Abstract
Proton nuclear magnetic resonance (1H-NMR)-based metabolomics enables the high-resolution and high-throughput assessment of a broad spectrum of metabolites in biofluids. Despite the straightforward character of the experimental methodology, the analysis of spectral profiles is rather complex, particularly due to the requirement of numerous data preprocessing steps. Here, we evaluate how several of the most common preprocessing procedures affect the subsequent univariate analyses of blood serum spectra, with a particular focus on how the standard methods perform compared to more advanced examples. Carr–Purcell–Meiboom–Gill 1D 1H spectra were obtained for 240 serum samples from healthy subjects of the Asklepios study. We studied the impact of different preprocessing steps—integral (standard method) and probabilistic quotient normalization; no, equidistant (standard), and adaptive-intelligent binning; mean (standard) and maximum bin intensity data summation—on the resonance intensities of three different types of metabolites: triglycerides, glucose, and creatinine. The effects were evaluated by correlating the differently preprocessed NMR data with the independently measured metabolite concentrations. The analyses revealed that the standard methods performed inferiorly and that a combination of probabilistic quotient normalization after adaptive-intelligent binning and maximum intensity variable definition yielded the best overall results (triglycerides, R = 0.98; glucose, R = 0.76; creatinine, R = 0.70). Therefore, at least in the case of serum metabolomics, these or equivalent methods should be preferred above the standard preprocessing methods, particularly for univariate analyses. Additional optimization of the normalization procedure might further improve the analyses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The high-throughput characterization of the biochemical processes underlying specific phenotypes or diseases is generally termed metabolic profiling or metabolomics. A typical methodology in metabolomics is the application of 1H-nuclear magnetic resonance (NMR) on biological fluids, such as blood serum or urine, yielding high-resolution spectral profiles, “fingerprints”, of a whole range of metabolites [1]. Typical examples illustrating the success of this methodology can be found in the fields of toxicology [2], diet [3], and cardiovascular disease [4]. While other methods, such as the different mass spectrometry technologies, often have the particular advantage of a larger sensitivity, the major benefits of NMR are the non-destructive character of the measurements and the relative robustness of the results, allowing quantification of the different metabolites [5].
However, the inherent nature of the technology and samples results in the obligatory introduction of spectral data preprocessing methods after acquisition. In the first step, overdominating peaks such as the residual water peak (even after presaturation) and urea in urine spectra are removed, followed by normalization, data reduction, and variable definition of the remainder of the spectra and, if required, data scaling of the resulting variables. Currently, several freely available software packages performing most or all of these steps are freely available, e.g., HiRes [6], MetaboAnalyst [7], Automics [8], and the R-package Metabonomic [9] in addition to commercially available softwares.
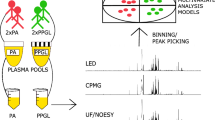
The necessity of the different preprocessing methods has already been fully discussed by others, e.g., in [10] and [11]. It typically comprises some form of data normalization, often followed by binning, variable definition, and data scaling. The preprocessing methods evaluated in this manuscript are schematically illustrated in Fig. 1. Data normalization is applied to reduce variation in the overall concentration/intensities of the analyzed biofluids, both due to natural variation (biological dilution effects) as to experimental factors (e.g., biofluid dilution in buffer, gain fluctuations NMR equipment). The standard method, integral normalization, normalizes the individual intensities of a specific spectrum by dividing them by the summed intensity of that spectrum. Integral normalization is also referred to as constant or total sum normalization. There are alternative methods as well, such as probabilistic quotient (PQ) normalization [12]. This algorithm usually starts with the spectra after integral normalization and uses the latter to generate a reference spectrum, typically a median spectrum. For each of the individual spectra, a series of quotients is generated by the element-wise division of the spectrum by the reference spectrum. Regions containing no or possibly interfering signals are ignored if possible, and the median of the remaining quotients is subsequently used as correction factor. Alternatively, the algorithm can be applied on already binned spectra.
Schematic overview. Once unwanted spectral regions are removed, there is usually a global difference in resonance intensities between the spectra (a). Data normalization aims at reducing these differences (b). If complete spectra are not to be used, they are divided into subsets by binning (c). For each of these bins (delineated by the dotted lines), a new variable is defined (d) and used in the subsequent analysis. Adaptive-intelligent binning was applied and, for each bin, a new variable was defined by taking the maximum intensity (horizontal lines in d) for each spectrum. Remark: Probabilistic quotient normalization can also be applied on the newly defined variables (not shown)
Spectral binning or bucketing tries to improve and facilitate the subsequent statistical data analysis by reducing the number of variables. For standard binning, this includes the division of the spectra into equally sized (typically 0.04 ppm) bins. There exist more advanced binning algorithms as well, e.g., adaptive-intelligent (AI) binning [13] and Gaussian binning [14], which, in general, try to focus on a better peak definition through the identification of conserved minima among the spectra and the application of these minima as bin edges. After the division of the spectra into several bins, the different original spectral intensities in each bin have to be transformed into a single new variable. For the remainder of this manuscript, this will be referred to as “variable definition.” The most common procedures for this purpose are the use of the sum/average (standard) or maximum of the individual spectral intensities within each bin as new variable.
An alternative to binning is the application of algorithms, sometimes library-based, capable of peak identification (“peak picking”)—often in combination with peak alignment—and listing, e.g., PARS [15], targeted profiling [16], Hough transform-based alignment [17, 18], and recursive segment-wise peak alignment [19]. Also here, in most cases, both maximal and averaged/summed intensities can be used for the subsequent analysis.
Finally, depending on the specific nature of the subsequent analytical methodologies, data scaling of the individual variables can be required, e.g., by performing unit variance or Pareto scaling. Since the performance of the specific data scaling procedures largely depends on the biological relevance of the individual variables in the context of the experiment and cannot be estimated by our design (cf. infra), this report does not describe its effect.
Despite the fact that more innovative methodologies are available, the standard preprocessing methods are still widely used. The aim of this article is therefore to evaluate the effects of these standard methods in blood-serum-based metabolomics experiments and to compare them with the effects of a selected set of more advanced methods. A similar evaluation has already been performed for urine spectra [11], but it has to be noted that blood serum (and plasma) strongly differs from urine due to a higher viscosity, lipid and protein content, and a more prominent peak overlap, which all complicate the analysis of serum spectra. On the other hand, urine has the particular disadvantage of being more prone to peak shifts induced by matrix-dependent differences such as pH, salt content, temperature, etc. [20].
Here, the different methods will be evaluated by analyzing their effects on the correlation of the resulting datasets with independently measured variables. Carr–Purcell–Meiboom–Gill (CPMG) spin echo pulse sequence experiments were used to obtain NMR spectra for 240 blood serum samples from apparently healthy subjects of the Asklepios study, a longitudinal population study focusing on aging, cardiovascular hemodynamics, and inflammation and their interplay in cardiovascular disease development [21]. The data were preprocessed with different data normalization (either integral/constant sum or PQ normalization), binning (no, equidistant, or AI binning), and variable definition (sum or max) procedures. Since PQ normalization can be applied prebinning and post-binning, both options were independently evaluated.
Subsequently, these differently preprocessed datasets were correlated with the triglyceride, glucose, and creatinine concentrations measured by standard analytical techniques. The rationale is that triglyceride, glucose, and creatinine can be considered as prototypes of different types of metabolites, exhibiting chemical shifts ranging from roughly 0.5 to 4 ppm, the most peak-dense region of serum spectra. In addition, the three metabolites include peaks of different intensities: triglycerides are characterized by different resonance intensities, including large peaks; glucose is generally represented by medium-intensity peaks, while for creatinine only relatively small peaks can be found. Furthermore, these metabolites can also easily be quantified by standard photometric laboratory procedures, providing independent measurements that compose the adequate testing ground to evaluate the performance of the different preprocessing methods with respect to subsequent univariate analyses.
Materials and methods
Samples and independent metabolite concentration measurements
For a complete overview of the subjects and the Asklepios study goals and methods, we refer to [21]. The Asklepios study complies with the Declaration of Helsinki. The ethical committee of the Ghent University Hospital approved the study protocol, and a written informed consent was obtained from each participant prior to enrollment into the study.
Serum samples from 240 male Asklepios study participants, aged 35–55 years old, were obtained from venous blood, which was sampled in Venosafe serum tubes (Terumo, Haasrode, Belgium), cooled at 4 °C, and centrifuged. Samples were transferred to Nunc CryoTube vials (Nunc A/S, Roskilde, Denmark) and stored long-term at −80 °C. Serum triglyceride concentrations were assayed by the esterase PAP kinetic colorimetric reaction (without glycerol correction). For serum glucose quantification, the standard hexokinase enzymatic method was used. Serum creatinine concentrations were measured by a rate-blanked kinetic Jaffé method. Quantification of the three metabolites was performed using commercial reagents on a Modular P automated analyzer (Roche Diagnostics, Mannheim, Germany). Median (interquartile range) concentrations for triglycerides, glucose, and creatinine were, respectively, 108.5 (85.4–149.8), 94 (88–99), and 0.94 (0.87–1.04) mg/dl.
NMR measurements
Samples were defrosted, and 250 μl of serum was diluted in 550 μl aliquots (10% D2O, 0.9% NaCl w/v). All measurements were performed at 30 °C throughout, on a Bruker Avance II spectrometer, operating at 1H frequency of 700.13 MHz, equipped with a 5 mm TXI-Z ATMA probe and a BACS-60 sample changer, under TopSpin 2.0. Water signal suppression was obtained by presaturation (2.22 s relaxation delay, 50 Hz B1 field strength). Each individual CPMG spin echo was repeated 20 times and resulted in a total delay of 20 ms to allow for T2 relaxation editing (based on [22]). Each spectrum was constructed by accumulating 64 scans (0.78 s acquisition time each) of 16,384 complex time domain points, resulting in a spectral width of 30 ppm. Uniform handling of all samples was maximized by automated tuning and matching as well as gradient shimming using the Topshim routine for each serum sample. The “baseopt” routine in the TopSpin 2.0 software was used during acquisition to obtain flatter baselines. Before Fourier transformation, zero filling was applied to the 131,072 real-time domain points, followed by apodization with a squared cosine bell window function. All spectra were referenced to the chemical shift of lactate (methyl signal, left peak of the doublet, 1.335 ppm).
Data preprocessing and analysis
Data normalization and binning procedures were implemented in Matlab, version 7.2 (The MathWorks, Natick, MA) as in house-written routines. Only the region 12 to −3 ppm was used for further analysis, and the water peak region was excluded (5.12 to 4.48 ppm). For PQ normalization, the peak-rich region between 4.4 and 0.5 ppm was used as normalization region. Equidistant binning was applied on the region of 10 to −0.5 using 0.04 ppm bins. For AI binning, the regions of 12 to 10 and −1 to −3 ppm were considered as noise regions, and the quality parameter was equal to 0.3. Noise bins were removed before further analysis. More information on the AI binning procedure is available in [13]. Standard statistical procedures were performed in R 2.10.0 (correlation analyses) and PASW statistics 18 (univariate general linear model). Statistical tests were two-sided, and the level of significance was 0.05.
Results
Starting from the raw CPMG data, there were 14 combinations of different preprocessing methods possible: three types of normalization (integral, PQ prebinning, PQ post-binning) × two types of binning (equidistant, AI binning) × two types of variable definition (average/sum, max) + two types of normalization (integral, PQ) for the complete spectra (no binning and therefore no variable definition either). This yielded 14 differently preprocessed datasets, which we will simply refer to as “preprocessed datasets” (Table 1). The resulting number of variables in these preprocessed datasets is generally comparable for all methods (ranging from 259 to 272), except when complete (i.e., not binned) spectra were used (number of variables equals number of data points = 62,718).
For each of the preprocessed datasets, we aimed to assess whether the values of the individual variables could be used as a proxy for the corresponding metabolite concentrations, thereby identifying the best combination of preprocessing methods. Hence, we independently measured the concentrations of three metabolites (triglycerides, glucose, and creatinine, Table 1) and assessed how well they correlated with the different variables of the preprocessed datasets. Therefore, for each of these metabolites and each preprocessed dataset, Pearson correlations were calculated between the independently measured concentrations of the selected metabolite and each variable of the preprocessed dataset. As each of these metabolites displays multiple resonances at distinct frequencies in a single spectrum, we subsequently selected the maximum correlation for that dataset for further analysis. This yielded three maximum Pearson correlation values per preprocessed dataset, one for each independently measured metabolite. The different correlation values (R) have been summarized in Table 1 and were ranked in order to clarify the preprocessing-specific effects. Visual inspection of Table 1 already reveals that the same methods have rather similar effects for each of the metabolites. This will be further elaborated throughout the results section, but first, it should be validated whether the corresponding peaks are indeed the metabolites under study.
Metabolite validation
The resonances corresponding with the maximal correlations were compared with published data [23], thereby evaluating whether the matching metabolites were indeed triglycerides, glucose, and creatinine. Except for several creatinine correlations, explicitly indicated in Table 1, we could indeed confirm that, with a very high probability, the resonances belonged to the three metabolites under study. The resonances corresponding to the correlation maxima are depicted in Fig. 2a (triglycerides), b (glucose and creatinine).
Maximum correlations for triglycerides, glucose, and creatinine. Partial averaged spectrum indicating regions (horizontal bars, length depends on binning method) of maximum correlations with triglycerides (TX, a), glucose (GX, b), or creatinine (CX, b) for the different preprocessing methods X, with X = 1 (AI binning, integral normalization, maximum intensities), 2 (AI binning, integral normalization, summed intensities), 3 (PQ normalization post-AI-binning, maximum intensities), 4 (PQ normalization post-AI-binning, summed intensities), 5 (PQ normalization before AI binning, maximum intensities), 6 (PQ normalization before AI binning, summed intensities), 7 (equidistant binning, integral normalization, maximum intensities), 8 (equidistant binning, integral normalization, summed intensities), 9 (PQ normalization post-equidistant-binning, maximum intensities), 10 (PQ normalization post-equidistant-binning, summed intensities), 11 (PQ normalization before equidistant binning, maximum intensities), 12 (PQ normalization before equidistant binning, summed intensities), 13 (integral normalization, no binning), or 14 (PQ normalization, no binning). For creatinine maximum correlations C8, C10, and C12, the matching spectral region (5.76 to 5.72 ppm) does not correspond with a known creatinine chemical shift and was therefore not depicted in this figure
Metabolite- and preprocessing-specific effects
The different methods and metabolites were statistically compared using a full-factorial analysis of variance (ANOVA; constructed as a univariate general linear model in PASW), with removal of non-significant terms (P > 0.05), yielding the model displayed in Table 2. Due to the large differences in R value variance between (and within) the different metabolites (Table 1), the complete set of R values was transformed into z scores before inclusion as dependent variable. Since the effects of binning and variable definition cannot be studied using complete spectra (not binned), the latter were not included in the model. In the remainder of the results section, the different significant effects are explored, and more specific interaction terms are provided where necessary.
Metabolite-specific effects
The largest effect is the difference between the different metabolites (P < 0.001, Table 2). The analysis consistently yields very good correlations for triglycerides (R = 0.95 to 0.99), while the results for creatinine have a rather large range (R = 0.19 to R = 0.70), from acceptable to completely unsatisfactory, particularly for those correlation maxima not corresponding with creatinine (Table 1). Generally, the correlations for glucose (R = 0.46 to R = 0.76) are better than the creatinine correlations, but inferior compared to triglycerides. Figure 2 depicts the corresponding peak heights, which are rather similar for triglycerides and glucose, but smaller for creatinine. Resonance intensity therefore only partially explains the differences in maximum correlations.
Effect of normalization
The ranking of the preprocessing methods according to their performance (maximal R values) in Table 1 illustrates the highly significant (Table 2, P < 0.001) difference between integral and PQ normalization, prebinning and post-binning. The standard method, integral normalization, scores inferior for each of the metabolites: both for triglycerides and glucose, integral normalization yields worse results irrespective of the other preprocessing methods (Table 1, triglyceride and glucose). For the smaller creatinine peaks, its negative effect is surpassed by that of variable definition; within both variable definition groups (max or sum), however, integral normalization again scores consistently worse than PQ normalization (Table 1, creatinine).
The effect of normalization also appears to depend on the type of binning (global interaction term, P = 0.031, Table 2). Inspection of the specific interaction terms reveals that this can be attributed to a better effect of PQ normalization post-binning compared to prebinning, but only for AI-binned spectra (specific interaction term AI binning × PQ normalization, post-binning vs. prebinning: P = 0.024). This can also be seen in Table 1 by a pairwise comparison of the R values for PQ normalization post-binning and prebinning, with other preprocessing methods equal. For example, one can look at AI-binned PQ-normalized spectra with maximum intensity variable definition for each metabolite (AI PQ max in Table 1): AI PQ (post) max scores better than AI PQ (pre) max for triglycerides (R = 0.981 vs. R = 0.980), glucose (R = 0.756 vs. R = 0.674), and creatinine (R = 0.696 vs. R = 0.625). This is also the case for spectra with summed intensity variable definition in combination with AI binning, but not always for equidistant-binned spectra.
Effects of binning
AI binning generally generates better results, since it consistently yields better correlations for each condition than its equidistant-binned counterpart (standard method; P < 0.001, Table 2). For example, when looking pairwise at PQ-normalized spectra, post-binning, and with maximum intensity variable definition (PQ (post) max in Table 1), AI binning performs better than equidistant binning for triglycerides (R = 0.981 vs. R = 0.980), glucose (R = 0.756 vs. R = 0.619), and creatinine (R = 0.696 vs. R = 0.573). The same conclusion is valid for all other comparisons between AI- and equidistant spectra, with only one exception (triglycerides, integral normalization with summed intensity variable definition). Furthermore, AI binning even yields comparable or better results than not binned spectra.
As demonstrated in Fig. 2, the spectral regions corresponding with maximum correlations in Table 1 often overlap for equidistant and AI-binned spectra, which suggests that the most likely cause for the better performance of AI binning is a better variable definition. This is particularly clear for the creatinine correlations in Fig. 2b, where only the singlet peaks identified by AI binning actually correspond with the full creatinine peak. For not binned spectra, only the (approximate) maximum of the singlet is selected, while for the other methods the bins include other metabolites as well.
Effect of variable definition
While the use of summed intensities for each bin (standard method), as variable definition method, generally yields better results (P = 0.001, Table 2), the effect of variable definition depends to an even larger extent on the metabolite under study (interaction term metabolite × variable definition: P < 0.001, Table 2). For example, when looking at triglycerides, within either the PQ normalization (post-binning and prebinning) or the integral normalization group, summed intensity variable definition almost always scores better than when maximum intensities were used (Table 1). On the other hand, in the case of creatinine, the use of summed intensities always yields inferior results (specific interaction term for creatinine: P < 0.001, Table 2). The results for glucose were less straightforward.
As the difference is very clear and large for the small creatinine peaks (pairwise R value differences >0.2), but only minor for the larger triglyceride peaks (pairwise R value differences <0.01), the use of maximum intensities appears to be the most appropriate general solution (Table 1). The latter conclusion contrasts the significant result from the ANOVA (P = 0.001, Table 2); however, one should take into account that this model was based on z values and thereby does not integrate the size of the deleterious effect when summed intensities for smaller peaks are used.
A last significant interaction can be found between the types of normalization and variable definition (P = 0.020, Table 2), which may be attributed to the positive effect of using maximum intensity (instead of summed intensity) variable definition after integral normalization (specific interaction term, P = 0.008). It is unclear how this interaction term can be explained, but, as the positive effect is small, particularly when compared with the large, negative effect of integral normalization, its practical implications are irrelevant.
A comment on data normalization
The effect of data normalization was the most important modifiable effect in Table 1. Therefore, we compared the global impact of integral and PQ normalization on the preprocessed datasets. In a first step, as explained before, all correlations (R) between the independently measured triglyceride concentration and the resonance intensities for all chemical shifts in a single preprocessed dataset were calculated. But instead of identifying the maximal R value, here, the R 2 values were calculated for all variables of the specific dataset and summarized in frequency polygons (equivalent to histograms, but with only the line connecting the midpoints of the tops of the virtual histogram bars depicted). This was performed for both AI-binned and equidistant-binned spectra, in each case with both maximum- and summed intensity variable definition (available as Electronic Supplementary Figure S1, panels a and b).
Both in Supplementary Figure 1a and b and for both variable definition types, it is clear that standard, integral normalization yields a larger amount of substantially high correlations compared to PQ normalization, except for the highest correlations (corresponding to the maximal R values in Table 1). For 240 samples, a R2-value > 0.1 already corresponds with a P value of <10−6, Supplementary Figure 1 therefore demonstrates that after integral normalization of a specific dataset, almost all of its variables are highly correlated with the triglyceride concentration. Since this is biologically more than doubtful, it most likely implies that a large amount of the correlations depicted in Supplementary Figure 1 are artificial, and might be explained by the fact that integral normalization is largely affected by the quantitatively very prominent lipid fraction in blood serum. This introduction of artificial dependencies most likely contaminates the original variables.
Based on Electronic Supplementary Figure S1, it is most likely that also for PQ normalization, an important amount of the correlations is artificially introduced by the normalization process itself, although to a far lesser extent than for integral normalization. This effect was also weakly present for correlations with glucose and virtually absent for creatinine. PQ normalization prebinning yielded results comparable to post-binning (data not shown).
Discussion
NMR spectroscopy is increasingly applied for the simultaneous measurement of a broad spectrum of metabolites in biofluids. The data presented in this manuscript support the concept that 1H-NMR-based metabolomics, with appropriate preprocessing, can be an excellent tool for determining the blood serum phenotype caused by or associated with disease. Indeed, disturbances in the metabolites studied (triglycerides, glucose, creatinine) are for example closely interrelated in the metabolic syndrome, a prediabetic condition predisposing to cardiovascular and renal disease [24].
The study of more metabolites, with independent measurements, would have further increased the ability to make generally applicable conclusions. However, the different chemical properties of the three metabolites under study, their variation in resonance intensities and frequencies, and our rather straightforward results lead us to suggest that these findings already give a clear indication of the effects of the different preprocessing methods. We demonstrated that, for univariate metabolomics data analysis, one should avoid the use of the standard preprocessing methods and prefer applying an intelligent data reduction algorithm, PQ normalization (post-binning), and the use of maximal intensities for each bin. When combined with other appropriate preprocessing procedures, binning improves results compared to the use of complete (i.e., not binned) spectra. Although some of our conclusions can most likely be generalized to other biofluids (e.g., introduction of artificial correlations), such as urine, an important limitation of this research is that our conclusions are only valid for univariate data analysis. For this manuscript, we only evaluated correlations, but we are certain that an optimal correlation, i.e., with a minimal deviation from the actual values, will also yield optimal results for other commonly used univariate methods, such as, for example, t tests.
Our design did not readily allow us to assess the impact of different preprocessing procedures on commonly used multivariate methods, such as principal component analysis (PCA) and partial least squares (PLS) analysis. This was, among others, caused by the large amount of variance orthogonal to the metabolites studied, which has a major effect on these methods. For example, it was impossible to obtain any predictive value when applying PLS regression to estimate creatinine levels, for any of the preprocessing methods evaluated, most likely because the relative variance of the corresponding peaks was too small (data not shown). On the other hand, while experiments involving spiked samples, e.g., [11], represent a departure from normal experimental conditions, they have the major benefit of eliminating these other sources of variance.
Another important reason why PCA- and PLS-based analyses are rather inappropriate in this setting is that the normalization procedure introduced artificial correlations with particularly the larger triglyceride and glucose peaks, making it unclear whether good performance of PLS or PCA might be attributed to proper data preprocessing, whether it is the consequence of the introduction of these artificial correlations.
Zhang and coworkers evaluated different preprocessing methods on urine serum spectra [11]. They concluded that an intelligent data reduction method (they used peak picking, in combination with maximal intensities) should be preferred and that, in the case of univariate analyses, integral normalization yields the best results—although they did not evaluate PQ normalization. Good performance of integral normalization in the case of urine spectra has been reported before [25], but it is clear that the major drawback of this method is the fact that metabolites with (several) large peaks will have a detrimental effect on the normalization factor, both in urine [10, 26] as in the lipid and protein-rich serum, while other methods, such as PQ normalization and histogram matching normalization [26], reduce this effect. However, our data demonstrate that additional optimization of the normalization procedures might be required to fully eliminate the introduction of artifacts in the variance–covariance structure of the resulting dataset. The application of endogenous standards, with independent measurements, might provide a good alternative, under the strict conditions that the corresponding peaks are non-overlapping with other metabolites and, more important, that the optimal normalization factor is resonance intensity independent.
Our results suggest that PQ normalization particularly scores better when applied post-AI-binning (or after the application of equivalent, intelligent data reduction procedures). We propose that this can be attributed to the fact that larger peaks are divided in several variables by equidistant binning, which will—again—introduce a bias in the calculation of the normalization factor.
Although the effect was smaller than for other preprocessing methods, AI binning scored consistently better than the standard method, equidistant binning (matched on other procedures), and scored comparable to or better than lack of binning. The most obvious reason for this is a better variable definition, also in combination with a maximum intensity variable definition. Maxima of the same metabolite tend to fall in the same bin (peak shift effect is reduced, also compared to not binned spectra), and higher intensities of other peaks are generally excluded.
This is important as the use of maxima yields generally good results and scores clearly better for smaller peaks. The latter might be explained by the fact that any remaining baseline or introduced artifact is proportionately small for larger peaks but contributes for a substantial part to the summed intensities of small peaks. The use of maximal intensities will counteract this effect for small peaks but yields marginally worse results for larger peaks, probably because of the fact that summed intensities somewhat reduce the random noise on the individual intensities. These observations lead us to suggest that, when combined with other appropriate preprocessing methods, variable definition by the use of maximum intensities should be generally preferred.
Independent of the preprocessing methods evaluated here, it should be noted that additional optimization of the experimental procedures could also significantly improve the presented results. Optimization studies of the NMR experimental setup, such as [22], combined with independent metabolite measurements, as performed in this study, might yield optimal conditions which further enhance the correlation between NMR metabolomics results and the true metabolite concentrations For example, a longer CPMG pulse train delay (than the 20 ms used here) might yield better macromolecular suppression and a better creatinine quantitation. However, this might also result in an inferior triglyceride quantitation, again illustrating the difficult tradeoff in the quest for an optimal “general” methodology.
Concluding remarks
In summary, this manuscript illustrates the importance of an appropriate choice of preprocessing procedures, since every procedure has a clear effect on the eventual data quality. If data preprocessing is to be followed by univariate data analyses, at least in the case of serum metabolomics, the standard preprocessing methods, i.e., integral data normalization and equidistant (or no binning), should be avoided and replaced by PQ normalization post-AI-binning or equivalent methodologies. Although PQ normalization introduced less artificial correlations than integral normalization, additional optimization of the normalization procedure might further improve results. If low-intensity metabolites also belong to the scope of the experiment, the use of maximal intensities yields the best overall results. It is unclear whether these methods also yield the best results for multivariate analyses; future experiments using other designs, e.g., with spiked-in metabolites, might reveal the answer.
References
Griffin JL (2003) Metabonomics: NMR spectroscopy and pattern recognition analysis of body fluids and tissues for characterisation of xenobiotic toxicity and disease diagnosis. Curr Opin Chem Biol 7:648–654
Holmes E, Nicholls AW, Lindon JC, Connor SC, Connelly JC, Haselden JN, Damment SJ, Spraul M, Neidig P, Nicholson JK (2000) Chemometric models for toxicity classification based on NMR spectra of biofluids. Chem Res Toxicol 13:471–478
Xu J, Yang S, Cai S, Dong J, Li X, Chen Z (2010) Identification of biochemical changes in lactovegetarian urine using 1H NMR spectroscopy and pattern recognition. Anal Bioanal Chem 396:1451–1463
Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q, Ebbels T, De Iorio M, Brown IJ, Veselkov KA, Daviglus ML, Kesteloot H, Ueshima H, Zhao L, Nicholson JK, Elliott P (2008) Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 453:396–400
Lindon JC, Holmes E, Nicholson JK (2005) An overview of metabonomics. In: Robertson DG, Lindon J, Nicholson JK, Holmes E (eds) Metabonomics in toxicity assessment. Taylor & Francis, Boca Raton
Zhao Q, Stoyanova R, Du S, Sajda P, Brown TR (2006) HiRes—a tool for comprehensive assessment and interpretation of metabolomic data. Bioinformatics 22:2562–2564
Xia J, Psychogios N, Young N, Wishart DS (2009) MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res 37:W652–W660
Wang T, Shao K, Chu Q, Ren Y, Mu Y, Qu L, He J, Jin C, Xia B (2009) Automics: an integrated platform for NMR-based metabonomics spectral processing and data analysis. BMC Bioinform 10:83
Izquierdo-Garcia JL, Rodriguez I, Kyriazis A, Villa P, Barreiro P, Desco M, Ruiz-Cabello J (2009) A novel R-package graphic user interface for the analysis of metabonomic profiles. BMC Bioinform 10:363
Craig A, Cloarec O, Holmes E, Nicholson JK, Lindon JC (2006) Scaling and normalization effects in NMR spectroscopic metabonomic data sets. Anal Chem 78:2262–2267
Zhang S, Zheng C, Lanza IR, Nair KS, Raftery D, Vitek O (2009) Interdependence of signal processing and analysis of urine H-1 NMR spectra for metabolic profiling. Anal Chem 81:6080–6088
Dieterle F, Ross A, Schlotterbeck G, Senn H (2006) Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem 78:4281–4290
De Meyer T, Sinnaeve D, Van Gasse B, Tsiporkova E, Rietzschel ER, De Buyzere ML, Gillebert TC, Bekaert S, Martins JC, Van Criekinge W (2008) NMR-based characterization of metabolic alterations in hypertension using an adaptive, intelligent binning algorithm. Anal Chem 80:3783–3790
Anderson PE, Reo NV, DelRaso NJ, Doom TE, Raymer ML (2008) Gaussian binning: a new kernel-based method for processing NMR spectroscopic data for metabolomics. Metabolomics 4:261–272
Torgrip RJO, Lindberg J, Linder M, Karlberg B, Jacobsson SP, Kolmert J, Gustafsson I, Schuppe-Koistinen I (2006) New modes of data partitioning based on PARS peak alignment for improved multivariate biomarker/biopattern detection in H-1-NMR spectroscopic metabolic profiling of urine. Metabolomics 2:1–19
Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM (2006) Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal Chem 78:4430–4442
Alm E, Torgrip RJO, Aberg KM, Schuppe-Koistinen I, Lindberg J (2009) A solution to the 1D NMR alignment problem using an extended generalized fuzzy Hough transform and mode support. Anal Bioanal Chem 395:213–223
Alm E, Torgrip RJ, Aberg KM, Schuppe-Koistinen I, Lindberg J (2010) Time-resolved biomarker discovery in (1)H-NMR data using generalized fuzzy Hough transform alignment and parallel factor analysis. Anal Bioanal Chem. doi:10.1007/s0021600934215
Veselkov KA, Lindon JC, Ebbels TM, Crockford D, Volynkin VV, Holmes E, Davies DB, Nicholson JK (2009) Recursive segment-wise peak alignment of biological (1)H NMR spectra for improved metabolic biomarker recovery. Anal Chem 81:56–66
Lindon JC, Nicholson JK, Everett JR (1999) NMR spectroscopy of biofluids. In: Webb GA (ed) Annual reports on NMR spectroscopy, vol 38. Academic, London
Rietzschel ER, De Buyzere ML, Bekaert S, Segers P, De Bacquer D, Cooman L, Van Damme P, Cassiman P, Langlois M, Van Oostveldt P, Verdonck P, De Backer G, Gillebert TC (2007) Rationale, design, methods and baseline characteristics of the Asklepios study. Eur J Cardiovasc Prev Rehabil 14:179–191
Lucas LH, Larive CK, Wilkinson PS, Huhn S (2005) Progress toward automated metabolic profiling of human serum: comparison of CPMG and gradient-filtered NMR analytical methods. J Pharm Biomed Anal 39:156–163
Nicholson JK, Foxall PJ, Spraul M, Farrant RD, Lindon JC (1995) 750 MHz 1H and 1H–13C NMR spectroscopy of human blood plasma. Anal Chem 67:793–811
Ritz E (2007) Metabolic syndrome: an emerging threat to renal function. Clin J Am Soc Nephrol 2:869–871
Schnackenberg LK, Sun J, Espandiari P, Holland RD, Hanig J, Beger RD (2007) Metabonomics evaluations of age-related changes in the urinary compositions of male Sprague Dawley rats and effects of data normalization methods on statistical and quantitative analysis. BMC Bioinform 8(Suppl 7):S3
Torgrip RJO, Aberg KM, Alm E, Schuppe-Koistinen I, Lindberg J (2008) A note on normalization of biofluid 1D H-1-NMR data. Metabolomics 4:114–121
Acknowledgments
The authors would like to thank Dimitri Broucke, Frida Brusselmans, and Fien De Block for their excellent technical assistance.
This work was supported by grants from the Research Foundation–Flanders (FWO Vlaanderen; G.0427.03; FWO09/PDO/056) and the Special Research Fund of Ghent University (BOF, 011D10004). The Research Foundation–Flanders is also greatly acknowledged for a Ph.D. fellowship to DS, a post-doctoral grant to TDM (FWO09/PDO/056), and a research grant to JCM (G.0064.07). The 700 MHz equipment of the Interuniversitary NMR Facility was jointly financed by Ghent University, the Free University of Brussels, and the University of Antwerp via the FFEU-ZWAP incentive of the Flemish Government.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary materials
Below is the link to the electronic supplementary material.
ESM 1
(PDF 730 kb)
Rights and permissions
About this article
Cite this article
De Meyer, T., Sinnaeve, D., Van Gasse, B. et al. Evaluation of standard and advanced preprocessing methods for the univariate analysis of blood serum 1H-NMR spectra. Anal Bioanal Chem 398, 1781–1790 (2010). https://doi.org/10.1007/s00216-010-4085-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-010-4085-x