Abstract
The reliable quantification of genetically modified organisms (GMOs) by real-time PCR requires, besides thoroughly validated quantitative detection methods, sustainable calibration systems. The latter establishes the anchor points for the measured value and the measurement unit, respectively. In this paper, the suitability of two types of DNA calibrants, i.e. plasmid DNA and genomic DNA extracted from plant leaves, for the certification of the GMO content in reference materials as copy number ratio between two targeted DNA sequences was investigated. The PCR efficiencies and coefficients of determination of the calibration curves as well as the measured copy number ratios for three powder certified reference materials (CRMs), namely ERM-BF415e (NK603 maize), ERM-BF425c (356043 soya), and ERM-BF427c (98140 maize), originally certified for their mass fraction of GMO, were compared for both types of calibrants. In all three systems investigated, the PCR efficiencies of plasmid DNA were slightly closer to the PCR efficiencies observed for the genomic DNA extracted from seed powders rather than those of the genomic DNA extracted from leaves. Although the mean DNA copy number ratios for each CRM overlapped within their uncertainties, the DNA copy number ratios were significantly different using the two types of calibrants. Based on these observations, both plasmid and leaf genomic DNA calibrants would be technically suitable as anchor points for the calibration of the real-time PCR methods applied in this study. However, the most suitable approach to establish a sustainable traceability chain is to fix a reference system based on plasmid DNA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Legislation in the European Union demands the labelling of food and feed products consisting of, or containing “more than 0.9 % genetically modified organisms” (GMOs), provided the GMO has been placed on the market in accordance with the Community legislation [1]. This enforces the necessity to develop and validate reliable quantification methods, as well as the need to produce certified reference materials (CRMs) to calibrate and control the application of these methods. In October 2004, the European Commission recommended to express the content of GM material in food and feed as the percentage of GM haploid genomes in relation to the total of haploid genomes in a DNA mixture obtained for a certain plant species [2]; this percentage is referred to as DNA copy number ratio.
Quantitative real-time PCR targeting specific DNA fragments is currently the method of choice for GMO identification and quantification. In particular, event-specific PCR, which targets the unique junction sequence between the host genomic DNA (gDNA) and the introduced DNA, is being widely used for determining the relative DNA amount in GM foods. This requires calibration curves based on calibrants with a precisely known content of the measured targets. The choice of calibrants is critical and determines the measurement unit in which the measurement results are expressed. Due to the limited availability of dedicated GMO calibrants, gDNA extracted from powder CRMs produced from GM seeds or extracted from other GM samples such as seeds or leaves are commonly used as calibrants. A critical issue connected to the calibrants derived from seed powder is that the application of the copy number-based GMO unit may be biased (for instance for maize, due to the different ratios of the maternal and paternal genome in the various seed tissues). It is difficult to check the genetic purity of a powder produced from seeds, as the genetic composition of all seeds used for the production cannot be controlled. Alternatively, it is possible to extract gDNA from leaves of verified GM plants. Calibrants may then be produced from genetically pure DNA extracted from leaves of single plant individuals, which are consistently either homozygous or heterozygous for the GM event (ratio of GM to reference of 2:2 and 1:2, respectively). For each plant individual, this ratio can be determined directly by PCR prior to a production of the reference material. However, even in this case, the genetic composition of a GMO plant is not known with absolute certainty and depends on the breeding processes applied. Plasmid DNA (pDNA) could be an alternative to gDNA. Plasmids have several advantages over extracted gDNA, such as the well-defined and fully characterised sequence, stability over long periods of time when frozen, straightforward manipulation in routine analysis, allowing also for a well-controlled and repeatable production. However, it has to be demonstrated that using pDNA as opposed to gDNA for quantitative PCR calibration does not introduce a measurement bias in such a way that it would be impossible to set a sound reference system.
The use of plasmids which contain cloned junction-specific and endogenous sequences—so-called dual-target plasmids—as calibrants for GM quantification has been demonstrated previously [3–11]. However, the validity of the conclusions drawn from these studies is limited, as they have been based on repeatability studies within the same laboratory. Reproducibility across different laboratories has been recently addressed in several studies by specifically designed interlaboratory comparisons for quantifying a particular GM event. Within these studies, the behaviour of pDNA and gDNA calibrants has also been compared; however, this has been largely based on measurement data provided by a very limited number of participating laboratories. A prerequisite for evaluating the suitability of a calibrant is a sufficiently large number of measurement data to allow for testing the statistical significance of various influencing parameters. Therefore, a first systematic study comprising a large set of data was conducted by the Institute for Reference Materials and Measurements (IRMM), and a detailed strategy for the certification of a MON810 CRM with respect to its DNA copy number ratio, using a pDNA calibrant, was published [12]. A suitability pre-study had been performed in the frame of this certification project. It showed that there was no significant difference between the PCR efficiencies of pDNA- and gDNA-derived calibration curves when the MON810-specific PCR method was applied. As no generalisation could be done and the selection of the suitable type of DNA calibrant (i.e. pDNA or gDNA) still remained an issue, further studies had to be carried out for CRMs intended for copy number certification and the associated event-specific real-time PCR method.
In the current study, the analytical behaviour of DNA calibrants for GM quantification has been further evaluated based on data obtained during interlaboratory comparisons. Three independent parallel comparisons, designed and coordinated by IRMM, aimed to investigate the possible impact of the type of DNA calibrant on the determination of the copy number ratio for various GM events [13–15]. It was intended to certify the following seed powder CRMs, already certified for their GM mass fractions [16–18], additionally for their copy number ratio using a suitable calibration system: ERM-BF415e (NK603 maize), ERM-BF425c (356043 soya), and ERM-BF427c (98140 maize). During the described interlaboratory comparisons, these materials were used as unknown samples. For each GM event, a set of samples consisting of two calibrants, namely gDNA extracted from plant leaves (gDNAl) and a multi-target pDNA with a known number of cloned DNA fragments per plasmid, i.e. ERM-AD415 [19], ERM-AD425 [20], and ERM-AD427 [21], was used to calibrate the event-specific real-time PCR method. The properties of the pDNA and gDNAl calibrants were statistically evaluated and compared with respect to PCR performance (e.g. efficiency and coefficient of determination), equivalence to the seed powder CRMs resembling food/feed samples, as well as to the measured DNA copy number ratio for each CRM, on a case by case basis. The results enabled the certification of the three powder CRMs for their copy number ratios and allowed defining a general strategy for the development and certification of new GMO CRMs.
Materials and methods
Experimental details
ERM-BF415e, ERM-BF425c, and ERM-BF427c are powder CRMs produced and certified by IRMM for their GM mass fraction, and containing (19.6 ± 0.9), (10.0 ± 1.1), and (20.0 ± 0.8) g/kg GM, respectively (expanded uncertainty with a coverage factor of 2). These GM powders are gravimetrically prepared mixtures of NK603 maize, 356043 soya, and 98140 maize, respectively, and corresponding non-GM seed powders with verified purity, similar particle size distribution, and corrected for their water content [16–18]. Three interlaboratory comparisons were conducted to certify the existing CRMs additionally for their DNA copy number ratios [13–15]. In order to verify the influence of the DNA extraction procedure on the measurement results, three different extraction methods were applied to each CRM: (1) cetyltrimethylammonium bromide (CTAB) method, followed by a Genomic tip-20/G purification step, and GENESpin kit (ERM-BF415e); (2) DNAExtractor kit with a Genomic-tip 20/G purification step, CTAB method combined with a Genomic tip-20/G purification step, and GENESpin kit (ERM-BF425c); (3) CTAB method, DNeasy plant mini kit, and GENESpin kit (ERM-BF427c). Event-specific real-time PCR methods validated by the European Union Reference Laboratory for GM Food and Feed (EURL-GMFF), using primer pairs and labelled TaqMan® probes specific for the GM event and plant taxon-specific endogeneous reference gene (Table 1), were applied as simplex assays. Based on the findings reported by Broothaerts et al. [22], the quantification method of the maize NK603 event described here targets a 79-bp fragment from the endogenous maize hmg gene instead of the 70 bp fragment of the maize endogenous adh1 gene used in the method validated by the EURL-GMFF. A similar real-time PCR assay targeting the same maize-specific hmg gene was used in the EURL-GMFF method for the quantification of the maize 98140 event. For the relative quantification of the soya 356043 event, a soya-specific real-time PCR method amplifies a 74-bp fragment of the le1 reference target sequence. pDNA and gDNA were used as calibrants for all real-time PCR measurements. The gDNA calibrant was extracted from leaves of hybrid NK603 maize, hybrid 98140 maize and hybrid 356043 soya plants, respectively. Each plant was individually tested for the presence of the specific transgene. Three pDNA calibrants developed by IRMM, namely ERM-AD415, ERM-AD425, and ERM-AD427, each of them consisting of a multi-target plasmid carrying the targeted junction-specific sequence and the taxon-specific sequence, were used specifically for the three GM events as calibrants.
Each study contained three method combinations of extraction and real-time PCR methods, measured by at least 12 different laboratories. Laboratories worldwide specialised in the field of GM detection were selected on the basis of experience and quality management system in place as follows: 21 laboratories were envisaged for each of the copy number certification studies dealing with the NK603 and 98140 maize events, whereas for the 356043 soya study, 32 laboratories were appointed. For the 98140 maize study, a total of 36 individual analyses were requested, which resulted in 144 real-time PCR experiments; for each of the NK603 maize and 356043 soya studies, a total of 42 analyses were carried out, and results from 168 real-time PCR experiments were collected and evaluated (Table 2).
For each independent analysis, two bottles from each powder material, i.e. ERM-BF415e, ERM-BF425c, and ERM-BF427c, were provided. From each bottle, three samples were extracted and analysed (n = 3), further referred to as “unknowns” (U1-U3 for bottle 1, and U4-U6 for bottle 2). In order to evaluate the intermediate precision from each laboratory, the extraction of DNA from the seed powder provided with the two bottles, as well as the real-time PCR measurements, was performed on two different days (further referred to as day 1 and 2). The DNA concentration was estimated by measuring the absorption at 260 nm and/or by PicoGreen® dsDNA Assay Kit (Molecular Probes® Invitrogen, Carlsbad, CA). The total DNA concentration of each extract was adjusted to 20 ng/μL with nuclease-free water; these “undiluted extracts” were d-fold diluted to cover a total of 4–5 concentration levels (the dilution factor d was in-house tested and differently chosen for each study: 1.5, 3.0, 4.0, and 6.0 (NK603 maize), 3.0, 6.0, 9.0, and 12.0 (356043 soya), and 1.5, 3.0, and 4.0 (98140 maize), respectively). The dilution series were analysed in triplicate by event-specific real-time PCR, using primer pairs and labelled TaqMan® probes specific for the GM event and reference gene (Table 1), and pDNA and gDNAl as calibrants.
Reagents, kits, and consumables
For each copy number certification study, the following materials were sent to the participating laboratories: pDNA calibrant CRM (1 tube), gDNAl calibrant (1 tube), matrix CRMs used as unknowns (2 bottles), transgene primers and probe (3 tubes), and endogene primers and probe (3 tubes). For the soya 356043 interlaboratory comparison, the participating laboratories were also provided with the DNAExtractor kit (Eurofins GeneScan GmbH, Freiburg, DE) with Genomic-tip 20/G (Qiagen, Benelux B.V., Venlo, NL), and the GeneSpin kit (Eurofins GeneScan GmbH, Freiburg, DE); Universal PCR Master Mix (Applied Biosystems, Foster City, CA), nuclease-free water (Promega Benelux), and plasmid dilution buffer (1.0 mM Tris, 0.01 mM EDTA, pH 8.0, containing 2 ng/μL ColE1 background plasmid DNA) were included as well. All DNA solutions, kits, and consumables, as well as the unknowns, were shipped to the participating laboratories on dry ice, ensuring that the temperature did not exceed 0 °C. The participating laboratories had to prepare buffers for the CTAB DNA extraction method, perform DNA extraction (including DNA quantification) and real-time PCR, and provide raw data to the IRMM.
Preparation of pDNA calibrants
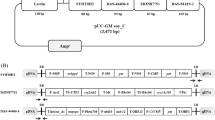
The plasmid DNA calibrants ERM-AD415, ERM-AD425, and ERM-AD427 were prepared by IRMM. Each plasmid DNA CRM is related to a specific GM event (Table 2) and contains a pUC18-derived multi-target plasmid into which two PCR target sequences were inserted. One of the two targets consists of a DNA fragment specific for the genetic modification of interest (i.e. maize NK603, soya 356043, and maize 98140); the other target sequence is a defined Zea mays or Glycine max taxon-specific DNA fragment. Certified are the numbers of cloned DNA fragments for the two PCR targets per plasmid [19–21]. The CRMs are available in tubes containing 500 μL of the plasmid DNA in 1.0 mM Tris, 0.01 mM EDTA, pH 8.0 buffer and 1 ng/μL ColE1 background plasmid DNA; each tube contains approximately 109 copies of the multi-target plasmid DNA, which corresponds approximately to 3.3 ng DNA per tube for ERM-AD425 and ERM-AD427, and 4.1 ng DNA per tube for ERM-AD415, respectively.
Preparation of gDNAl calibrants
Genomic DNA was extracted from plant leaves from heterozygous NK603 and 98140 maize, and homozygous 356043 soya, respectively. Prior to the large-scale extraction of gDNA from leaf tissues, the GM positive status of each individual plant was confirmed by real-time PCR. To do that, gDNA was extracted from leaves collected separately from each plant, by using the following protocols: (1) rapid DNA extraction method [26] (maize) and (2) DNeasy Plant Mini Kit (Qiagen, Benelux B.V., Venlo, NL).
The large-scale gDNA extraction from 1 g of verified GM leaves was performed using the protocol for automated DNA extraction and the Chemagic DNA Plant Kit (Chemagen Biopolymer-Technologie AG, Baesweiler, DE), and applying the manufacturer’s instructions for a 96-needles head. A volume of 400 μL of gDNA solution was provided to the participating laboratories to be used for preparing the gDNAl calibration curves in the real-time PCR experiments (for the content in copy number of the targets, see the “Real-time PCR” section).
Total DNA quantification was performed at IRMM on a FluoStar Galaxy reader (BMG Labtech GmbH, Offenburg, DE) using the PicoGreen® dsDNA Assay Kit (Molecular Probes® Invitrogen, Carlsbad, CA), or on a Nanodrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) for gDNA and pDNA, respectively.
DNA extraction from CRM powders
Genomic DNA was extracted from seed powder CRMs (Table 2), according to the published extraction methods and specifications included in the commercially available kits. A chemical extraction method, namely the CTAB protocol adopted from ISO 21571:2005, was applied in all three studies to extract gDNA, starting from 100 mg maize and 200 mg soya powder, respectively. Genomic DNA isolated by CTAB extraction from ERM-BF415e was subjected to further purification using a Genomic-tip 20/G column kit (Qiagen, Benelux B.V., Venlo, NL) according to the manufacturer’s instructions. This additional purification step was also applied to ERM-BF425c in combination with the DNAExtractor kit protocol (Eurofins GeneScan GmbH, Freiburg, DE), starting from 200 mg soya seed powder. In addition, another extraction was carried out for both NK603 and 98140 maize seed powders, using the GENESpin DNA kit (GeneScan Analytics GmbH, Freiburg, DE) and a sample intake of 200 mg. A further extraction with the DNeasy plant mini kit (Qiagen, Benelux B.V., Venlo, NL) was only applied to ERM-BF427c, using a 100-mg sample intake. Depending on the extraction protocol, the DNA pellets were finally dissolved in nuclease-free water, whereas the membrane-purified DNA was eluted in Tris-based elution buffer. All DNA samples were diluted to 20 ng/μL initial concentrations, which have been also estimated as copies/μL, by considering the average genome size of the respective species. For the conversion of DNA concentrations from ng/μL into copies/μL related to the endogeneous target sequence, the following parameters were taken into account: the equivalent mass for a given number of base pairs (bp) [27], event-specific zygosity and size of the haploid plant genome. The average sizes of 2.43 × 109 and 1.12 × 109 bp were assumed for the haploid maize and soya genomes, respectively [27]. For the transgenic target, the parental origin of the donor of the GM trait (only for maize) [28] as well as the certified GM mass fraction expressing the GM content were additionally included. These calculations resulted in the following estimated initial DNA concentrations for the unknown powder sample:
-
1)
Endogeneous target: 8 copies/μL (ERM-BF415e and ERM-BF427c) and 18 copies/μL (ERM-BF425c), respectively;
-
2)
Transgenic target: 97 copies/μL (ERM-BF415e and ERM-BF427c) and 176 copies/μL (ERM-BF425c), respectively.
Real-time PCR
The DNA copy number ratios of the matrix CRMs were determined by event-specific real-time PCR, according to the EURL-GMFF validated methods and previously described protocols [13–15]. Details about primer and probe sequences and amplicon sizes are given in Table 1. Simplex reactions using the TaqMan Universal PCR Master Mix were performed by the participating laboratories according to the manufacturer’s instructions (Applied Biosystems), leaving the choice of the instrument detection system to the user. The real-time PCR experiments were carried out in 96-well microtiter plates, in a total volume of 25 μL (NK603 maize and 356043 soya) and 50 μL (98140 maize), with all runs performed for 45 cycles; the baseline and threshold of the individual real-time PCR measurements were set automatically. For the NK603 maize and 356043 soya, the set-up was adapted for 384-well plates for one laboratory.
To evaluate the performance of using pDNA compared to gDNAl as calibrants, the DNA copy numbers of the specific transgene and endogene fragments for each matrix CRM were estimated. To do that, calibration curves were set up separately for the target genes, based on serially diluted calibrants. High- and low-range dilution series were prepared by the participating laboratories from pDNA in TE buffer (1.0 mM Tris, 0.01 mM EDTA, pH 8.0); only in the case of soya, the plasmid dilution buffer was supplemented with ColE1 background pDNA (2 ng/μL). The gDNAl calibrant was provided to the laboratories at concentrations of approximately 1.6 × 104 cp/μL (maize) and 105 cp/μL (soya), respectively; the working stock was then diluted in nuclease-free water. The endogeneous calibration curves for maize DNA copy number quantification were made of five dilution points, with concentration levels ranging from 2 × 102 to 105 copies/μL (pDNA calibrant) and 2 × 102 to 1.6 × 104 copies/μL (gDNAl calibrant); for quantifying the GM maize DNA, the calibration curves were also based on five concentrations from 10 to 104 copies/μL (pDNA calibrant) and 10 to 2 × 103 copies/μL (gDNAl calibrant). In the case of the GM soya event, serially diluted pDNA and gDNAl were used to construct target-specific, five points-based calibration curves in a similar way, with concentrations ranging from 2 × 102 to 2 × 105 copies/μL (pDNA calibrant) and 2 × 102 to 105 copies/μL (gDNAl calibrant) for the endogene, and concentrations ranging from 5 to 104 copies/μL (pDNA calibrant) and 5 to 2 × 103 (gDNAl calibrant) for the transgene. All dilutions points together with a non-template control (NTC) were analysed in triplicate (maize) or quadruplicate (soya) on the real-time PCR plate.
One set of analysis generated, therefore, two data sets, i.e. one from calibration with pDNA and one from calibration with gDNAl. Each of these sets is comprised of a total of eight calibration curves consisting of the detection of the endogeneous and transgenic targets, spread over 2 days (i.e. one series of endogeneous and transgenic calibrations per day). The overall set-up resulted in a total of four target-specific real-time PCR plates; for each plate, two parallel calibrations using either pDNA or gDNAl were used.
Acceptance criteria for data sets
Data acceptance criteria for the current studies had been established based on a previously published approach [29]. Acceptance criteria were set to ensure that only reliable data sets are statistically analysed, knowing that this results in the exclusion of many data sets. The description of a sequential analysis-based algorithm that allows for the straightforward selection of sets of analysis derived from interlaboratory comparisons according to pre-defined criteria as outlined in the related certification reports [13–15]. These criteria are mainly focused on the calibrants and their fitness for purpose, and include the calibration performance and working interval. Technical criteria, such as the NTC, dilution of the unknown powder samples, and Ct variability among replicate measurements were also considered. Besides this, the consistency between unknown dilutions within a dilution series as well as between the independent DNA extracts per day was addressed with respect to the measured DNA copy number ratio.
A maximum relative standard deviation (RSD) of 25 % (within the limits of variation for real-time PCR detection methods) [30] was set to reflect acceptable repeatability conditions for one-day analysis, as well as reproducibility conditions for a complete set of analysis. Sets of analysis that did not fulfil all acceptance criteria were excluded. The results obtained on the two days of analysis were considered separately for the two calibrants. This means that the rejection of a set of analysis calibrated with pDNA did not trigger the exclusion of the set of analysis that was calibrated with gDNAl and vice versa. An overview of all rejections per set of analysis for all three certification studies is given in Table 3.
With respect to the technical acceptance criteria, only sets of analysis for which no signal was detected for NTCs were accepted. Sets of analysis were rejected if any inconsistency in the dilution series was noticeable as an abnormal trend of reported threshold cycles (Ct; a higher dilution factor was expected to result in increasing Ct). This criterion was applied to all types of DNA samples, i.e. calibrants and seed powder CRMs used as unknowns. Furthermore, mean values of replicate measurements were not accepted if the Ct difference between one replicate and the mean Ct of the remaining replicates within the same dilution point was higher than 1.5. Dilution points were also excluded if the amplification signal was absent. This alone did not trigger the rejection of the complete set of analysis, unless the remaining number of acceptable dilution points within the dilution series was insufficient.
The calibration performance was assessed by different parameters, according to the minimal performance requirements set by the method validation guidelines of the EURL-GMFF [30]. In particular, these parameters included the linearity and slope of the calibration curves. Firstly, only calibration curves with a coefficient of determination (R 2) of at least 0.98 were accepted [16]. Secondly, control limits for PCR efficiency (ε) were applied. For both calibrants, transgene and endogene mean PCR efficiencies and their standard deviations were calculated [29]. Control limits were calculated per calibrant and targeted gene as the mean plus or minus one standard deviation. The resulting highest and lowest control limits applied in the three studies and their corresponding slopes are listed in Table 4. These values were consistent with the performance of the respective real-time PCR methods used for GM quantification observed within in-house performed studies and from results obtained by collaborations with external laboratories.
The unknown powder samples were analysed undiluted and d-fold diluted. DNA extracts and dilutions exhibiting a Ct value outside the working interval of the calibration curve were excluded for further calculations. Using the two calibrants, the mean Ct values for the undiluted DNA extracts and their dilutions for each unknown sample were converted into transgene and endogene copy numbers of which copy number ratios were calculated; only the copy number ratios of the undiluted extracts were used to calculate the certified value. However, the copy number ratio of an undiluted extract was only considered for calculation if at least one more copy number ratio from its corresponding dilution series could be calculated. Moreover, for a set of analysis to be accepted, a minimum of two “unknowns” per day needed to fulfil this criterion, i.e. a maximum of six copy number ratios and a minimum of four copy number ratios per data set and calibrant were accepted.
Statistical analyses
To evaluate the analytical behaviour of gDNAl and gDNA, the properties of the calibration curves obtained with each type of calibrant were investigated. In the first step, the PCR efficiency for the target-specific PCR method and the linearity of the corresponding regression lines calculated on the basis of serial dilutions of the pDNA and gDNAl calibrants were compared and statistically analysed. In a second step, the suitability of the calibrant was investigated by comparing the PCR efficiencies and coefficients of determination of the calibration curves with the curves obtained from the dilutions of gDNA extracted from the seed powder materials (gDNAs). In the third step, the distribution of the copy number ratios measured with pDNA and gDNAl calibrants was checked, whereby the results from each data set (i.e. analysis performed during 1 day) were treated individually. In all studies, no deviation from unimodality and normal distribution of the data was observed.
The probability plots showed normal distributions for the PCR efficiency, linearity, and copy number ratio values using pDNA and gDNAl calibrants, for all unknowns. Values of the GM event-specific copy number ratios were, furthermore, plotted per extraction method and calibrant to investigate the occurrence of any extraction-derived effect. The number of accepted sets of analysis per DNA extraction method and calibrant was, however, low and a normality test was, therefore, not performed [31]. Alternatively, DNA copy number ratios within the intervals created by the subtraction and addition of the standard deviation were compared.
Single factor ANOVA was used to investigate the similarity of the calibrants. It was conducted with PCR efficiencies, coefficients of determinations, and DNA copy number ratios as dependent variables originating from all copy number certification studies. The data were subsequently grouped per calibrant and certification study, and single factor ANOVA was performed for each calibrant selected as categorical factor.
Results
Impact of the extraction method on the copy number ratio
The DNA copy number ratios obtained via each extraction method by real-time PCR calibrated with pDNA calibrants ERM-AD415, ERM-AD425, and ERM-AD427, respectively, had overlapping mean ± s intervals. This was also true for the DNA copy number ratios obtained when gDNAl extracted from leaves was used as a calibrant for the specific GM quantification. The comparisons of the copy number ratios determined by event-specific PCR methods with a given DNA extraction method (Fig. 1) showed that all three DNA extraction methods within a study perform equally well. Therefore, the data of the three different DNA extraction methods applied in each study were pooled per calibrant. In all three studies, a similar effect was observed from plotting the copy number ratio for each matrix material: the mean DNA copy number ratio obtained when the measurements were calibrated with gDNAl was higher than the value obtained with the corresponding pDNA calibrant (Fig. 1). Single factor ANOVA confirmed that the calibrant had a significant influence on the DNA copy number ratio (Table 5).
Comparison of DNA copy number ratios grouped per DNA extraction method (CTAB (diamond), CTAB-Tip20 (triangle), GeneSpin (small square), DNAExtractor (line), and Qiagen/DNeasy (big square)). The corresponding mean value ± s is plotted relative to the GM event for each calibrant. N stands for the number of sets of analysis used for evaluation, from each calibrant and extraction method
PCR efficiencies of the calibrants and matrix materials
As there is a significant influence of the type of calibrant on the measured DNA copy number ratio (Table 5 and Fig. 1), the data obtained using the two calibrants could not be pooled. Consequently, a calibrant-defined copy number ratio was calculated for each GM event. In order to select the most suitable calibrant, the PCR amplification kinetics of the pDNA and gDNAl calibrants were compared to that of the gDNAs extracted from the corresponding ground seed material. The first parameter was the PCR efficiency ε estimated for both transgenic and endogenous targets, using the three DNA types (i.e. pDNA, gDNAl, and gDNAs). These PCR efficiencies were only compared if the respective data sets passed the selection criteria defined beforehand. These selection criteria were applied in order to avoid interferences of technically questionable results.
The means of the PCR efficiencies covered a rather wide range in the case of NK603 maize and 356043 soya materials. For both transgenic and endogenous targets, the PCR efficiencies never exceeded 100 % indicating no problem of PCR inhibition and all slightly lower than 100 % except of the NK603 target where efficiencies below 90 % were reported for the three tested types of calibrants suggesting suboptimal PCR conditions (Table 6). Comparing the targets, PCR efficiencies were higher for the endogenous gene, i.e. hmg and lectin, than for the transgene, i.e. NK603 and 356043. A difference between the PCR efficiencies corresponding to the pDNA and the gDNAl calibrations was observed: 2.8 % for the hmg, 4.6 % for the NK603, 7.0 % for the lectin, and 6.0 % for the 356043 targets, respectively. For all these target sequences, the PCR efficiencies of the pDNA and gDNAl calibrants were found to be significantly different (Table 7) using a simplex real-time PCR detection method.
For the 98140 maize, the variation of the PCR efficiency observed for each type of DNA was smaller (Table 6). The PCR efficiencies of the gDNAs calibration curves were always larger than the PCR efficiencies obtained with the pDNA and gDNAl calibrants. The PCR efficiencies estimated for both endogeneous and transgenic targets were similar for the pDNA and gDNAl calibrants (96.9 and 95.2 % vs. 96.5 and 95.6 % for the pDNA and gDNAl calibration curves of the taxon- and event-specific detection methods, respectively). No significant differences between the PCR efficiencies of the pDNA and gDNAl calibrants were found for the hmg and 98140 targets (Table 7) on simplex real-time PCR detection methods.
Similar observations were made in all three studies for the PCR efficiencies of gDNAs, and pDNA and gDNAl calibrants. For each GM event investigated, the PCR efficiencies were compared and showed a large overlap for both transgenic and endogeneous targets, for all three DNA types (Table 6). When F tests were applied, differences of the PCR efficiencies of the unknown seed powder samples and corresponding calibrants were, however, noted in each certification study. In a few cases only, the differences of the PCR efficiencies of one type of calibrant to the unknown powder samples were statistically not significant. For instance, no significant difference between the PCR efficiencies of both NK603 (p = 0.25, α = 0.05) and hmg (p = 0.59, α = 0.05, Kruskal–Wallis test) targets for pDNA and gDNAs was observed. All other comparisons led to significantly different results for both transgenic and endogeneous real-time PCR targets. Similarly, with the exception of the PCR efficiencies of gDNAl and gDNAs for the transgenic target 98140, the PCR efficiencies of the calibrants and unknown samples differed significantly for the hmg and 98140 target sequences. For the 356043 soya event, statistical analysis proved that the type of DNA did have an effect on the PCR efficiency. The PCR efficiencies of pDNA, gDNAl, and gDNAs were significantly different for both lectin and 356043 target sequences.
Comparison of the coefficients of determination of the DNA calibration curves
The second parameter studied was the coefficient of determination (R 2) that provides information about the fitting of data to a linear calibration curve model for both targets, using the three types of DNA, i.e. pDNA, gDNAl, and gDNAs (Table 6). Data were grouped per type of DNA in order to assess the similarity of the calibration curves for each GM event in terms of linearity. Sets of analysis exhibiting a R 2 value below 0.98 for one of the calibration curves [16] were not accepted for this evaluation as it may reflect erroneous dilutions or inappropriate PCR amplification.
A comparison of the coefficients of determination of the pDNA and gDNAl calibration curves showed no major influence of the type of the DNA calibrant in the case of the hmg endogenous target, for both NK603 and 98140 maize events (Table 6). In both cases, the distribution of the coefficient of determination displayed a larger variation for the transgenic target. No major differences were observed between the three types of DNA for the lectin and 356043 targets, respectively, in the case of the GM soya event. However, R 2 values were slightly higher for the NK603 maize and 356043 soya calibrants than for the corresponding gDNAs extracted from seed powder, indicating some difficulties encountered by the laboratories to correctly quantify low number of DNA targets in the diluted samples (Table 6). Nevertheless, the coefficients of determination of gDNAs dilution curves were closer to those of the pDNA calibrant for all endogeneous and transgenic targeted sequences.
Comparison of the DNA copy number ratios
The mean DNA copy number ratios were determined for each matrix material, i.e. ERM-BF415e, ERM-BF425c, and ERM-BF427c, and compared regarding the use of the corresponding pDNA or gDNAl as calibrants (Table 8). For these comparability studies, copy number ratios obtained from accepted data sets within one day for a particular certification study were compared. As there was no detectable influence of the DNA extraction method on the GM quantification, it was possible to combine the measurement results of the three corresponding extraction methods. The data sets generated by either pDNA or gDNAl calibration followed a normal distribution for the soya and maize events. As previously observed, the DNA copy number ratios for ERM-BF415e, ERM-BF425c, and ERM-BF427c, respectively, obtained by the two types of calibrants resulted in overlapping values (Table 5 and Fig. 1). However, the mean DNA copy number ratios were statistically different (Table 8). Single factor ANOVA analysis confirmed that the data sets calibrated with pDNA were significantly different from the data sets calibrated with gDNAl. Therefore, the DNA copy number data calibrated with pDNA or gDNAl could not be pooled. For all three studies, calibration with gDNAl provided values that were 12–35 % higher than those obtained with pDNA (Table 8). The standard deviations of the results using the two calibrants were, however, in all cases similar.
Discussion
An integrated analysis is provided here for three interlaboratory comparison studies performed in order to assess the suitability of two types of calibrants, namely pDNA and gDNAl, for GM quantification in particular for the certification of seed powder CRMs for their DNA copy number ratios. Moreover, three parameters were evaluated to check that the PCR amplification kinetics in the sample extract was not significantly different from those of the calibrant. Previous publications [8, 12, 29] have already addressed some of these issues, also regarding the variations of the between-laboratory performance. However, they were limited to one specific GM event. Taking into account the PCR-intrinsic performance variability of GMO quantification methods, a systematic study comprising various GM events was needed to allow, in a generalised manner, for a straightforward decision which calibrant is best suited.
Based on the interlaboratory comparison data, the robustness of GM quantification by real-time PCR toward the chosen DNA extraction was carefully investigated. Previous studies have shown that the extraction method can have an influence on the quality of isolated gDNA and, therefore, on the subsequent real-time PCR measurements [29, 32, 33]. The current method validation practice [34], which includes a single DNA extraction method, restricts the assessment of any potential effects of the extraction method on the measurement results to a particular combination of extraction and detection methods. Besides the DNA extraction method typically applied for the validation of the particular GM measurement procedure, two more extraction methods were chosen for each study in order to rule out any influences caused by the extraction method, which might lead to a shift in the estimated copy number ratio [29]. It turned out that for each investigated GM event, the chosen DNA extraction method had no impact on the DNA copy number ratio (Fig. 1). This suggests that DNA extracted from the seed powder materials did not dramatically differ in quality with the three extraction methods applied. The combination of extraction method and event specific real-time PCR gave similar results for each type of DNA calibrant applied to the studied GM events. Higher copy number ratios were, however, obtained in all cases when gDNAl was used for calibration.
There are ongoing discussions for GMO quantification, concerning the type of calibrant to be used and the measurement unit for which the reference materials for quality control should be certified. One prerequisite for a suitable calibrant is the similar behaviour to the sample investigated, which can be assessed by comparing the calibration curves with the curves generated for a diluted typical food/feed sample. As no significant difference was previously observed between the properties of pDNA and gDNA, both calibrants have been used for calibration in several studies [11]. Such studies may, however, not be representative due to the limited number of calibration curves compared.
In the studies reported here, statistical analyses based on large sets of data show that pDNA and gDNAl calibrants behave differently with respect to the PCR efficiencies from DNA target to target, with the exception of the PCR efficiencies of the 98140 maize target sequences, where a similar behaviour was observed instead (Table 6). Differences in PCR efficiencies of pDNA and gDNAl calibrants were previously reported in the literature and appeared to be caused by the solution used for the dilution of the calibrants [12]. Furthermore, one hypothesis is that the conformational state of the DNA template may influence the specific PCR amplification, especially during the early stage of PCR when the template is the dominant target. This may explain the different behaviour of pDNA and gDNAl (supercoiled vs. highly ordered conformational types) in terms of PCR efficiency. The individual PCR efficiency of the amplification of each target sequence has a significant impact on the GM quantification by real-time PCR. Therefore, the effect of a small difference in PCR efficiencies of the transgenic and endogenous targets on GM quantification by real-time PCR can generate a rather large difference in copy number due to the exponential nature of the PCR amplification. Consequently, such significant differences in PCR efficiencies of the calibrants may explain the difference of GM copy number ratios observed for all three events studied here, when the mean values are compared (Table 8). The lowest difference (12 %) is obtained for the 98140 maize, likely due to the small difference in PCR efficiencies of the pDNA and gDNAl calibrants (Table 6).
The PCR efficiencies for the two types of DNA calibrants were also compared with the PCR efficiencies for the gDNAs extracted from unknown seed powder samples (Table 6). The PCR efficiencies estimated from the dilution series of gDNAs were in all three studies higher than the PCR efficiencies of the corresponding gDNAl calibrant and, with the exception of the 98140 maize, smaller than those of the corresponding pDNA calibrant. Consequently, the DNA copy number ratio might be overestimated because the PCR efficiency of the analytical sample is higher than the PCR efficiency of the calibration curve [35] in the cases of ERM-BF415e (NK603 maize), ERM-BF425c (356043 soya), and ERM-BF427c (98140 maize) and gDNAl calibration as well as ERM-BF427c and pDNA calibration. On the other side, underestimation of the GM content expressed in DNA copy number ratios is expected when the PCR efficiency for the sample is lower than the PCR efficiency for the calibration curve, which is the case for ERM-BF415e and ERM-BF425c and the corresponding pDNA calibration.
For all three GM events, the differences in PCR efficiencies between the pDNA calibrant and gDNAs from unknown samples were smaller than those between the gDNAl calibrant and the unknowns for the transgenic target sequence, with one exception (98140 maize). The opposite situation was observed for the endogeneous target (data not shown). Smaller differences between the PCR efficiencies of the calibrant and the test sample should result in DNA copy number ratios closer to the true value [12]. Based on the smallest difference in PCR efficiencies, one would choose pDNA for the calibration of the event-specific real-time PCR method applied to the 98140 maize model. No generalisation can be, however, made as the differences in PCR efficiencies of the calibrant and unknown samples were not consistent for the NK603 maize and 356043 soya events. One would have to investigate the suitability of the calibrant on a case by case basis, also taking into account other parameters. Importantly, both pDNA and gDNAl calibrants provided good linearity of the calibration curve. Comparable behaviour of both types of calibrants relative to that of the unknown samples was furthermore observed for all three GM events (Table 6).
A significant impact of the calibrant on the measured copy number ratio was observed in all three interlaboratory comparisons. The measurement results generated from the pDNA calibration were lower compared to those obtained with gDNAl calibration curves (Table 8) for all GM events investigated; the standard deviation was in all cases fairly small, indicating a good reproducibility of both pDNA- and gDNA-based calibration curves. The best-suited calibrant for GM quantification is expected to yield a measured copy number ratio close to the true value. But particularly for GM maize, the GM content of the CRMs certified on the basis of GM mass fraction cannot be accurately converted into copy number ratio [3, 36, 37] and an estimate of the true value is therefore difficult. This is due to the fact that the relationship between the % GM copies and % mass of GM material is influenced by a series of complex biological factors (e.g. zygosity, ploidy of the tissue(s), endoreduplication, and for a heterozygous seed material, parental origin of the donor of the GM trait). This relationship differs from one material to the next and, apart from the situations where the investigated test material is known to have the same origin as the calibrant, the GM mass fraction will be biased due to the not precisely known biological factors. However, the biology-based inaccuracies in converting the GM content of the CRMs certified for the GM mass fraction into copy numbers are not an issue for the homozygous GM soya seeds. A theoretical DNA copy number ratio of 1 can be predicted for ERM-BF425c. In this case, the measured DNA copy number ratio using the pDNA calibrant ERM-AD425 might be underestimated by about 15 % (Table 8). On the other hand, when the gDNAl calibrant is used, the measured DNA copy number ratio might be overestimated by 15 %. These observations confirm that the differences between the PCR efficiencies for test sample and calibrant have an influence on the final GM ratio estimation. The measurement results obtained for the 354063 soya matrix by means of two calibrations are affected to the same extent, but with opposite deviations from the theoretical value. This makes the choice of the calibrant rather challenging. Consequently, the user has to carefully select the calibration tools and reliably apply them in a consistent manner to the GM quantification procedure. Comparable results from different laboratories can only be expected if they apply the same calibration system.
To date, no independent parallel comparisons between GMO measurements using both plasmid calibrants and the more common gDNAl extracted from leaves have been published. This work aimed at setting a reference system which is applicable for longer periods of time in a reliable manner. The specific assays described in this study show that both pDNA and gDNAl are equally suitable to calibrate the real-time PCR methods applied herein, but that the resulting data are generally different for each of the targeted sequences. It could not be proven that one of the calibrants behaves more similar to the gDNAs extracted from seed powder. It should be kept in mind that GMO testing laboratories quantifying a particular GM event in a matrix, which is often not made of pure seed powder but may consist of a processed food, would ideally need a DNA calibrant of the same nature as the DNA extracted from this sample in order to avoid a bias. To date, no studies have been performed to demonstrate the equivalent analytical behaviour of gDNA extracted from seed powders and gDNA extracted from a variety of processed food test samples. However, the study described above demonstrates already that genomic DNA from seeds and leaves behave differently for the GM events investigated, even if they have the same DNA inserts.
No immediate conclusion on the selection of the best-suited calibrant can be drawn due to the high variability of the PCR measurement results reported by the participating laboratories. The variation between data sets and the variation within data sets reported in interlaboratory comparisons illustrate the importance of having a sufficient number of data sets for studying the analytical behaviour of different calibrants. Although large data pools were available in order to generate a considerable number of calibration curves and to compare their responses, the hypothesis that pDNA is the superior calibrant was not supported by these studies. However, the use of pDNA as calibrant has several advantages over gDNAl, such as complete characterisation of sequence and structure, straightforward production and reproduction of additional batches, unproblematic storage and distribution, and high stability. In addition to that, the study presented here provides sufficient experimental evidence that pDNA is suitable for the calibration of GMO measurements with respect to the DNA copy number ratio, and generalises the applicability of plasmids as event-specific quantification systems.
The event-specific multi-target pDNA calibrants used in this study have been created for the first time for the three targeted events. As these plasmids have a known and confirmed number of DNA fragments cloned into each plasmid, they resemble a sustainable possibility to measure the GM content as DNA copy number ratio. The remaining question is in how far the DNA copy number ratio measured by real-time PCR deviates from the “true value” when the pDNA calibration is applied, as both types of calibrants described here, i.e. pDNA and gDNAl, did not lead to the theoretical value expected for the homozygous soya (see the results of the 356043 soya study).
Nevertheless, due to the potential as sustainable metrological traceability anchor point, well characterised properties, as well as the additional practical reasons mentioned above, it is reasonable to decide for tailored pDNA as the calibrant of choice. The use of such pDNAs as calibrants can enforce the harmonised use of validated event-specific PCR methods among the laboratories and to streamline GMO analysis, and eventually support the harmonization between the legislative requirements and detection methodologies. In conclusion, our study shows that the most suitable metrological approach is to set a reference system based on pDNA as primary calibrant for DNA copy number ratio measurements, together with the approved GM quantification method published by the EURL-GMFF.
References
Regulation (EC) No 1830/2003 of the European Parliament and of the Council of 22 September 2003 concerning the traceability and labelling of genetically modified organisms and the traceability of food and feed products produced from genetically modified organisms and amending Directive 2001/18/EC (2003). Official Journal of the European Union L:24–28
Commission Recommendation of 4 October 2004 on technical guidance for sampling and detection of genetically modified organisms and material produced from genetically modified organisms as or in products in the context of Regulation (EC) No 1830/2003 (2004). Official Journal of the European Union L:18–26
Burns M, Corbisier P, Wiseman G, Valdivia H, McDonald P, Bowler P, Ohara K, Schimmel H, Charels D, Damant A (2006) Comparison of plasmid and genomic DNA calibrants for the quantification of genetically modified ingredients. Eur Food Res Technol 224:249–258
Debode F, Marien A, Janssen E, Berben G (2010) Design of multiplex calibrant plasmids, their use in GMO detection and the limit of their applicability for quantitative purposes owing to competition effects. Anal Bioanal Chem 396:2151–2164
Taverniers I, Van Bockstaele E, De Loose M (2001) Use of cloned DNA fragments as reference materials for event specific quantification of genetically modified organisms (GMOs). Mededelingen (Rijksuniversiteit te Gent 66:469–472
Taverniers I, Windels P, Van Bockstaele E, De Loose M (2001) Use of cloned DNA fragments for event-specific quantification of genetically modified organisms in pure and mixed food produc. Eur Food Res Technol 213:417–424
Taverniers I, Van Bockstaele E, De Loose M (2004) Cloned plasmid DNA fragments as calibrators for controlling GMOs: different real-time duplex quantitative PCR methods. Anal Bioanal Chem 378:1198–1207
Lievens A, Bellocchi G, De Bernardi D, Moens W, Savini C, Mazzara M, Van den Eede G, Van den Bulcke M (2010) Use of pJANUS-02-001 as a calibrator plasmid for Roundup Ready soybean event GTS-40-3-2 detection: an interlaboratory trial assessment. Anal Bioanal Chem 396:2165–2173
Yang L, Pan A, Zhang K, Guo J, Yin C, Chen J, Huang C, Zhang D (2005) Identification and quantification of three genetically modified insect resistant cotton lines using conventional and TaqMan real-time polymerase chain reaction methods. J Agric Food Chem 53:6222–6229
Zhang H, Yang L, Guo J, Li X, Jiang L, Zhang D (2008) Development of one novel multiple-target plasmid for duplex quantitative PCR analysis of roundup ready soybean. J Agric Food Chem 56:5514–5520
Taverniers I, Windels P, Vaitilingom M, Milcamps A, Van Bockstaele E, Van den Eede G, De Loose M (2005) Event-specific plasmid standards and real-time PCR methods for transgenic Bt11, Bt176, and GA21 maize and transgenic GT73 canola. J Agric Food Chem 53:3041–3052
Charels D, Broeders S, Corbisier P, Trapmann S, Schimmel H, Emons H (2007) Toward metrological traceability for DNA fragment ratios in GM quantification. 3. Suitability of DNA calibrants studied with a MON 810 corn model. J Agric Food Chem 55:3268–3274
de Andrade E, Jeynov B, Corbisier P, Kortekaas AM, Trapmann S, Vincent S, Emons H (2011) Certification of a Maize NK603 Reference Material for its DNA Copy Number Ratio. Certified Reference Material ERM®-BF415e. ISBN 978-92-79-19074-2. EUR 24699 EN. Luxembourg: Publications Office of the European Union
Meyer W, de Andrade E, Broeders S, Charels D, Corbisier P, Merveillie A, Trapmann S, Emons H (2011) Certification of a Soya 356043 Reference Material for its DNA Copy Number Ratio. Certified Reference Material ERM®-BF425c. ISBN 978-92-79-19070-4. EUR 24695 EN. Luxembourg: Publications Office of the European Union
Caprioara-Buda M, Trapmann S, Gancberg D, Corbisier P, Merveillie A, Contreras Lopez MC, Emons H (2011) Certification of Maize 98140 Reference Material ERM®-BF427c. ISBN 978-92-79-19279-1. Luxembourg: Publications Office of the European Union
Trapmann S, Conneely P, Corbisier P, Gancberg D, Gioria S, Van Nyen M, Schimmel H, Emons H (2005) The Certification of Reference Materials of Dry-Mixed Maize Powder with different Mass Fractions of NK603 Maize Certified Reference Materials ERM®-BF415 (ERM®-BF415-a/ERM®-BF415-b/ERM®-BF415-c/ERM®-BF415-d/ERM®-BF415-e/ERM®-BF415-f). ISBN 92-894-8063-7. EUR 21270 EN. Luxembourg: Office for Official Publications of the European Communities
Gancberg D, Conneely P, Contreras Lopez M, Corbisier P, Gioria S, Van Nyen M, Vincent S, Trapmann (2007) Certification of Reference Materials of Soya Powder with different Mass Fractions of 356043 Soya, ERM®-BF425 (ERM®-BF425a, ERM®-BF425b, ERM®-BF425c, ERM®-BF425d). ISBN 978-92-79-07464-6. EUR 23029 EN. Luxembourg: Office for Official Publications of the European Communities
Gancberg D, Corbisier P, de Andrade Silva E, Mazoua S, Merveillie A, Tumba M-F, Trapmann S (2009) Certification of reference materials of maize seed powder with different mass fractions of genetically modified 98140 maize. Certified Reference Material ERM®-BF427 (ERM®-BF427a, ERM®-BF427b, ERM®-BF427c, ERM®-BF427d). ISBN 978-92-79-10174-8. EUR 23537 EN. Luxembourg: Office for Official Publications of the European Communities
Jeynov B, de Andrade E, Broothaerts W, Corbisier P, Mazoua S, Merveillie A, Trapmann S, Emons H (2011) Certification of Plasmid DNA Containing NK603 Maize DNA Fragments. Certified Reference Material ERM®-AD415. ISBN 978-92-79-19071-1. EUR 24696 EN. Luxembourg: Publications Office of the European Union
Corbisier P, Broeders S, Meeus N, Mazoua S, Meyer W, Trapmann S, Emons H (2011) Certification of Plasmid DNA Containing 356043 Soya DNA fragments. Certified Reference Material ERM®-AD425. ISBN 978-92-79-18993-7. EUR 24687 EN. Luxembourg: Publications Office of the European Union
Caprioara-Buda M, Corbisier P, Gancberg D, Broeders S, Trapmann S, Mazoua S, Matayron G, Emons H (2011) Certification of Plasmid DNA Containing 98140 Maize DNA Fragments. Certified Reference Material ERM®-AD427. ISBN 978-92-79-19277-7. EUR 24722 EN. Luxembourg: Publications Office of the European Union
Broothaerts W, Corbisier P, Schimmel H, Trapmann S, Vincent S, Emons H (2008) A single nucleotide polymorphism (SNP839) in the adh1 reference gene affects the quantitation of genetically modified maize (Zea mays L.). J Agric Food Chem 56:8825–8831
Event-Specific Method for the Quantitation of Maize Line NK603 Using Real-Time PCR - Validation Report and Protocol (2005) EU-RL GMFF. http://gmo-crl.jrc.ec.europa.eu/summaries/NK603report_mm.pdf. Accessed April 2010
Event-specific Method for the Quantification of Maize 98140 Using Real-time PCR - Validation Report and Protocol (2011) EU-RL GMFF. http://gmo-crl.jrc.ec.europa.eu/summaries/DP-098140-6_validated_Method.pdf. Accessed January 2011
Event-specific Method for the Quantification of Soybean Event DP-356043-5 Using Real-time PCR - Validation Report and Protocol (2009) EU-RL GMFF. http://gmo-crl.jrc.ec.europa.eu/summaries/DP-356043_validated_Method_correctedversion1.pdf. Accessed April 2010
Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19:1349
Arumuganathan K, Earle ED, Nuclear DNA (1991) Content of some important plant species. Plant Mol Biol Rep 9:208–218
Zhang D, Corlet A, Fouilloux S (2008) Impact of genetic structures on haploid genome-based quantification of genetically modified DNA: theoretical considerations, experimental data in MON 810 maize kernels (Zea mays L.) and some practical applications. Transgenic Res 17:393–402
Charels D, Broeders S, Corbisier P, Trapmann S, Schimmel H, Linsinger T, Emons H (2007) Toward metrological traceability for DNA fragment ratios in GM quantification. 2. Systematic study of parameters influencing the quantitative determination of MON 810 corn by real-time PCR. J Agric Food Chem 55:3258–3267
Definition of Minimum Performance Requirements for Analytical Methods of GMO Testing (2008). http://gmo-crl.jrc.ec.europa.eu/doc/Min_Perf_Requirements_ Analytical_methods.pdf. Accessed September 2009
Schwarze J (ed.) (2009) Grundlagen der Statistik 2: Wahrscheinlichkeitsrechnung und induktive Statistik. Verlag Neue Wirtschafts-Briefe, GmbH & Co. KG, Herne 1986
Peano C, Samson MC, Palmieri L, Gulli M, Marmiroli N (2004) Qualitative and quantitative evaluation of the genomic DNA extracted from GMO and non-GMO foodstuffs with four different extraction methods. J Agric Food Chem 52:6962–6968
Smith DS, Maxwell PW (2005) Use of quantitative PCR to evaluate several methods for extracting DNA from corn Xour and cornstarch. Food Control 18:236–242
European Union Reference Laboratory for GM Food & Feed official website. http://gmo-crl.jrc.ec.europa.eu/statusofdoss.htm
Cankar K, Stebih D, Dreo T, Zel J, Gruden K (2006) Critical points of DNA quantification by real-time PCR—effects of DNA extraction method and sample matrix on quantification of genetically modified organisms. BMC Biotechnol 6:1–15
Holst-Jensen A, De Loose M, Van den Eede G (2006) Coherence between legal requirements and approaches for detection of genetically modified organisms (GMOs) and their derived products. J Agric Food Chem 54:2799–2809
ENGL explanatory document on the use of “Percentage of GM-DNA copy numbers in relation to target taxon specific DNA copy numbers calculated in terms of haploid genomes” as a general unit to express the percentage of GMOs (2007). http://engl.jrc.ec.europa.eu/docs/HGE-release.pdf. Accessed in September 2009
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Caprioara-Buda, M., Meyer, W., Jeynov, B. et al. Evaluation of plasmid and genomic DNA calibrants used for the quantification of genetically modified organisms. Anal Bioanal Chem 404, 29–42 (2012). https://doi.org/10.1007/s00216-012-6104-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-012-6104-6