Abstract
Introduction
Oral cancer is a sixth commonly occurring cancer globally. The use of tobacco and alcohol consumption are being considered as the major risk factors for oral cancer. The metabolic profiling of tissue specimens for developing carcinogenic perturbations will allow better prognosis.
Objectives
To profile and generate precise 1H HRMAS NMR spectral and quantitative statistical models of oral squamous cell carcinoma (OSCC) in tissue specimens including tumor, bed, margin and facial muscles. To apply the model in blinded prediction of malignancy among oral and neck tissues in an unknown set of patients suffering from OSCC along with neck invasion.
Methods
Statistical models of 1H HRMAS NMR spectral data on 180 tissues comprising tumor, margin and bed from 43 OSCC patients were performed. The combined metabolites, lipids spectral intensity and concentration-based malignancy prediction models were proposed. Further, 64 tissue specimens from twelve patients, including neck invasions, were tested for malignancy in a blinded manner.
Results
Forty-eight metabolites including lipids have been quantified in tumor and adjacent tissues. All metabolites other than lipids were found to be upregulated in malignant tissues except for ambiguous glucose. All of three prediction models have successfully identified malignancy status among blinded set of 64 tissues from 12 OSCC patients with an accuracy of above 90%.
Conclusion
The efficiency of the models in malignancy prediction based on tumor induced metabolic perturbations supported by histopathological validation may revolutionize the OSCC assessment. Further, the results may enable machine learning to trace tumor induced altered metabolic pathways for better pattern recognition. Thus, it complements the newly developed REIMS-MS iKnife real time precession during surgery.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Oral cancer ranks sixth among all cancer in the world (Fernández et al. 2015). It accounts 3.8% of all cancer cases and 3.6% of cancer deaths (Shield et al. 2017). The Oral Squamous Cell Carcinoma (OSCC) is defined as a malignant neoplasm originating in the stratified epithelium of the oral tissues. The use of tobacco and alcohol is being considered as one of the major risk factors for oral cancer (Jané-Salas et al. 2012). The morbidity and mortality associated with oral cancer can be prevented to a greater extent by early detection (Jafari et al. 2013). However, early lesions are difficult to diagnose because they are usually asymptomatic and clinically varying in appearance (Rajeev et al. 2015). Slaughter et al. first explained the multifocal growth of oral squamous carcinoma in 783 patients as “Field cancerization” (Slaughter et al. 1953). According to field cancerization, microscopic patches of abnormal tissue surrounding the tumors gets proliferated due to preconditioning of epithelial area by carcinogenic agents (Curtius et al. 2018). Among all available cancer diagnostic techniques, biopsy and histopathological analysis are treated as gold standards (Seetharam and Ramachandran 1998; Rajeev et al. 2015). However, factors like inter-observer variability in grading, sample bias or patient’s cohort differences results in field cancerization without morphological changes and are being passed undetected (Curtius et al. 2018). Advanced molecular methods like polymerase chain reaction (PCR), fluorescent in situ hybridization (FISH) and flow cytometry have improved in understanding of pathogenesis, nature of lesion and prognosis. Further, optical spectroscopic techniques like fluorescence spectroscopy (FS), elastic scattering spectroscopy (ESS) have detected critical substances correlated with the biochemical changes occurring in tissues. Fluorophores mainly amino acids like tryptophan, tyrosine, structural proteins, co-enzymes, NADH, porphyrins etc. were exploited in tissues status profiling (Sreeshyla et al. 2014).Two techniques, NMR spectroscopy and mass spectrometry (MS) has been widely explored to identify metabolic changes occurred in malignant tissues (Chen and Yu 2019).
Both of the above techniques have long been employed in profiling the metabolic composition in available biofluids or tissues that were sampled from clinics (Jiménez and MacIntyre 2017). The metabolic composition or phenotype of an individual indicates the underlying complex gene environment interactions. Although there exists physiological variabilities among each tissues types, the snapshot of metabolic activity recorded on local phenotypes have a wide potential in evaluation of different stages of disease and other pathological conditions. Further, the gap of isolated metabolic phenotypes were bridged using systematic statistical modelling up to the system level (Nicholson et al. 2012). Clinical metabolomics employ Chemometrics like multivariate statistics to reduce the complex spectroscopic data into a relatable biochemical patterns of disease and their interventions. The metabolic similarities among each samples are mapped, and the correlating metabolites are extracted with linear projection methods like PCA and PLSDA. Further in validation, the multiparametric data with known histopathological endpoints are used as a training set to generate predictive models capable of separating samples from different histopathological classes (Nicholson et al. 2012). A similar approach of predictive PLS based abnormal breast cancer cell detection were explored by Zhu et al. (2017). CAMO Unscrambler X software uses “Jack-knife” estimation of parameter uncertainty to visualize the prediction results (Martens and Martens 2000).
During assessment of multivariate analysis on megavariate metabolomic data Rubingh et al. observed that latent variables (LV) and their selection plays critical role in performance of data analysis. They observed significant perturbation in results of cross validation, jack-knifing model parameter and permutation tests of predictive models (Rubingh et al. 2006). In NMR spectroscopy, latent variables represent spectral signals generated by underlying metabolite concentrations. Thus, effective predictive modelling depends on spectral binning as well as targeted metabolic profiling (Chang et al. 2007). In an earlier study, our group had effectively identified several metabolic perturbations of malignant tumor tissues form non-malignant bed and margins tissues specimens using HRMAS NMR spectroscopy. The predictive model generated on spectral region of 4.25–1.80 ppm which divided into 245 continuous bins of 0.01 ppm width. The OPLS-DA model had 97.4% of diagnostic accuracy, along with 100% sensitivity and 92.3% specificity. Confounder metabolites were identified as acetate, glutamate, taurine, TCho and lactate (Srivastava et al. 2011). Although OPLS-DA model did showed good separation between malignant and benign tissues, the result from spectral binning may have possibility of overfitting and thus deceptive (Chang et al. 2007). Due to possibility of metabolite signal overlapping, alternative method of peak-picking PLS algorithm were proposed (Ammann et al. 2006).
As technologies for spectral processing updated, it allowed us to quantify the underlying metabolites signals. Bruker Amix software’s deconvolution allows to quantify the lipid signals, while Chenomx scan be used to identify each metabolites present in 1H NMR tissue spectra (Wishart 2008; Chenomx 2015). Unfortunately the quantitation of lipids and metabolites were restricted due to non-uniformity of TSP signals generated from isotropic susceptibility difference between intracellular and supernatant water compartment and were depends on sample spin rate (Hong et al. 2009). The unreliability of TSP further increased due to its tendency to interact with macromolecules especially proteins (Van 2013). The limitation can be resolved by a software generated artificial signal QUANTAS with a constant scaling factor. The QUANTAS allows us to introduce a uniform reference signal to all samples depending upon their spectral parameter and can be quantified from the sample having most uniform TSP signal. An analogous signal customCSI were allowed to introduce by Chenomx software in order to quantify the metabolites (Vitols and Mercier 2006; Chenomx 2015). The applicability of the results of the present work is in identification of biomarkers that trace altered metabolic pathways of OSCC along with its specificity in determination of clinical status. Thus, it provides support to less histopathologically specific in vivo chemical characterization of tissues by Rapid Evaporative Ionization Mass Spectrometry (REIMS) in real time (Nicholson et al. 2012).
This NMR based metabolomic study is an attempt to make a more robust and effective model which could blindly predict the malignancy in OSCC cases. Therefore, refined chemometric models were generated from metabolic profile which consists majority of the earlier recorded NMR spectral data of 36 OSCC patients (159 tissue specimens) with additional combination of newly recorded data from 7 OSCC patients (21tissue specimens). These models were tested to predict malignancy among a set of 64 blinded tissues (42 tumor and adjacent oral tissues; 22 neck tissues including arteries, veins, nerves and lymph nodes) obtained from twelve patients undergoing surgery having OSCC along with neck invasions. The validation of the results were further reinstated by histopathological analysis of the same tissues specimens. This study may contribute in establishing the correlation of metabolite composition to clinical status of various tissues under the influence of OSCC.
2 Materials and methods
2.1 Subjects and study protocol
Nineteen OSCC patients, including 12 patients having neck invasions, who were enrolled in this study had undergone surgical treatment at King George’s Medical University (KGMU), Lucknow. The mean age of patients who were suffering from oral cancer was 44 years. None of the patients were dehydrated and/or anemic (mean Hb 9.8 g/dl) at the time of surgery. Informed consent was obtained from all the patients and protocol was approved by KGMU Ethical Committee.
Tissue samples comprising of tumor and adjacent benign oral mucosa along with tissues from neck region which includes nerve, vein, artery and lymph nodes excised during surgical resection were obtained from each patient. Sampling was purely based on visual screening by the surgeon. All tissue specimens were stored in high quality plastic vials and snap-frozen in liquid Nitrogen at the time of surgery and further stored at − 80 °C till proton HRMAS NMR spectra recorded.
2.2 Chemicals
All the chemicals including D2O (with 99.96% deuterium-enriched and 0.03% sodium-3-(trimethylsilyl)-2,2,3,3-d4-propionate—TSP) was purchased from Sigma–Aldrich Inc. (St. Louis, MO, USA). All other chemicals used were of reagent grade with the highest purity commercially available, unless otherwise mentioned. The retailer, chemical purity and batch were assured to be matched with the chemicals used in earlier work of Srivastava et al. (2011).
2.3 1H HRMAS NMR spectroscopy
All tissue specimens were thawed and washed with D2O and the dissected inner core were packed in 4 mm ZrO2 rotor of 50 µl capacity, to this, 20 µl of D2O having 0.03% TSP was added as Chemical Shift Index (CSI). The 1H HRMAS NMR spectra recorded with Bruker Avance 400 MHz spectra having 4 mm HRMAS 1H-13C dual probehead operating on 400.13 MHz. Rotational sidebands eliminated with sample spin of 4.0 kHz. With reference to earlier work performed by Srivastava et al. (2011), Proton NMR spectra of all tissue specimens were recorded with 1D NOESY pulse sequence, keeping mixing time τm: 100 ms, total relaxation time: 3.99 s and 8250.8 Hz of spectral width. Each spectra recorded with 128 transients plus 2 dummy scans that took a total recording time of 9 min. Similarly, 1D CPMG pulse sequence with water suppression comprising an echo time of 0.1 ms each (0.2 ms) flanked between 180° pulses with 800 cycles of repetitions having a total echo time of 160 ms were recorded for each tissue samples. The relaxation time of 3.99 s includes 2 s of delays and 1.99 s of acquisition time. Both NOESY and CPMG experiments accompanied with water presaturation of 2 s using weak transmitter pulse at the transmitter frequency of 400.1318806 MHz prior to the execution of hard pulses for recording of the NMR spectral data. Further, sample temperature maintained at 283 K during spectral acquisition using nitrogen gas cooling system (Srivastava et al. 2011).
2.4 Histopathology
After HRMAS NMR spectral recording, both 21 oral tissues from 7 new OSCC patients and 64 blinded tissues specimens from 12 patients have OSCC with neck invasion were subjected to histopathological analysis. To do so, each tissue samples fixed in 10% formalin, then embedded into paraffin. After that all samples sent to histopathological analysis at KGMU, Lucknow (Srivastava et al. 2011). Result of histopathological studies show, among 64 blinded tissues from 12 patient having OSCC with neck invasion, 22 out of 42 tumor and adjacent oral tissues have confirmed well differentiated SCC. Further, four secondary lymph nodes and a carotid sheath identified with OSCC invasion among 22 neck tissues. (Overall details of tissues used in present study are provided in the Supplementary Table ST1.)
2.5 NMR spectra, processing and assignments
The 1H HRMAS NMR spectra of all tissues from both NOESY and T2 filtered CPMG experiments were processed with Bruker Topspin 3.5 spl6 software. Each of these spectra were then divided into 1501 repetitive continuous bins of 0.005 ppm size corresponding region of 0.5 to 8.4 ppm (using Amix, version 4.4.1, Bruker BioSpin, Switzerland) and water region (4.70–5.10 ppm) removed in order to perform metabolites signal intensity based predictive analysis (Paul et al. 2018).
Lipid signals assigned and quantified in NOESY 1H HRMAS NMR spectra (or NOESY Spectra) using software Amix deconvolution algorithm (Wishart 2008). In absence of uniform TSP signal, quantification of lipid signal performed with respect to software generated artificial signal QUANTAS having constant scaling factor of 15e−04 a.u. (Farrant et al. 2010; Bharti and Roy 2012). Concentration assigned to the QUANTAS signal were calibrated with respect to TSP signal of known concentration among model tissue set. Assigned concentration of QUANTAS is 0.8344 mM. Since NMR Spectroscopy only sensitive to mobile lipids (Rizwan and Glunde 2014), present work only measure quantities of these lipid species, which are (1) linoleic acid, (2) Sum of Higher Polyunsaturated Fatty Acids (SHi-PUFA) (Alexandri et al. 2017), (3) Mono Unsaturated fatty Acids (MUFA), (4) Saturated Fatty Acids (SFA), (5) Triglyceride (TG), (6) Total Fatty Acids (TFA) and (7) Free Fatty Acids (FFA) from signals of NOESY spectra recorded on tissues specimens. The mathematical expression for quantitation of lipid species is documented in Supplementary Table ST2.
Similarly, metabolite signals were identified and quantified in CPMG 1H HRMAS NMR spectra (or CPMG spectra) using Chenomx (version 8.1) (Chenomx 2015). For unambiguous characterization of various metabolites having overlapping signal viz. guanidoacetate, 2-oxoglutarate, Asn, 4-aminobutyrate, choline, MeOH, Ser etc., earlier recorded two-dimensional COSY and HSQC spectra (Srivastava et al. 2011) were reanalyzed and assignment were further reinstated through matching results with BMRB and HMDB database. Quantification of metabolites performed with respect to Chenomx software introduced Custom CSI signal (Chenomx 2015) and calibrated with respect to TSP of known concentration. Assigned concentration of Custom CSI signal is 0.0644 mM. While quantification, all parameters of custom CSI signal were maintained unaltered.
All tissue spectra were analyzed for residual TSP signals, the signal intensity of TSP most commonly occurring among all tissue spectra was assigned as effective TSP signal. The quantitation of above all lipid and metabolites were performed with respect to the effective TSP signal. Further any deviation in quantitation by above analytical methods were measured as limit of quantitation (LOQ) and Limit of detection (LOD) (Mikkelsen and Cortón 2016) from sets of prepared samples with known concentration of metabolites and TSP (As mentioned in Supplementary Procedure SP1).
2.6 Predictive modelling and optimization
Processed spectral data from 1H HRMAS NMR spectroscopy of malignant and benign tissues can be categorized as: NOESY spectra, CPMG spectra and Metabolite concentration including lipids. Each data categories were independently explored in predictive modeling and tissue analysis. The metabolic separation between malignant and benign tissues of model set were mapped with score and loading plots of Principle Component Analysis (PCA) using ‘The Unscrambler X’ software package (version 10.4, Camo ASA, Norway) (Srivastava et al. 2011). Predictive models were generated with Partial Least square methods (PLS) on each data category. For spectral data categories, OSCC filtration were performed and therefore OPLS-DA models were developed to reduce data ambiguity due to any diurnal changes occupied among the tissues (Gavaghan et al. 2002). For metabolites concentration based modelling, both PLS-DA and OPLS-DA models were compared for their efficacy in prediction of malignancy status among unknown 64 tissues from blinded set of 12 patients suffering from OSCC along with neck invasion.
The blinded set of 64 tissues from 12 OSCC patient with neck invasion have diverse tissue type like nerve, artery, veins, lymph node as well as facial tissue, muscle and tongue cancer. These diversification among tissues may experience isolated and unique metabotype (Nicholson et al. 2012) that may interfere with PLS-DA and OPLS-DA models based malignancy prediction. Therefore all prediction models were optimized by dividing the model set of 180 tissues (n = 108 benign, n = 72 malignant) from 43 OSCC patients were divided into training set of 133 tissues of 35 patients and test set of 47 tissues from 8 patients (details provided in the Supplementary table ST 4). In order to maintain tissues variability similar to blinded set, all patients with muscle tissues were selected in test set. The trial prediction models was generated from the training set of 133 malignant and benign tissue which were tested form malignancy 47 tissues from 8 patients in test set. From CPMG and NOESY spectral data, OPLS-DA model generated and tested in malignancy prediction. Successively unsupervised PCA identified corresponding confounder metabolite peaks with respect to malignant and benign tissues. Further performance of OPLS-DA model enhanced by selecting suitable latent variable (LV) (Szymańska et al. 2012) as mentioned in Supplementary Procedure SP2–3.
Similarly, the significance of OSC filtration in quantitative metabolomics were validated from AUROC of each metabolite, performance parameter of OPLS-DA model and predictive accuracy of unknown tissues of test set (as mentioned in Supplementary Procedure SP4). Then PLS-DA model also tested for efficacy of discreet metabolites excluding data of SHi-PUFA, FFA, TFA and TCho (in Supplementary Procedure SP5). Finally three approaches, VIP (Variable Importance Projection) score, HCA (Hierarchical Cluster Analysis) dendrogram splitting height and probabilities in NPP (Normal Probability Plot) used as criteria for metabolite selection in order to study LV based performance enhancement of quantitative PLS-DA prediction model (as mentioned in Supplementary Procedure SP6). All modelling, prediction and validation were performed with Unscrambler X (version 10.4, CAMO), Simca-P (version 14.1, MKS Umetrics) (Moazzami et al. 2015) and Metaboanalyst (version 4.0) (Chong et al. 2018).
2.7 Tissue analysis and classification
Predictive (OPLS-DA) models were generated from CPMG spectral data on a set of 180 tissues (n = 108 benign, n = 72 malignant) from 43 patients using results of above optimization. The generated models employed for malignancy analysis on corresponding CPMG spectral data recorded on unknown set of 42 tissues from 12 OSCC patients. Comparing the histopathological status of OSCC tissues, effective cut-off value separating malignant and benign tissue regions were identified. Positive and negative likelihood for each tissue determined on the basis predicted with deviation scores (Kinross 2019). Any tissues have higher prediction score were assigned malignant, the accuracy prediction measured from number of true positive and true negative tissues case in respect to their histopathological status were validated. Similarly, NOESY spectral data were harnessed in determining the pathological status of 42 blinded tissues from 12 patients using prediction model generated from model set of tissues. Each of above model were also tested on set of neck tissue in order to determine OSCC invasion.
Each of 42 tumor and adjacent tissues as well as 22 neck tissues from 12 patients in blinded set was profiled for the metabolite and lipid species concentration. The profiled lipids and metabolites concentration data of each tissue were then subjected to pathological prediction with the predictive model generated from corresponding concentration data of model set tissues as mention above. Further, the above model was tested to determine the OSCC in neck tissues which includes secondary lymph nodes. The metabolic profile of OSCC invasion free neck tissues mainly arteries, veins, nerves and lymph nodes were also compared using unsupervised multivariate analysis and confounder metabolites determined from variable importance projection (VIP) scores. Further the influence of OSCC invasion characterized by addition of secondary lymph nodes and carotid sheath in the VIP score of PLS-DA model on metabolite concentrations.
2.8 Analysis of lipid and metabolic perturbations and their pathways
2.8.1 Lipid and metabolite profiling
Significance of 48 metabolites including 7 lipid species in differentiating malignant tissue from benign tissues was tested with Student’s t test along with their box plot (using metaboanalyst, version 4.0, www.metaboanalyst.ca) (Chong et al. 2018). The variation in lipid profile from benign tissue to malignant tissue in model set were evaluated with univariate Box–whisker plot (using Microsoft Office, version 2016, Microsoft Corporation USA) (Beskeen et al. 2016). Contribution of each lipid species in overall lipid content were graphed with Pie-Doughnut chart (Microsoft office, version 2016) for model set. Also the correlation between TFA and TG among benign tissues and malignant tissues in model set measured individually along with regression coefficient R2 (Unscrambler X, version 10.4).
2.8.2 Pathway analysis
Using Metaboanalyst (version 4.0), a set 25 confounder metabolites were identified from PLS-DA VIP score of 48 metabolites measured among 180 tissues of model set. Pathway and Enrichment analysis module of Metaboanalyst (version 4.0) (Chong et al. 2019) were then applied on confounder metabolite concentration data from the model set of 180 tissues in order to highlight the dysregulated network of pathways (Chong et al. 2018). Finally tracking the identified pathways, a generalized network of pathways locating majority of metabolites were drawn using “An atlas of biochemistry and molecular biology” as reference (Michal and Schomburg 2012).
3 Result
3.1 Oral SCC tissues spectra and profiling
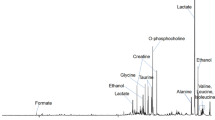
Figure 1 represent a stack plot of CPMG spectra of OSCC and adjacent tissues from model set. From bottom to top spectra are from benign bed, benign margin, benign muscle, other benign tissue, malignant bed, malignant margin, tumor tissue and other malignant tissue. CPMG spectra of all benign tissues were dominated by lipid signal. Only benign muscle tissues demonstrated signals from small molecular metabolites like Cr, Lac, Gln and Total Choline (TCho). The CPMG spectra of malignant tissues show strong cluster of metabolites signal. A similar pattern of lipids and metabolites signals observed in NOESY spectra of all above tissues (Supplementary Figure SF1). Additionally, signals of TG were observed in NOESY spectra. Overall 44 metabolites including lipids were successfully quantified from the NOESY spectra (Supplementary Figure SF2) and CPMG spectra (Supplementary Figure SF3). Supplementary Table ST3 listed the identified metabolite with their HMDB ID. The table also included the Sum of Higher Polyunsaturated Fatty Acids (SHi-PUFA), Free Fatty Acids (FFA), Total Fatty Acids (TFA) and TCho in order to maintain metabolic significance as well as to imitate the signal overlap in NMR spectra by similar metabolites.
A similar stack plot of bar diagram for 48 metabolites concentration bars for above set of tissues are shown in Supplementary Figure SF4a. The full concentration bar diagram shows very tall bar of Lac concentration among all malignant tissues. Other metabolites basically amino acids like Glu, Gln, Ala, Lys and Tau were also have significant concentration among malignant tissues. While benign tissue show a complete metabolite scarcity. Only benign muscle tissues have significant concentration bars of Cr, Glc, Lac and Tau. Enlarging the bar diagram as in Supplementary Figure SF4b through reduction of concentration window range to 0.275 mM allow us to profile less concentrated metabolites. The concentration windows show accumulation of large number of metabolites in malignant bed tissues. Although benign bed tissues too had a number of metabolite concentration bars, yet their concentrations were not comparable with malignant bed tissues. Benign margin have specific lipid composition pattern which consisting of large TFA bar, medium SFA, MUFA, TG and FFA bars, minute SHi-PUFA and linoleic acid bar for concentrations. Similar pattern, or benign pattern, of lipid composition bars were seen in all benign tissues except benign muscles. However among all malignant tissues, these lipid composition pattern were highly altered. All malignant tissues have negligible amount of TG, linoleic acid and SHi-PUFA. Estimated Limit of quantitation (LOQ), Limit of detection (LOD) (Long and Winefordner 1983) in observations of Supplementary Procedure SP1 showed a insignificant deviations by both QUANTAS based and CustomCSI based metabolite quantitation.
3.2 Observations from latent variable based predictive model optimization
Supplementary Figure SF5 and SF6 show the results of optimization process applied to CPMG, NOESY spectral data. In CPMG spectra, confounder metabolite signals responsible for effective separation of malignant and benign tissues were found in the region of 1.42–4.22 ppm. These signals are generated by Lac, Ala, Glu, TCho, Tau, α-H amino acids and lipids. In NOESY spectra, major confounders are lipid signals which appeared between 1.41 and 4.69 ppm. The region also includes signal from TCho.
Supplementary Figure SF7–9 demonstrates the significance of OSC filtration in fully quantitative analysis. Although there is an increase in sensitivity of Glc and lipids after OSC filteration, accuracy of malignancy prediction by metabolite concentrations based OPLS-DA model was found to be less than that PLS-DA model. In Supplementary Figure SF10, discreet metabolites excluding SHi-PUFA, FFA, TFA and TCho were further explored for malignancy prediction of test set of tissues. Lastly, results of latent variable as metabolite selection given in Supplementary Figure SF11–16, Supplementary Table ST5–7 along with detailed observations. Among three approach of latent variable as metabolite selection, VIP score based metabolite selection was found to be most effective.
3.3 Observation from blinded set of tissues analysis
MRI of patient 1 from blinded set imaged an engrossing tumor grown in buccal cavity under the tongue (Supplementary Figure SF17). Seven tissues received were histopathologically identified as artery, vein, facial, margin 1, lymph node, nerve and tumor tissues. Following three blocks inscribed the results of spectroscopic analysis of 64 OSCC tissues from 12 patients including above tissues from patient 1.
3.3.1 Block 1: based on CPMG spectral analysis
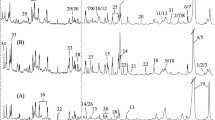
Figure 2 represents a stack plot of CPMG spectra recorded on tumor and adjacent oral tissues (at the top half) and neck tissues including artery, vein, nerve and lymph node (at the bottom half). Clusters of metabolite signals visible in CPMG spectra of tumor periphery tissue while the CPMG spectra of adjacent tissues are dominated with lipid signals. CPMG spectra of all invasion free neck tissues from patient 1, including artery, lymph node and nerve were dominated from lipid signals, except vein which has spectral profile similar to benign muscle tissues.
Representative stack plots of HRMAS 1H CPMG NMR spectra recorded on tissues specimens received from patient1 along with respective histopathological status. The CPMG spectra of tumor and adjacent oral tissues as (top): a tumor periphery (malignant), b lateral margin (benign), c facial tissues (benign). The CPMG spectra of neck tissues as (bottom): d nerve (no invasion), e lymph node (no invasion), f vein (no invasion) and g artery (no invasion)
Supplementary Figure SF18 shows the malignancy prediction among 42 tissues in blinded set from 12 patients using different CPMG spectra-based OPLS-DA prediction model. Each model is validated with AUROC, permutation test, R2 and Q2 values. The model with spectral width of 1.42–4.67 ppm is most effective in segregation of blinded tissues into layers of benign tissues and malignant tissues which were further reinstated by histopathological analysis. Figure 3a, b represent corresponding 3D PCA, OPLS-DA score plot. Both 3D score plots showed clear separations between malignant and benign tissues. Further the 3D PCA score plot had a minor separation between benign muscle tissues and rest of the benign tissues. PCA loading plot in Fig. 3c show upregulation of Ala, Glu, Ac, Cr, TCho, Tau, Lac and Glc in malignant tissues, while all lipid species were downregulated. Figure 3d outlines the malignant and benign tissues from each of 12 patients. Validation parameters gives OPLS-DA model are 0.997 and 0.990 for sensitivity and specificity, 0.914 and 0.868 for R2 and Q2, observed statistics were preserved for more than 45 permutation test (Supplementary Figure SF18(2)). Further the prediction model successfully identified malignant carotid-sheath and all 4 secondary lymph nodes from neck (Fig. 3e). However, two false positive cases of benign margins as well as false predictions of invasion free arteries, nerve among neck tissues were also observed. These tissue specimens had significant metabolite signals.
a Unsupervised 3D PCA score plot on CPMG spectral data which display a separation in distribution of 72 malignant tissues (including bed, margin, tumor and other) and 108 benign tissues (including bed, margin, muscle and other), b 3D score plot of generated OPLS-DA model, c loading plot for contribution lipids and metabolites. d CPMG spectra-based malignancy prediction among tumor and adjacent oral tissues using above prediction model (Result obtained in form of red horizontal line of prediction score with box of positive and negative deviations to uncertainty.). e Application of CPMG spectra-based predictive analysis in determination of OSCC invasion among neck tissues in addition to secondary lymph node identification. Color coding as follows: for tumor and adjacent oral tissues: red: malignant histopathology; blue: benign histopathology. For neck tissues: maroon: OSCC invasion histopathology; light green: no invasion histopathology
3.3.2 Block 2: based on NOESY spectral analysis
Similarly stack plot of NOESY spectra of tissues from patient 1 in Supplementary Figure SF19 showed dominance of lipid signals in artery, facial, margin 1, lymph node and nerve, while vein spectra show signals of Glu, TCho, Cr and Glu. In NOESY spectra of tumor periphery, lipid signals shrunk and large numbers of metabolites signals were observed.
Results of NOESY spectra-based predictive analysis on blinded tissues depicted in Supplementary Figure SF20. Predictive model generated from NOESY spectral data of width 1.42–4.69 ppm found most effective with three false positive. Figure 4a, b represent 3D PCA, OPLS-DA score plots respectively. Each of these plots show significant separation of benign and malignant tissues. Corresponding loading plot in Fig. 4c show correlation of lipids to benign tissue while TCho, Cr, Glu, Ala, Gly and Lac to malignant tissues. Figure 4d outlines the malignant and benign tissues from each of 12 patients. The R2 and Q2 for OPLS-DA model are 0.831 and 0.806 respectively while sensitivity and specificity are 0.996 and 0.991 respectively (Supplementary Figure SF20(2)). Large deviation in some tissue developed due to presence of spectral noise in their respective spectra. Further the investigation of OSCC invasion among neck tissues successfully identifies malignant carotid sheath and all the four secondary lymph nodes (Fig. 4e). However, there are several false positive predictions including arteries, veins and nerves.
a Unsupervised 3D PCA score plot on NOESY spectral data which display a separation in distribution of 72 malignant tissues (including bed, margin, tumor and other) and 108 benign tissues (including bed, margin, muscle and other), b 3D score plot of generated OPLS-DA model, c loading plot for contribution lipids and metabolites. d NOESY spectra-based malignancy prediction among tumor and adjacent oral tissues using above prediction model. (Result obtained in from of Red horizontal line of Prediction score with box of positive and negative deviations to uncertainty). e Application of NOESY spectra-based predictive analysis in determination of OSCC invasion among neck tissues in addition to secondary lymph node identification. Color coding as follows: For Tumor and adjacent oral tissues: red: malignant histopathology; blue: benign histopathology. For neck tissues: maroon: OSCC invasion histopathology; light green: no invasion histopathology
3.3.3 Block 3: full set metabolites concentration analysis:
3.3.3.1 Metabolic profile of tissues obtained from 12 OSCC patients
64 tissues received from 12 patients suffering from Oral SCC were profiled for metabolic and lipidomic composition as shown in Supplementary Figure SF21 (patients 1–12). The benign pattern of lipid composition of Supplementary Figure SF4b were also observed in all the benign adjacent tissues from 12 patients. The histopathologically malignant tissues of each of 12 patients show a similar deranged lipid composition pattern with relatively higher amount of FFA. Further all of these tissues showed very high Lac concentration bars as well as significant bars of Glu, Tau, Ala and TCho concentration. All invasion free neck tissues displayed a dominative but benign pattern of lipid composition. Upon concentration scale window enlargement, each tissues type of neck tissues displayed their unique metabolic signature. In arteries, visible number of metabolites bars were observed with higher Glc/Lac ratio. These bar heights were found to be reduced for veins along with low Glc/Lac ratio. In few cases, veins also had large metabolite bars especially for Cr, Glc and Lac. Nerves had equal amount of Glc and Lac and small amount Tau. Among all OSCC free neck tissues types, lymph nodes have higher intensity of lipid bars. However these lipid profiles were observed to have disproportionated in histopathologically identified secondary lymph nodes. All secondary lymph node tissues had higher concentration of metabolites with highest Lac concentrations.
3.3.3.2 Tissue analysis using quantitative prediction model
Both 3D PCA and 3D PLS-DA score plot in Fig. 5a, b show a visible separation between malignant and benign tissues. The distribution of malignant tissues and benign tissues including benign muscle tissues in metabolite concentration based 3D PCA score plot (Fig. 5a) were arranged similar to CPMG spectra-based 3D PCA score plot (Fig. 3a). Loading plot in Fig. 5c demonstrates upregulation of metabolites in malignant tissues and downregulation of lipid species. Figure 5d represents malignancy prediction of 42 tissue specimens from blinded set of 12 patients using metabolite concentration-based PLS-DA model. The PLS-DA model with R2 and Q2 of 0.833 and 0.832, sensitivity and specificity for AUROC are 0.996 and 0.991 [Supplementary Figure SF22(2)]. In accuracy, metabolite concentration-based PLS-DA has zero false positive and one false negative prediction i.e. patient9 margin1. Among neck tissues, concentration-based PLS-DA prediction model successfully distinguished secondary lymph nodes from rest of the tissues, however it could not able to predict OSCC invasion in carotid-sheath. While, the prediction via OPLS-DA model on metabolite concentrations of model set of 180 tissues form 43 OSCC patients generates two false positive and three false negative cases among 42 tissues of blinded set of 12 patients and one misclassification from neck tissues [Supplementary Figure SF22(1)]. The PLS-DA model generated from discreet metabolites were also equally sensitive to malignant tissue as the model with addition of TCho, TFA, FFA and SHi-PUFA data (Supplementary Figure SF23).
a Unsupervised 3D PCA score plot on metabolites concentration data which display a separation in distribution of 72 malignant tissues (including bed, margin, tumor and other) and 108 benign tissues (including bed, margin, muscle and other), b 3D score plot of generated PLS-DA model, c loading plot for contribution lipids and metabolites. The tissues with red names have higher malignancy prediction score and were malignant histopathological status. Blue named tissues have benign histopathological status. d Metabolite concentration-based malignancy prediction among tumor and adjacent oral tissues using above prediction model. (Result obtained in form of Red horizontal line of Prediction score with box of positive and negative deviations to uncertainty). e Application of metabolite concentration-based predictive analysis in determination of OSCC invasion among neck tissues in addition to secondary lymph node identification. Color coding as follows: for tumor and adjacent oral tissues: red: malignant histopathology; blue: benign histopathology. For Neck tissues: maroon: OSCC invasion histopathology; light green: no invasion histopathology
3.4 Neck tissues metabolomics in patients suffering from OSCC
Multivariate analysis of invasion free neck tissues including arteries, veins, nerves and lymph nodes were randomly distributed each 3D PCA score plots generated from CPMG, NOESY spectral data and metabolite concentration data (Supplementary Figure SF24a). With inclusion of secondary lymph nodes and carotid sheath, a separation appeared in 3D PCA score plots of all three data categories as shown in Supplementary Figure SF24b, between invasion free neck tissues cluster arteries, veins, nerves and lymph nodes and the OSCC invaded cluster of secondary lymph nodes and carotid sheath.
VIP score plot for invasion free neck tissues showed upregulation in most of amino acids in arteries, while Glc, Lac, Tau Gln, Cr and homocysteine (Hcys) in veins. The lymph node has upregulation of FFA and PCho. Nerves had moderate metabolite profile (Supplementary Figure SF23c). Upon addition of secondary lymph nodes in metabolite concentration data, VIP score list gets altered (Supplementary Figure SF23d). The VIP score depicted an upregulation of all metabolites in secondary lymph nodes while invasion free lymph nodes had higher amount of TFA. Since present study consisted of single carotid sheath, it couldn’t be included in VIP scores. The comparative stack plot of invasion free lymph node and secondary lymph node (having OSCC invasion) based on CPMG and NOESY spectra (in Supplementary Figure SF25a,b) showed normal lymph nodes were dominated by lipid signals while secondary lymph node spectra were accompanied with large number of metabolite signals. The stack plot of metabolite concentration bars in Supplementary Figure SF25c showed reduction of lipid species among secondary lymph nodes although the benign lipid pattern (as shown in Supplementary Figure SF4) were maintained. Secondary lymph nodes also had very high concentration of Lac, Tau and Glu.
The results for studies of metabolites selection in PLS-DA model efficiency is given in Supplementary Figure 26 (1–3). The effect of tissue homogeneity in model sets were visible in PLS-DA model efficiency. Accuracy of PLS-DA for VIP score based metabolite selection was found to be less than that of NPP score based metabolite selection.
3.5 Metabolic perturbations in malignant tissues and their pathway correlations
Figure 6a provides the results of Student’s t-test on 48 metabolites including 7 lipid species quantified in model set. Corresponding box plots for each 48 metabolites are provided in Supplementary Figure SF27. Box plots of all metabolites shows high concentration of lipid species in benign tissues and high metabolites concentration among malignant tissues of model set. Overall 2D t-test divides the metabolites into three classes. Class 1 comprises the lipid species which are observed in excess in benign tissues and have positive t.stat values. Class 2 comprises the rest of the metabolites except carnitine (Car), Cr, ethanol (Et), Glu, inosine (Ino), isopropanol (IsPrOH). Class 2 metabolites are in higher concentration in malignant tissues and have negative t.stat values. Both Class 1 and Class 2 metabolite and lipid species act as confounder for malignant and benign tissues separation and are represented by pink dot in t-test (Fig. 6a). Class 3 consists of non-influential or ambiguous metabolites mainly Car, Cr, Et, Glc, Ino and IsPrOH. These insignificant metabolites further classified into firstly, metabolically significant but equally abundant Glc and Car, secondly non-uniformly distributed Cr and finally trace metabolites like Et, Ino, IsoPrOH. Detailed values of Student’s t-test on 48 metabolites including t-stats and probabilities provided in Supplementary Table ST8.
a 2D Student’s t test plot of 48 metabolites quantified in 180 tissues of Model set (benign, n = 108 and malignant, n = 72) received from 43 patient suffering from OSCC. The 48 metabolites are: (1) 1,3-dihydroxyacetone; (2) 2-oxoglutarate; (3) 4-aminobutyrate; (4) acetate; (5) adenine; (6) alanine; (7) asparagine; (8) aspartate; (9) betaine; (10) carnitine; (11) choline; (12) creatine; (13) ethanol; (14) fumarate; (15) glucose; (16) glutamate; (17) glutamine; (18) glycine; (19) guanidoacetate; (20) histidine; (21) homocysteine; (22) inosine; (23) isoleucine; (24) isopropanol; (25) lactate; (26) leucine; (27) lysine; (28) methanol; (29) methionine; (30) o-acetylcarnitine; (31) o-phosphocholine; (32) phenylalanine; (33) serine; (34) taurine; (35) threonine; (36) tyrosine; (37) uracil; (38) valine; (39) myo-inositol; (40) sn-glycero-3-phosphocholine; (41) TCho; (42) SHi-PUFA; (43) linoleic acid; (44) MUFA; (45) SFA; (46) TG; (47) TFA; ( (48) FFA. b. VIP score plot upon PLS-DA on 48 metabolite concentration of Model tissues set. c. Representative pathway analysis plot from metabolite concentration of Model set tissues. d. Generalized pathway network locating identified metabolites of malignant tissues according to “An atlas of biochemistry and molecular biology” (Michal and Schomburg 2012)
Box–Whisker plots in Supplementary Figure SF28 shows overall reduction of lipid species in malignant tissues of model set. Triglycerides and PUFA concentration were highly reduced in malignant tissues. Overall reduction in total fatty acid were also observed in malignant tissues. The mean, median, SD, box parameters are presented in Supplementary Table ST9. Pie-doughnut chart in Supplementary Figure SF29a, b showed increase of FFA fraction from benign tissues (71%) to malignant tissues (92%). While TG-TFA correlation is very strong in benign tissues, it completely gets disappeared in malignant tissues of model set as shown in Supplementary Figure SF29c, d.
VIP score in Fig. 6b represents 15 major confounder metabolite in OSCC positive tissues among model set of 180 tissues from 43 patients. Lac, Glu, Gly, Ala and TCho were highly upregulated and only TFA was observed downregulated in malignant tissues. The pathway analysis predicted 33 metabolic pathways as shown in Fig. 6c. The pathways of glutathione, Arg-Proline, Gln-Glu metabolism have very high probability of occurrence in malignant tissues. While pathway frequencies for PUFA metabolisms (include linoleic and linolenic acid), Gly-Ser-Thr, Tau-hypotaurine and Ala-Asp-Glu metabolisms were severely altered in comparison to benign tissues. Noticeably pathways for glycolysis-gluconeogenesis and pyruvate metabolism were observed to have probabilities above 80 in −log(p) units. Further, Enrichment analysis demonstrates Warburg effect maximum probability (p value) as shown in Supplementary Figure SF30. The fatty acid metabolisms are least enriched. Supplementary Table ST10 represents identified metabolism of Fig. 6c. According to the atlas of biochemical pathways (Michal and Schomburg 2012), 42 metabolites were found to be connected into a single network with glucose as a major source of substrate for metabolism Fig. 6d.
Finally, results of all the above predictions as tabulated along with comparisons to histopathological status and details of 12 patients are presented in Supplementary Table ST11.
4 Discussion
The human body has more than 500 histologically distinct cell types which perform specific functions. Each of these cell types has a unique pattern of gene expression, proteome, and metabolome (Nicholson et al. 2012). Thus, a diverse metabolic phenotype among different somatic tissues especially in benign bed, margin and muscle tissues appeared in present study. During metabolic investigation, appearance of adipose nature among majority of benign margins (Paul et al. 2018), vascular nature of bed tissues (Marti 2005), and high creatine content in muscle tissues (Chen et al. 2008) confirmed the above proposition. The interaction of these metabolic phenotypes only occurred through the vascular and lymphatic system (Nicholson et al. 2012). In the absence of any external signal such as growth factor, normal cell does not proliferate autonomously. Thus, it able to conserve cellular resources to a larger extent (DeBerardinis et al. 2008). Several derangements in metabolic profile appeared in malignant tissues are likely associated with several cancer-related conditions. First of all, any altered metabolite concentration indicates an underlying mutation of metabolic enzyme thus affecting all the pathways relating to the metabolites. It decreases the flux of metabolites from major pathways which upregulates its compensating pathways in order to generate a support for anabolic metabolism. These change of flux through alternative pathways enhances macromolecular synthesis in proliferating tumor cell (Sullivan et al. 2016). Under inadequate vascularization, growing tumor accompanies the ischemic and hypoxic condition (Proskuryakov and Gabai 2010). This hypoxic environment associated with aggressive cancer causes necrotic cell death. Tata et al. detected the increased amount of oleic acid or MUFA and arachidonic acid in necrotic breast cancer tumor tissues using DESI-MS (Tata et al. 2016). Numerous studies reports association of tumors with networks of irregular and poorly functional capillaries generated from angiogenesis. These capillaries exhibit high permeability resulting in a flow pattern of fluid that results in concentration gradient at tumor microenvironment level (Shieh 2011). The visible changes in metabolic profile of malignant bed, margin, and muscle along with tumor tissues received from 53 OSCC patients in present study has been identified from spectral intensities of different metabolites and their concentrations measured from 1H HRMAS NMR spectra. The efficacy of identified biomarkers tested for predicting malignancy status of 64 blinded tissues obtained from 12 patients suffering from OSCC with neck invasion.
Out of 48 metabolites quantified in tumor and adjacent tissues of model set, Glc shows highest probability and have equal abundance in both malignant and benign tissues by box-whisker plot (De Winter 2013). Such behavior explained the ubiquitous nature of Glc that serve several metabolic requirements in human body as it acts as fuel in ATP generation. Under normal condition, it produces 36 ATP molecule via oxidative phosphorylation. Some tissues also utilizes excess of glucose in lipogenesis using acetyl-CoA from Krebs cycle (Zhang et al. 2019). Glycolysis also provides flux of glycerol necessary to support TG/FFA cycle (Reshef et al. 2003). Wigfield et al. reported a mechanisms of Lac upregulation under hypoxic conditions of HNSCC along with high rate of glycolysis (Wigfield et al. 2008). Falling of pyruvate flux would alters the formation of lipid deposits. These lipid deposits are further consumed by proliferating cells for synthesis of signaling lipid molecules and membrane phospholipids (Park et al. 2018). The mobile lipids sensitive NOESY spectra (Delikatny et al. 2011; Khanna et al. 2018) based OPLS-DA model on OSCC tissues predicted malignancy among blinded set of tissues at an accuracy of 93.75%. Although lipid is the major confounder, its low abundance in oral tissue along with mild changes in malignant tissues causes relatively low accuracy.
PCA of CPMG spectra displayed an additional upregulation of Lac, Cr, TCho, Glu, Gln, Gly and Ala in malignant tissue as in our previous study (Srivastava et al. 2011). Constructed OPLS-DA model has 91.19% predictive accuracy, which improved further to 93.75% accuracy and 100% of sensitivity upon addition of anomeric Glc and Ala spectral region. According to Felig, plasma amino acid maintained a well-defined concentrations in normal subject which have little intra- or inter-individual variation, except in muscle tissues which harbor 50% of total body pool of amino acids (Felig 1975). Present study showed maximum pooling of Cr in muscle tissues and thus appeared as an ambiguous metabolite in Student t-test. The ambiguous abundance of other metabolite, Glc, can be explained on the basis Warburg effect. The growing biosynthetic demands of proliferating cells are usually met by Warburg effect (Vander Heiden et al. 2009). It describes the upregulation of glycolytic metabolism over oxidative metabolism, on the basis of its capability to provide necessary equivalents of carbons and reducing cofactors required by proliferating cells (Folmes et al. 2011). For example, glycolysis plays a key role in regulation of choline metabolism (Valli et al. 2015). In addition to glycolysis, glutaminolysis also provides biosynthetic intermediate and energy support to proliferating cells. Their growing demand as well as accumulating Lac in tumor tissue induces angiogenesis (Polet and Feron 2013). Similarly, non-toxic Ala that has significant catabolic role are circulated via glucose-alanine cycle (Felig 1975). Taurine, another confounder metabolite, upregulated in tumor tissues maintains osmotic balance in hypoxic environment (Srivastava et al. 2010).
The quantification of tissue metabolites provides the opportunity to study the effect of malignancy upon them, and provide an independence from NMR parameters like number of protons, their splitting pattern, etc. The similarity in result of concentration-based multivariate analysis to that of CPMG spectra, proved the conservation of the data integrity during transformation of metabolite and lipid NMR spectral signals to their respective concentrations. However, sensitivity enhancement of metabolites concentration by orthogonal signal correction (Gavaghan et al. 2002) results in overfitting in Glc, MUFA and SFA which reduces the concentration-based OPLS-DA model’s predictive accuracy (Beckwith-Hall et al. 2002). Further there is an achievement of similar accuracy in malignancy prediction of blinded set of 64 tissues even after narrowing list of metabolites as variable selection especially with normal probability distribution has the highest accuracy in malignancy prediction of blinded 64 tissues. It proves a possibility of tuning the prediction model through tracking of all malignancy effected metabolites.
Quantitative metabolic profiling also enhanced in emerging the distinctive metabolic signatures of neck tissues especially in VIP score. Arterial metabolic profile majorly showed extensive amount of small molecular metabolites or nutrients, which experienced extensive perturbations as in profile of veins (Ivanisevic et al. 2015). In addition to massive amount of Lac in veins, significant amount of Gln, Tau, Cr and Glc suggests a high metabolic activities were occurring in neighboring tissues. VIP scores of nerves indicates a moderation in metabolic activities especially for Glc and Gly (Bhakoo et al. 1996). The metabolic profile of normal lymph nodes signifies the immunological properties of phospholipids (Olszewski 2019). The disturbance and shift of above metabolic pattern due to introduction of secondary lymph nodes accompanied with least amount of TFA confirms the presence of active cellular proliferation that have high fatty acid catabolism (Ubellacker and Morrison 2019).
Prognostic importance of glucose in cancer had long been reported. Further PET imaging agents mainly “18F-2-fluoro-2-deoxy-D-glucose” were in the phase of clinical trials for non-invasive tumor imaging (Penson 2009). Evaluation of tumor status could be performed on the visible metabolites in the network of clinical sugar cycle. Since intact tissue NMR spectra is intimate with in vivo tumor status, assignment of ex vivo and in vivo spectral signals provides precise information of biochemical status of malignancies (Srivastava et al. 2011). In the present work, 1H HRMAS NMR spectroscopy simultaneously detected lipids and metabolites content in tumor and adjacent tissues maintaining tissue integrity for further histopathological studies. The set of 48 metabolites were detected, in which 42 metabolites were correlated into a single network of metabolic pathways. These detection and quantification of tissue metabolites that identifies as substrates and products for underlying metabolic pathways allows to model any epigenetic alterations by selecting highly effected metabolites located in the pathways. Prognostic accuracy in identification of malignancies among blinded set of tissues also demonstrated decision making efficacy of predictive multivariate models. Further, the class separation obtained among malignant and benign tissues in NPP showed the fusibility of NMR spectral chemometrics to the results of mass spectrometry (Forshed et al. 2007). Thus outcome of the present work may provide support in the avenue of precision medicine (Beger et al. 2016), especially in phenome centres, where HRMAS NMR based clinical biomarker diagnostics have layered above real-time surgical diagnostics through nanospray Mass Spectrometry (Bonner et al. 2019) are being performed. Our results may strengthen REIMS-MS iKnife diagnostics with its metabotype data in building database and enable the machine learning to unbiased tumor specific pattern for precision excision during surgery (Ashizawa et al. 2017) in near future.
5 Conclusion
Present work demonstrated the application of 1H HRMAS NMR spectroscopy in quantitation of lipid and metabolite species in intact tissue specimens. It further showed the feasibility of spectral intensity-based as well as concentration-based metabolomics in prediction of malignancy among blinded tissues of patients suffering OSCC along with neck invasion. Thus, the present work may be helpful in machine learning based modelling OSCC biomarkers to plethora of metabolites distributed in tumor and neighboring tissues and hence serve as support for real-time surgical diagnostics in developing modern phenome centers for precise tumor excision.
Abbreviations
- OSCC:
-
Oral squamous cell carcinoma
- HRMAS:
-
High Resolution Magic Angle Spinning
- CPMG Spectra:
-
CPMG 1H HRMAS NMR Spectra
- NOESY Spectra:
-
NOESY 1H HRMAS NMR Spectra
- OPLS-DA:
-
Orthogonal Partial Least Square Discriminant Analysis
- PLS-DA:
-
Partial Least Square Discriminant Analysis
- QUANTAS:
-
QUANTification by Artificial Signal
- CustomCSI:
-
Custom Chemical Shift Index
- SHi-PUFA:
-
Sum of Higher Polyunsaturated Fatty Acids
- MUFA:
-
Monounsaturated Fatty Acid
- SFA:
-
Saturated Fatty Acids
- TG:
-
Triglycerides
- FFA:
-
Free Fatty Acids
References
Alexandri, E., Ahmed, R., Siddiqui, H., Choudhary, M. I., Tsiafoulis, C. G., & Gerothanassis, I. P. (2017). High resolution NMR spectroscopy as a structural and analytical tool for unsaturated lipids in solution. Molecules,22(10), 1663.
Ammann, L. P., Merritt, M., Sagalowsky, A., & Nurenberg, P. (2006). Peak-finding partial least squares for the analysis of 1H NMR spectra. Journal of Chemometrics: A Journal of the Chemometrics Society,20(6–7), 231–238.
Ashizawa, K., Yoshimura, K., Johno, H., Inoue, T., Katoh, R., Funayama, S., et al. (2017). Construction of mass spectra database and diagnosis algorithm for head and neck squamous cell carcinoma. Oral Oncology,75, 111–119.
Beckwith-Hall, B. M., Brindle, J. T., Barton, R. H., Coen, M., Holmes, E., Nicholson, J. K., et al. (2002). Application of orthogonal signal correction to minimise the effects of physical and biological variation in high resolution 1H NMR spectra of biofluids. Analyst,127(10), 1283–1288.
Beger, R. D., Dunn, W., Schmidt, M. A., Gross, S. S., Kirwan, J. A., Cascante, M., et al. (2016). Metabolomics enables precision medicine: “A white paper, community perspective”. Metabolomics,12(9), 149.
Beskeen, D. W., Cram, C. M., Duffy, J., Friedrichsen, L., & Reding, E. E. (2016). Illustrated Microsoft Office 365 and Office 2016: Introductory. Boston: Cengage Learning.
Bhakoo, K. K., Williams, I. T., Williams, S. R., Gadian, D. G., & Noble, M. D. (1996). Proton nuclear magnetic resonance spectroscopy of primary cells derived from nervous tissue. Journal of Neurochemistry,66(3), 1254–1263.
Bharti, S. K., & Roy, R. (2012). Quantitative 1H NMR spectroscopy. TrAC Trends in Analytical Chemistry,35, 5–26.
Bonner, F. W., Maslen, L., Lindon, J. C., Lewis, M. R., & Nicholson, J. K. (2019). Conception, implementation and operation of large-scale metabolic phenotyping centres: phenome centres. In: The handbook of metabolic phenotyping (pp. 385–405). Amsterdam: Elsevier.
Chang, D., Weljie, A., & Newton, J. (2007). Leveraging latent information in NMR spectra for robust predictive models. In Proceedings of the Pacific symposium on biocomputing 2007 (pp. 115–126). Hackensack, NJ: World Scientific.
Chen, J.-H., Wu, Y. V., DeCarolis, P., O'Connor, R., Somberg, C. J., & Singer, S. (2008). Resolution of creatine and phosphocreatine 1H signals in isolated human skeletal muscle using HR-MAS 1H NMR. Magnetic Resonance in Medicine,59(6), 1221–1224.
Chen, X., & Yu, D. (2019). Metabolomics study of oral cancers. Metabolomics,15(2), 22.
Chenomx, N. M. R. (2015). Suite. Edmonton, AB: Chenomx Inc.
Chong, J., Soufan, O., Li, C., Caraus, I., Li, S., Bourque, G., et al. (2018). MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Research,46(W1), W486–W494.
Chong, J., Wishart, D. S., & Xia, J. (2019). Using metaboanalyst 4.0 for comprehensive and integrative metabolomics data analysis. Current Protocols in Bioinformatics,68(1), e86.
Curtius, K., Wright, N. A., & Graham, T. A. (2018). An evolutionary perspective on field cancerization. Nature Reviews Cancer,18(1), 19.
De Winter, J. C. F. (2013). Using the Student's t-test with extremely small sample sizes. Practical Assessment, Research & Evaluation,18(10), 1–12.
DeBerardinis, R. J., Lum, J. J., Hatzivassiliou, G., & Thompson, C. B. (2008). The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metabolism,7(1), 11–20.
Delikatny, E. J., Chawla, S., Leung, D. J., & Poptani, H. (2011). MR-visible lipids and the tumor microenvironment. NMR in Biomedicine,24(6), 592–611.
Farrant, R. D., Hollerton, J. C., Lynn, S. M., Provera, S., Sidebottom, P. J., & Upton, R. J. (2010). NMR quantification using an artificial signal. Magnetic Resonance in Chemistry,48(10), 753–762.
Felig, P. (1975). Amino acid metabolism in man. Annual Review of Biochemistry,44(1), 933–955.
Fernández, A., Córdova, P., Badenier, O., & Esguep, A. (2015). Epidemiological characterization of oral cancer. Literature review. Journal of Oral Research,4(2), 137–145.
Folmes, C. D. L., Nelson, T. J., & Terzic, A. (2011). Energy metabolism in nuclear reprogramming. Biomarkers in Medicine,5(6), 715–729.
Forshed, J., Idborg, H., & Jacobsson, S. P. (2007). Evaluation of different techniques for data fusion of LC/MS and 1H-NMR. Chemometrics and Intelligent Laboratory Systems,85(1), 102–109.
Gavaghan, C. L., Wilson, I. D., & Nicholson, J. K. (2002). Physiological variation in metabolic phenotyping and functional genomic studies: use of orthogonal signal correction and PLS-DA. FEBS Letters,530(1–3), 191–196.
Hong, Y. S., Coen, M., Rhode, C. M., Reily, M. D., Robertson, D. G., Holmes, E., et al. (2009). Chemical shift calibration of 1H MAS NMR liver tissue spectra exemplified using a study of glycine protection of galactosamine toxicity. Magnetic Resonance in Chemistry,47(S1), S47–S53.
Ivanisevic, J., Elias, D., Deguchi, H., Averell, P. M., Kurczy, M., Johnson, C. H., et al. (2015). Arteriovenous blood metabolomics: A readout of intra-tissue metabostasis. Scientific Reports,5, 12757.
Jafari, A., Najafi, S. H., Moradi, F., Kharazifard, M. J., & Khami, M. R. (2013). Delay in the diagnosis and treatment of oral cancer. Journal of Dentistry,14(3), 146.
Jané-Salas, E., López-López, J., Roselló-Llabrés, X., Rodríguez-Argueta, O.-F., & Chimenos-Küstner, E. (2012). Relationship between oral cancer and implants: Clinical cases and systematic literature review. Medicina Oral, Patologia Oral y Cirugia Bucal,17(1), e23.
Jiménez, B., & MacIntyre, D. (2017). NMR metabolic phenotyping in clinical studies. In J. C. Lindon, G. E. Tranter, & D. W. Koppenaal (Eds.), Encyclopedia of spectroscopy and spectrometry (3rd ed., pp. 140–145). Oxford: Academic Press.
Khanna, R., Kumar, K., & Roy, R. (2018). A case study of primary malignancy of buccal mucosa using proton HR-MAS NMR spectroscopy on tissue specimens. Journal of Oral Biology and Craniofacial Research,8(1), 68–72.
Kinross, J. M. (2019). Metabolic phenotyping in clinical practice. In The handbook of metabolic phenotyping (pp. 461–489). Amsterdam: Elsevier.
Long, G. L., & Winefordner, J. D. (1983). Limit of detection. A closer look at the IUPAC definition. Analytical Chemistry,55(7), 712A–724A.
Martens, H., & Martens, M. (2000). Modified Jack-knife estimation of parameter uncertainty in bilinear modelling by partial least squares regression (PLSR). Food Quality and Preference,11(1–2), 5–16.
Marti, H. H. (2005). Angiogenesis—a self-adapting principle in hypoxia. In M. Clauss & G. Breier (Eds.), Mechanisms of angiogenesis (pp. 163–180). Basel: Birkhäuser.
Michal, G., & Schomburg, D. (2012). Biochemical pathways: an atlas of biochemistry and molecular biology. New York: Wiley.
Mikkelsen, S. R., & Cortón, E. (2016). Bioanalytical chemistry. Hoboken: Wiley.
Moazzami, A. A., Frank, S., Gombert, A., Sus, N., Bayram, B., Rimbach, G., et al. (2015). Non-targeted 1 H-NMR-metabolomics suggest the induction of master regulators of energy metabolism in the liver of vitamin E-deficient rats. Food & Function,6(4), 1090–1097.
Nicholson, J. K., Holmes, E., Kinross, J. M., Darzi, A. W., Takats, Z., & Lindon, J. C. (2012). Metabolic phenotyping in clinical and surgical environments. Nature,491, 384.
Olszewski, W. L. (2019). Peripheral lymph: formation and immune function. Boca Raton: CRC Press.
Park, J. K., Coffey, N. J., Limoges, A., & Le, A. (2018). The heterogeneity of lipid metabolism in cancer. In A. Le (Ed.), The heterogeneity of cancer metabolism (pp. 33–55). Cham: Springer.
Paul, A., Kumar, S., Raj, A., Sonkar, A. A., Jain, S., Singhai, A., et al. (2018). Alteration in lipid composition differentiates breast cancer tissues: A 1H HRMAS NMR metabolomic study. Metabolomics,14(9), 119.
Penson, R. T. (2009). Sugar fuels cancer. Cancer,115(5), 918–921.
Polet, F., & Feron, O. (2013). Endothelial cell metabolism and tumour angiogenesis: Glucose and glutamine as essential fuels and lactate as the driving force. Journal of Internal Medicine,273(2), 156–165.
Proskuryakov, S. Y., & Gabai, V. L. (2010). Mechanisms of tumor cell necrosis. Current Pharmaceutical Design,16(1), 56–68.
Rajeev, R., Alexander, B., & Karthikeyan, S. (2015). Diagnostic methods for early detection of oral cancer: An overview. International Journal of Current Innovation Research,1(8), 197–200.
Reshef, L., Olswang, Y., Cassuto, H., Blum, B., Croniger, C. M., Kalhan, S. C., et al. (2003). Glyceroneogenesis and the triglyceride/fatty acid cycle. Journal of Biological Chemistry,278(33), 30413–30416.
Rizwan, A., & Glunde, K. (2014). Imaging of tumor metabolism: MR spectroscopy. In Functional imaging in oncology (pp. 147–180). Berlin: Springer.
Rubingh, C. M., Bijlsma, S., Derks, E. P. P. A., Bobeldijk, I., Verheij, E. R., Kochhar, S., et al. (2006). Assessing the performance of statistical validation tools for megavariate metabolomics data. Metabolomics,2(2), 53–61.
Seetharam, S. S., & Ramachandran, C. R. (1998). Fine needle aspiration cytology as a diagnostic test for oral squamous cell carcinoma. Oral Diseases,4(3), 180–186.
Shieh, A. C. (2011). Biomechanical forces shape the tumor microenvironment. Annals of Biomedical Engineering,39(5), 1379–1389.
Shield, K. D., Ferlay, J., Jemal, A., Sankaranarayanan, R., Chaturvedi, A. K., Bray, F., et al. (2017). The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012. CA: A Cancer Journal for Clinicians,67(1), 51–64.
Slaughter, D. P., Southwick, H. W., & Smejkal, W. (1953). “Field cancerization” in oral stratified squamous epithelium. Clinical implications of multicentric origin. Cancer,6(5), 963–968.
Sreeshyla, H., Shashidar, R., & Sudhee, U. (2014). Diagnostic aids in oral precancer and cancer. Indian Journal of Multidisciplinary Dentistry,4(2), 928–934.
Srivastava, S., Roy, R., Gupta, V., Tiwari, A., Srivastava, A. N., & Sonkar, A. A. (2011). Proton HR-MAS MR spectroscopy of oral squamous cell carcinoma tissues: An ex vivo study to identify malignancy induced metabolic fingerprints. Metabolomics,7(2), 278–288.
Srivastava, S., Roy, R., Singh, S., Kumar, P., Dalela, D., Sankhwar, S. N., et al. (2010). Taurine—a possible fingerprint biomarker in non-muscle invasive bladder cancer: A pilot study by 1H NMR spectroscopy. Cancer Biomarkers,6(1), 11–20.
Sullivan, L. B., Gui, D. Y., & Vander Heiden, M. G. (2016). Altered metabolite levels in cancer: implications for tumour biology and cancer therapy. Nature Reviews Cancer,16(11), 680.
Szymańska, E., Saccenti, E., Smilde, A. K., & Westerhuis, J. A. (2012). Double-check: validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics,8(1), 3–16.
Tata, A., Woolman, M., Ventura, M., Bernards, N., Ganguly, M., Gribble, A., et al. (2016). Rapid detection of necrosis in breast cancer with desorption electrospray ionization mass spectrometry. Scientific Reports,6, 35374.
Ubellacker, J. M., & Morrison, S. J. (2019). Metabolic adaptation fuels lymph node metastasis. Cell Metabolism,29(4), 785–786.
Valli, A., Rodriguez, M., Moutsianas, L., Fischer, R., Fedele, V., Huang, H.-L., et al. (2015). Hypoxia induces a lipogenic cancer cell phenotype via HIF1α-dependent and-independent pathways. Oncotarget,6(4), 1920.
Van, Q. N. (2013). Current NMR strategies for biomarker discovery. In Proteomic and metabolomic approaches to biomarker discovery (pp. 87–117). Amsterdam: Elsevier.
Vander Heiden, M. G., Cantley, L. C., & Thompson, C. B. (2009). Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science,324(5930), 1029–1033.
Vitols, C., & Mercier, P. (2006). Correcting lineshapes in NMR spectra. Edmonton: CHENOMX.
Wigfield, S. M., Winter, S. C., Giatromanolaki, A., Taylor, J., Koukourakis, M. L., & Harris, A. L. (2008). PDK-1 regulates lactate production in hypoxia and is associated with poor prognosis in head and neck squamous cancer. British Journal of Cancer,98(12), 1975.
Wishart, D. S. (2008). Quantitative metabolomics using NMR. TrAC Trends in Analytical Chemistry,27(3), 228–237.
Zhang, Q., Zhao, G., Yang, N., & Zhang, L. (2019). Fasting blood glucose levels in patients with different types of diseases. Progress in Molecular Biology and Translational Science,162, 277–292.
Zhu, Y., Chen, S., Chen, C., & Chen, L. (2017). Partial least squares (PLS) methods for abnormal detection of breast Cells. In International conference of pioneering computer scientists, engineers and educators. Berlin: Springer.
Acknowledgements
The authors are thankful to Division of SAIF, CSIR-Central Drug Research Institute, Lucknow where the 1H HRMAS NMR measurement were conducted. Mr. Anup Paul would like to thank UGC (SRF Award No. 18-12/2011(ii)EU-V) for financial assistance.
Author information
Authors and Affiliations
Contributions
AAS, AA, KG and RR designed the study. AA, KG and AAS performed the surgery and sampled the tissue specimens. SS and RR conducted the experiments. AP, and RR processed and analyzed the spectral data. NH performed the histopathology. Initial draft written by AP. AAS and RR edited and revised the paper. Project administration of the study have carried under AAS, SJ and RR. All authors carefully read and agree to be accountable for all aspects of the work.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no potential conflict of interest. The disclosure of potential conflict of interest in the prescribed format has been obtained from all the author.
Ethical approval
The study was ethically approved and the work was performed in strict accordance with the guidelines of Institutional Ethical Committee of King George’s Medical University (KGMU) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The subjects were explained the study procedure and written and informed consent were obtained from them prior to the study. The authors: Anup Paul, Shatakshi Srivastava, Raja Roy, Akshay Anand, Kushagra Gaurav, Nuzzat Hussain, Sudha Jain and Abhinav A. Sonkar are aware of the ethical policy.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Paul, A., Srivastava, S., Roy, R. et al. Malignancy prediction among tissues from Oral SCC patients including neck invasions: a 1H HRMAS NMR based metabolomic study. Metabolomics 16, 38 (2020). https://doi.org/10.1007/s11306-020-01660-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-020-01660-8