promotes chemically induced oral cancer in mice through activation of the 5-lipoxygenase pathway of arachidonic acid metabolism. Cancer Prev Res (Phila) 4(11):1863–1872
Mann EA, Spiro JD, Chen LL et al (1994) Phospholipid metabolite expression by head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg 120(7):763–769
Brasitus TA, Dudeja PK, Dahiya R (1986) Premalignant alterations in the lipid composition and fluidity of colonic brush border membranes of rats administered 1,2 dimethylhydrazine. J Clin Invest 77(3):831–840
Setou M (2010) Imaging mass spectrometry: protocols for mass microscopy. Springer, New York
Gross JH (2004) Mass spectrometry: a textbook. Springer-Verlag, New York
Alexandrov T, Becker M, Guntinas-Lichius O et al (2013) MALDI-imaging segmentation is a powerful tool for spatial functional proteomic analysis of human larynx carcinoma. J Cancer Res Clin Oncol 139(1):85–95
Thomas A, Patterson NH, Marcinkiewicz MM et al (2013) Histology-driven data mining of lipid signatures from multiple imaging mass spectrometry analyses: application to human colorectal cancer liver metastasis biopsies. Anal Chem 85(5):2860–2866
Sugiura Y, Setou M (2010) Imaging mass spectrometry for visualization of drug and endogenous metabolite distribution: toward in situ pharmacometabolomes. J Neuroimmune Pharmacol 5(1):31–43
Joyner JC, Keuper KD, Cowan JA (2013) Analysis of RNA cleavage by MALDI–TOF mass spectrometry. Nucleic Acids Res 41(1):e2
Rubakhin SS, Hatcher NG, Monroe EB et al (2007) Mass spectrometric imaging of the nervous system. Curr Pharm Des 13(32):3325–3334
Jackson SN, Woods AS (2009) Direct profiling of tissue lipids by MALDI–TOFMS. J Chromatogr B Anal Technol Biomed Life Sci 877(26):2822–2829
Sugiura Y, Konishi Y, Zaima N et al (2009) Visualization of the cell-selective distribution of PUFA-containing phosphatidylcholines in mouse brain by imaging mass spectrometry. J Lipid Res 50(9):1776–1788
Schone C, Hofler H, Walch A (2013) MALDI imaging mass spectrometry in cancer research: combining proteomic profiling and histological evaluation. Clin Biochem 46(6):539–545
Shimma S, Sugiura Y, Hayasaka T et al (2007) MALDI-based imaging mass spectrometry revealed abnormal distribution of phospholipids in colon cancer liver metastasis. J Chromatogr B Anal Technol Biomed Life Sci 855(1):98–103
Ishikawa S, Tateya I, Hayasaka T et al (2012) Increased expression of phosphatidylcholine (16:0/18:1) and (16:0/18:2) in thyroid papillary cancer. PLoS One 7(11):e48873
Sugiura Y, Shimma S, Setou M (2006) Thin sectioning improves the peak intensity and signal-to-noise ratio in direct tissue mass spectrometry. J Mass Spectrom Soc Jpn 54:45–48
Schwartz SA, Reyzer ML, Caprioli RM (2003) Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J Mass Spectrom 38(7):699–708
Sugiura Y, Setou M (2009) Selective imaging of positively charged polar and nonpolar lipids by optimizing matrix solution composition. Rapid Commun Mass Spectrom 23(20):3269–3278
Enomoto H, Sugiura Y, Setou M et al (2011) Visualization of phosphatidylcholine, lysophosphatidylcholine and sphingomyelin in mouse tongue body by matrix-assisted laser desorption/ionization imaging mass spectrometry. Anal Bioanal Chem 400(7):1913–1921
Hayasaka T, Goto-Inoue N, Sugiura Y et al (2008) Matrix-assisted laser desorption/ionization quadrupole ion trap time-of-flight (MALDI–QIT-TOF)-based imaging mass spectrometry reveals a layered distribution of phospholipid molecular species in the mouse retina. Rapid Commun Mass Spectrom 22(21):3415–3426
Morita Y, Ikegami K, Goto-Inoue N et al (2010) Imaging mass spectrometry of gastric carcinoma in formalin-fixed paraffin-embedded tissue microarray. Cancer Sci 101(1):267–273
Lv W, Yang T (2012) Identification of possible biomarkers for breast cancer from free fatty acid profiles determined by GC–MS and multivariate statistical analysis. Clin Biochem 45(1–2):127–133
Aust S, Bachmayr-Heyda A, Pils D et al (2013) Determination of tumor-infiltrating CD8+ lymphocytes in human ovarian cancer. Int J Gynecol Pathol 32(3):269–276
Bedoya AM, Jaramillo R, Baena A et al (2012) Location and density of immune cells in precursor lesions and cervical cancer. Cancer Microenviron 6(1):69–77
Tanaka H, Zaima N, Yamamoto N et al (2011) Distribution of phospholipid molecular species in autogenous access grafts for hemodialysis analyzed using imaging mass spectrometry. Anal Bioanal Chem 400(7):1873–1880
Hanada M, Sugiura Y, Shinjo R et al (2012) Spatiotemporal alteration of phospholipids and prostaglandins in a rat model of spinal cord injury. Anal Bioanal Chem 403(7):1873–1884
Gupta RA, Dubois RN (2001) Colorectal cancer prevention and treatment by inhibition of cyclooxygenase-2. Nat Rev Cancer 1(1):11–21
Zhang S, Du Y, Tao J et al (2008) Expression of cytosolic phospholipase A2 and cyclooxygenase 2 and their significance in human oral mucosae, dysplasias and squamous cell carcinomas. ORL J Otorhinolaryngol Relat Spec 70(4):242–248
Shibata M, Kodani I, Osaki M et al (2005) Cyclo-oxygenase-1 and -2 expression in human oral mucosa, dysplasias and squamous cell carcinomas and their pathological significance. Oral Oncol 41(3):304–312
Morita Y, Sakaguchi T, Ikegami K et al (2013) Lysophosphatidylcholine acyltransferase 1 altered phospholipid composition and regulated hepatoma progression. J Hepatol in press
Ide Y, Waki M, Hayasaka T et al (2013) Human breast cancer tissues contain abundant phosphatidylcholine(36:1) with high stearoyl-CoA desaturase-1 expression. PLoS One 8(4):e61204
Acknowledgments
This study was supported in part by a Grant-in-Aid for Scientific Research (no. 22500406) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, by a Grant-in-Aid for Young Scientists (S) from the JSPS to MS, by a SENTAN grant from the Japan Science and Technology Agency, and by a Grant-in-Aid from the Ministry of Health, Labour and Welfare.
Author information
Authors and Affiliations
Corresponding author
Additional information
Published in the topical collection Biomedical Mass Spectrometry with guest editors Mitsutoshi Setou, Toshimitsu Niwa, and Akira Ishii.
Rights and permissions
About this article
Cite this article
Uchiyama, Y., Hayasaka, T., Masaki, N. et al. Imaging mass spectrometry distinguished the cancer and stromal regions of oral squamous cell carcinoma by visualizing phosphatidylcholine (16:0/16:1) and phosphatidylcholine (18:1/20:4). Anal Bioanal Chem 406, 1307–1316 (2014). https://doi.org/10.1007/s00216-013-7062-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-013-7062-3