Abstract
Bare FePtCu nanoparticles (NPs) are first prepared for laser desorption/ionization mass spectroscopy (LDI-MS) analysis as affinity probes to selectively trap oppositely charged analytes from a sample solution. Our present results demonstrate bare FePtCu NPs to be a potentially useful matrix for surface-assisted laser desorption/ionization mass spectroscopy (SALDI-MS), for the analysis of small proteins and peptides. The upper detectable mass range of peptides was approximately 5 kDa, and the detection limit for peptides approximately 5 fmol. Sulfonate group-modified FePtCu nanoparticles (FePtCu-SO −3 NPs), with ionization being independent of the solution pH, can interact with a positively charged analyte, and the analyte-bound NPs can be separated from the reaction supernatant by centrifugation or an external magnetic field. An oligopeptide, Gly-Gly-Tyr-Arg (GGYR) from an oligopeptide mixture containing Asp-Asp-Asp-Asp (DDDD), Gly-Gly-Gly-Gly (GGGG) and GGYR, was detected using SALDI-MS with FePtCu-SO3(−) NPs employing electrostatic interaction. Furthermore, FePtCu-SO −3 NPs can detect lysozyme (Lyz) in human serum through the electrostatic attraction between positively charged Lyz and FePtCu-SO3(−) NPs at pH 8, while detection of negatively charged albumin in human serum is not possible.

Sulfonate group-modified FePtCu nanoparticles as a selective probe for LDI-MS
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) is extensively used in biochemical analysis and is an indispensable tool in proteomic research [1–7]. In a MALDI experiment, analyte molecules are mixed with a large molar excess of organic matrix, and the analyte molecules are thereby embedded throughout the matrix and separated from one another. Using the matrix increases the efficiency of energy transfer from the pulse laser to the analyte, and thus enhances the desorption/ionization efficiency. In this way, the desorption of analyte molecules is assisted by the matrix, making possible analysis of compounds with a molecular weight in excess of 300,000 Da. MALDI also has a higher tolerance to salts and buffers than electrospray ionization (ESI), another soft ionization technique. Despite the success of MALDI, three major problems associated with the use of organic matrixes are frequently encountered: (1) matrix optimization, (2) uneven analyte distribution, and (3) matrix interference. In particular, the MALDI mass spectra have a high chemical background noise due to the formation of clusters of matrix ions or degradation products of the matrix. The matrix background peaks appears in the low-mass region, i.e., below m/z 700, and MALDI is thus normally employed for macromolecules such as proteins synthetic polymers, and for oligonucleotide analysis.
Inorganic matrixes are more useful than organic matrixes such as α-cyano-4-hydroxycinnamic acid (CHCA) and 2,5-dihydroxybenzoic acid (DHBA) for the analysis of small molecules in LDI-MS [8]. This concept was originally introduced by Tanaka et al. using 30 nm cobalt powder suspensions in glycerol [1]. Surface-assisted laser desorption/ionization (SALDI), developed by Sunner et al. [9], utilizes graphite particles of micron-scales as the LDI-assisted material. More recently, to meet the requirement for analysis of small organic molecules, the applicability of SALDI without a liquid matrix such as glycerol has been studied using various nanomaterials including carbon nanotubes (CNT) [10], ZnO [11], TiO2 [12], Fe2O3/TiO2 [13], platinum [14], and gold nanoparticles (AuNPs) [15–18]. These strategies have included the use of desorption/ionization on porous silicon (DIOS) [19–21].
For analyses of complex biological samples, sample preparation also plays a critical role in obtaining good-quality mass spectra. In LDI-MS including SALDI and MALDI, signal suppression from multiple components in biological samples is a significant disadvantage. Thus, sample separation and enrichment of the analytes of interest from sample mixtures is an important step in detecting these analytes in the LDI-MS. Nanoparticles have enormous potential in sample pretreatment, especially for trace analyte enrichment, because functionalization of the nanoparticle surface provides the additional advantage of high capacity. Recently, various nanoparticles such as Au [22, 23], Ag [24, 25], silica [26], silanized iron oxide [27], and magnetic NPs coated with titania [13], zirconia [28], and alumina [29] have thus been employed for the separation and the enrichment of biomolecules from mixtures in LDI-MS analysis.
FePt NPs or FePtCu NPs are reportedly applicable to data storage and are the only permanent magnetic nanocomposites as yet established [30–33]. However, we consider FePt NPs and FePtCu NPs to also be promising affinity probes for LDI-MS, since these NPs can be modified with thiol-terminated molecules via the formation of Pt–S and Fe–S bonds [31]. The analyte-bound NPs can be separated from the reaction supernatant by centrifugation as well as an external magnetic field. However, there are no applications for affinity probes comprised of FePt NPs or FePtCu NPs in LDI-MS analysis of biomolecules. FePt NPs also generally interact with alkyl carboxylic acid (RCOOH), and alkylamine (RNH2): –COOH can covalently link with Fe, forming iron carboxylate (–COO–Fe), and –NH2, as an electron donor, preferentially binds to Pt via a coordination bond. Such interactions of FePt with amine- and carboxylic acid-containing biomolecules may allow selective affinity probes, i.e., for specific analytes, to be derived from complex samples.
In the present study, bare FePtCu NPs were newly prepared for LDI-MS analysis as affinity probes to selectively trap oppositely charged analytes from the sample solution. This work is, to our knowledge, the first demonstration that sulfonate group-modified FePtCu nanoparticles (FePtCu-SO3(−) NPs) are a potentially useful SALDI matrix which can be successfully utilized, without an elution step, for the separation and detection of a target oligopeptide, from an oligopeptide mixture, and lysozyme (Lyz) from human serum.
Experiment
Materials
Iron dichloride (FeCl2∙4H2O), chloroplatinic acid (H2PtCl6∙H2O), copper sulfate, (CuSO4∙5H2O), hydrazine (N2H4∙H2O), 0.1 M hydrochloric acid (HCl), ammonia water (25%), citric acid, nonafluoro-1-butanesulfonic acid (PFBS), trifluoroacetic acid (TFA), human insulin, dextromethorphan hydrobromide, caffeine, chicken egg-white Lyz, ethanol (99.6%), methanol (HPLC grade), and acetone (99.9%) were purchased from Wako Pure Chemical Industries Ltd. (Osaka, Japan). CHCA, amitriptyline hydrochloride, and ammonium citrate dibasic were purchased from Sigma-Aldrich (St. Louis, MO). Water (LC-MS grade) was purchased from Kanto Chemical Industries Ltd. (Tokyo, Japan). Cytochrome C was purchased from Calbiochem. Co. Asp-Asp-Asp-Asp (DDDD), Gly-Gly-Gly-Gly (GGGG), Gly-Gly-Tyr-Arg (GGYR), and Gly-Gly-His (GGH) were purchased from Peptide Institute, Inc. (Osaka, Japan). Human serum was purchased from CHEMICON International. 2-Mercaptoethanesulfonic acid sodium salt (HSCH2CH2SO3Na; >95%) was obtained from Tokyo Chemical Industries Ltd. (Tokyo, Japan).
Fabrication of Bare FePtCu NPs
FePtCu nanoparticles were prepared by reduction of metallic salts using hydrazine in aqueous medium, followed by modification with the method of Gibot et al. [34]. FeCl2∙4H2O (2 mmol), H2PtCl6∙H2O (2 mmol), and CuSO4∙5H2O (0.48 mmol) were dissolved in 100 mL of water at 70 °C, to achieve concentrations of 20 mM, 20 mM, and 4.8 mM, respectively. An aliquot (10 mL) of a freshly prepared aqueous solution of N2H4 (0.4 M, reduction agent) was rapidly added to the metal solution under vigorous stirring (~600 rpm). The mixture was then maintained at 70 °C for three hours under vigorous stirring. FePtCu NPs thus prepared were rinsed several times with water until the electric conductivity of metal ions in the supernatant was less than 1 mS/m, and then finally rinsed several times with pure ethanol.
To avoid complete aggregation of NPs, vigorous stirring was very important at the injection of reduction agent into the aqueous solution of metal salts. If the FePtCu NPs are synthesized under gentle stirring, a macroscopic size of large aggregates is formed. Such large aggregates cannot any disperse in the aqueous solution of peptides and proteins, resulting in no use as the NPs for the affinity studies.
Surface modification of FePtCu NPs with thiol sulfonate compound
The FePtCu NPs (100 mg) were re-suspended in 50 mL of ethanol by ultrasonic sonication for 10 min. An aqueous solution of HS (CH2)2SO3Na (1 M, 5.6 mL) was rapidly added to this solution under vigorous stirring (~800 rpm) at room temperature. Next, water (10 mL) was added to the solution and the mixture was then maintained at room temperature for 24 h under vigorous stirring. The sulfonate-modified FePtCu NPs [FePtCu-SO3(−)] were rinsed several times with water until the electric conductivity of metal ions in the supernatant was less than 1 mS/m, then finally rinsed several times with acetone and ethanol. FePtCu-SO3(−) NPs were dried to obtain NPs powders. The FePtCu-SO −3 NPs powders were stored at approximately −30 °C in a freezer before use for LDI-MS.
SALDI-MS
LDI mass spectra were acquired in linear mode on an AXIMA-CFR time-of-flight mass spectrometer (Shimadzu/Kratos, Manchester, UK) with a pulsed nitrogen laser (λ = 337 nm). One hundred laser shots were used to acquire the mass spectra. The analyte ions were accelerated at 20 kV under delayed extraction conditions. We do not know the actual laser fluence value of the instrument, although we do have the relative values, denoted by laser power (LP). The peak intensities of analytes are denoted by millivolts in mass spectra of Figs. 3, 4, 5, 6, and 8 in the present paper. The two-layer sample preparation method was employed for SALDI-MS with the FePtCu NPs: the first step was spotting of the FePtCu NPs solution (1 μL, 2 mg/mL), prepared by dispersing FePtCu NPs in water with ultrasonication for 5 min, onto a stainless steel plate, followed by drying; the second step was typically deposition of a 1.0 μL sample solution on the plate. To use citrate buffer as the proton source, the sample aqueous solution was mixed with a citrate aqueous solution [triammonium citrate (50 mM)/citric acid (100 mM), 3:1 (v/v)] for SALDI-MS analysis.
Sample extraction with FePtCu-SO3(−) NPs and LDI-MS
Oligopeptides
The pH values of stock aqueous solutions (100 μM or 1 μM) of peptides were adjusted to achieve a given pH value with HCl or ammonia water. A 100-μL solution of FePtCu-SO3(−) NPs (2 mg/mL) was added to this 1 mL stock solution of peptides, and the mixture was vortexed for 10 min. The peptide-bound NPs were separated from the supernatant by centrifugation (~6,000 rpm) for 1 min or by using an external magnetic field for 10 minutes. The peptide-bound NPs were further washed three times with water, having the same pH as the extraction solution. Finally, the 100-μL residue solution including 0.1% NBA and peptide-bound NPs was analyzed by SALDI-time-of-flight (TOF) MS.
Lysozyme in human serum
Human serum containing Lyz (10 μM and 1 μM) was prepared at pH 8. A 200-μL solution of FePtCu-SO3(−) (2 mg /mL) was added to the serum solution (1 mL), and the mixture was vortexed for 60 min. The Lyz-bound NPs were separated from the reaction supernatant by centrifugation (~6000 rpm) for 1 min. Then, 100 μL of water were added to this residue (i.e., the Lyz-bound NPs) followed by vortexing for 5 min. The Lyz-bound NPs were further washed three times with water at pH 8. The 1-μL suspension of Lyz-bound NPs and 1 μL of CHCA matrix (20 mg/mL, water/methanol mixture: 3:7) were applied to the MALDI steel plate and dried at room temperature. The Lyz bound to the NPs was analyzed by MALDI-time-of-flight (TOF) MS with the CHCA matrix. For comparison, human serum containing 10 μM of Lyz was analyzed by MALDI-TOF MS without the above-described sample extraction.
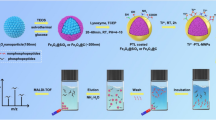
A schematic summary of these experimental procedures is shown in Fig. 1.
FT-IR attenuated total reflectance (FT-IR-ATR) spectroscopy
FT-IR-ATR spectroscopy was performed on a Jasco FT-IR 4200 spectrometer with an ATR attachment (ATR8100H) equipped with a mercury-cadmium-telluride (MCT) detector.
Electron microscopy
SEM imaging and EDX analysis of the FePtCu were carried out with a JEOL JSM-7401FNT at 15 kV. The TEM was performed at the Institute of Neurology and Genetics, Cyprus, on a JEOL-1010A instrument with an acceleration voltage of 120 kV. Drops of the solutions were placed on formvar-coated grids and were allowed to remain on these grids for a few minutes to facilitate water evaporation. The solutions were diluted with sufficient amounts of water before being examined by TEM.
Results and discussion
Characterization of FePtCu NPs
Figure 2 shows TEM and SEM images of FePtCu NPs. The size distribution of the NPs was broad, ranging from 20 to 100 nm. The EDX analysis indicated that the NPs consisted of three components, Fe, Pt, and Cu, but that the chemical component distribution of NPs is size-dependent (see supporting information). The smaller NPs (~30 nm) have a Pt rich component, while the large NPs (~100 nm) have a Fe-rich component. Thus, the surfactant free FePtCu prepared in this study was inhomogeneous in terms of chemical components and size, but this inhomogeneity was not a serious problem in using the NPs for performing SALDI. In fact, the use of sodium dodecylsulfate (SDS) surfactant for this method of synthesis produced FePtCu NPs with an average size of about 20 nm and the chemical compositions of these NPs were almost the same. However, the use of SDS surfactant-capped FePtCu NPs as the SALDI matrix was disadvantageous in terms of background noise, in the low-mass region, from the surfactant (observations not shown).
It should be noted that there is no direct evidence to show that the individual nanoparticles are actually FePtCu NPs. But we consider that the NPs are not simply a mixture of individual Pt, Cu, and Fe NPs from the following reasons.
-
1.
FePtCu NPs prepared in this study were rinsed several times with water. At that time, we separated the NPs from the supernatant by an external magnetic field. From this operation, individual Pt and Cu NPs without magnetic property are not included in the NPs, indicating that the NPs contains Fe component.
-
2.
From the EDX analysis, the Cu component of the NPs was almost the same, irrespective of the size of NPs (supporting information). That is, all NPs have the Cu component.
-
3.
The atom weight percent of individual Pt, Cu, and Fe from the EDX for the SEM image (supporting information) were 61.76, 16.41, and 21.83%, respectively. In the conversion to the mole ratio, Pt (M = 195.1), Cu (M = 63.55) and Fe (M = 55.85) were roughly estimated to be 0.32 (61.76/195.1), 0.26 (16.41/63.55), and 0.39 (21.83/55.85), respectively. The result of almost equal mole existence for these metals might reflect that the NPs are actually FePtCu NPs, but the ratio of individual metals depends on the size of NPs from the EDX image.
FePtCu NPs had magnetic properties. Hence, the FePtCu particles could be isolated from sample solutions using an external magnetic field and could be used as concentrating probes for the separation and purification of specific analytes from sample solutions. It is noteworthy that in place of FePt NPs, we used FePtCu NPs as the SALDI matrix, because of the weak magnetic property of FePt NPs with this preparation method.
Surface modification of FePtCu NPs with HS(CH2)2SO3Na was confirmed by absorbance from the ethylene chain around 2,900 cm−1 and the sulfonate group around 900 cm−1 using FT-IR-ATR (data not shown).
SALDI performance using bare FePtCu NPs
Peptides and proteins
Figure 3 displays the SALDI mass spectrum of angiotensin I (500 fmol) obtained using bare FePtCu NPs mixed with citrate buffer as the proton source. The mass spectrum shows the protonated molecular ion of angiotensin I at m/z 1,297. The detection limit of this peptide was 5 fmol, which is compatible with the value obtained using Fe2O3 NPs (~20 fmol) [27]. Small proteins like insulin (500 fmol) can also be detected, but larger proteins such as cytochrome C (500 fmol) were not detectable using FePtCu NPs. In contrast to the Fe2O3 SALDI [27], a series of iron–analyte adducts was not observed in the mass spectrum. The above results indicate that FePtCu NPs can be used as a SALDI matrix.
It is worth mentioning that the only reason for the multimetallic particles is not their magnetic properties. The use of the multimetallic particles increases the SALDI efficiency compared to the use of individual Pt, Cu, and Fe nanoparticles as the follows: the SALDI-MS of using Cu NPs could not detect angiotensin I [35]. In contrast, the use of the FePtCu NPs made it possible to detect the peptides with the detection limit of 5 fmol. The SALDI-MS of using the aggregates of spherical Pt NPs could not detect insulin [14], in contrast to the detection of insulin using FePtCu.
Small molecules (<500 Da)
Figure 4 shows the SALDI mass spectra of two small drug molecules: (a) dextromethorphan hydrobromide (1 pmol) and (b) amitriptyline hydrochloride (1 pmol). Compared to the MALDI mass spectra obtained with the CHCA matrix, it is clear that the matrix interferences are relatively low for the SALDI mass spectra using FePtCu. As might be expected, the detection of small salt ions is relatively easy with SALDI using FePtCu. However, detection of the proton adduct form of oligopeptides was difficult using SALDI-MS with FePtCu employing a typical proton source of TFA. Citrate buffer is known to be an effective proton source for SALDI-MS of peptides [27, 35–37], but the use of this buffer may be unfavorable for SALDI-MS of small peptides due to the appearance of strong molecular ion peaks of citrate in the low-mass region. In this study, therefore, nonafluoro-1-butanesulfonic acid (PFBS) was used as the proton source in place of citrate buffer. Figure 5 shows the SALDI mass spectra of GGH (100 pmol) obtained using different proton source agents, i.e., (a) TFA, (b) citrate buffer and (c) PFBS. Using PFBS, the mass spectrum contains only the protonated molecular ion of GGH at m/z 270, while the sodium or potassium–analyte adduct ions become the dominant peaks with the use of TFA. When using citrate buffer, the protonated molecular ion of GGH appears in the mass spectrum, but the sodium- and potassium-citrate adduct ions also are quite apparent at m/z 215 and 231, respectively. These results demonstrate that the addition of PFBS can significantly reduce metal ion adducts and thereby enhance the yield of the protonated molecular ion.
Extraction of target molecules using FePtCu NPs and detection by LDI-MS
Peptide mixtures
FePtCu-SO3(−) NPs were prepared by surface modification with HS(CH2)2SO3Na to extract the target oligopeptide through complementary electrostatic interactions. First, to confirm oligopeptide extraction efficiency employing the electrostatic interaction, the SALDI ion abundance of GGYR with an isoelectric point (pI) of 9.4 was examined as a function of the extraction solution pH. The ionization state of FePtCu-SO3(−) NPs is essentially independent of the solution pH from 3 to 10 because of the strong electrolyte action of the sulfonate group. At pH values of 4, 6, and 8, which are all below the pI, the SALDI mass spectra of GGYR (500 fmol) showed the ion signal of GGYR in the mass spectrum, whereas there was no signal at pH 10 which is above the pI (Fig. 6). This is because GGYR has a net positive charge at pH values below the pI and can be extracted efficiently with negatively charged NPs. As we confirmed the oligopeptide extraction efficiency employing the electrostatic interaction using FePtCu-SO3(−) NPs, the ability of the NPs to selectively extract oligopeptides from a mixture was studied using a 1 μM oligopeptide mixture of DDDD (pI = 3.2), GGGG (pI = 6.1), and GGYR (pI = 9.4). Figure 7a shows the SALDI mass spectra of the oligopeptide mixture of DDDD, GGGG, and GGYR under different conditions of pH 2.5, 8, and 10. With the strongly negatively charged NPs, positively charged oligopeptides were detectable at pH values below the pI. For example, at pH 8 only GGYR has a net positive charge and thus the ion signal of GGYR alone appeared in the mass spectrum, while there were no signals of DDDD and GGGG which have a net negative charge at pH 8.
Lysozyme in human serum
As Lyz is a high-pI enzyme that hydrolyzes the polysaccharide walls of bacteria, it is widely distributed in body tissues and secretions. Nevertheless, Lyz is present in low concentrations in urine and serum. Increased Lyz concentrations in urine and serum have been recognized as being associated with leukemia [38], renal diseases [39], and meningitis [40]. We therefore applied the extraction approach using FePtCu-SO3(−) NPs to extract Lyz (pI = 11) from human serum. Human albumin is the most abundant of proteins, and the suppression of albumin binding to nanoparticles is hence important for selectively detecting Lyz in human serum. FePtCu-SO3(−) NPs would be expected to detect Lyz in human serum through an electrostatic attraction between the positively charged Lyz and the FePtCu-SO3(−) NPs, while there would be no interaction with the negatively charged albumin in human serum at pH 8. Figure 8 shows the MALDI mass spectra of Lyz (a) before and (b) after extraction. Due to the presence of numerous plasma components, Lyz in 10 μM Lyz-spiked human plasma was barely detectable prior to affinity extraction (Fig. 8a). Following the extraction employing FePtCu-SO3(-) NPs, the proton adduct Lyz around m/z 14304 for the single-proton adduct, as well as at m/z 7153 and 4769 for the multiple proton adducts, is prominent in the MALDI spectrum (Fig. 8b). The Lyz from the 1 μM Lyz-spiked human plasma was undetectable even after affinity extraction (not shown). These results demonstrate that FePtCu-SO3(−) NPs can serve as a useful matrix for the selective detection of positively charged proteins in a complex mixture.
Conclusion
We have demonstrated that sulfonate-modified FePtCu nanoparticles (NPs) can be used as a SALDI matrix and as affinity probes. The strongly negatively charged surfaces of FePtCu NPs are capable of concentrating traces of positively charged species from aqueous solutions. A given oligopeptide from an oligopeptide mixture and lysozyme from human serum can be enriched using sulfonate-modified FePtCu NPs and directly analyzed by applying LDI-MS. Bare FePtCu NPs can be modified with various thiol-terminated molecules, via the formation of Pt–S or Cu–S bonds, to serve as affinity probes. These surface-modified FePtCu NPs are thus useful for LDI-MS analysis. Employing surface-modified FePtCu NPs combines rapid identification of analytes by SALDI-MS and MALDI-MS, thereby providing a convenient, effective, and specific bioassay for selectively trapping positively charged analytes from a sample solution without the need for an elution step.
References
Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yoshida T (1988) Rapid Commun Mass Spectrom 2:151–153
Karas M, Bachmann D, Hillenkamp F (1985) Anal Chem 57:2935–2939
Nielen MW (1999) Mass Spectrom Rev 18:309–344
Hanton SD (2001) Chem Rev 101:527–570
Deisewerd K (2003) Chem Rev 103:395–425
Karas M, Kruger R (2003) Chem Rev 103:427–439
Knochenmuss R, Zenobi R (2003) Chem Rev 103:441–452
Peterson DS (2007) Mass Spectrom Rev 26:19–34
Sunner J, Dratz E, Chen Y-C (1995) Anal Chem 67:4335–4342
Ugarov MV, Egan T, Khabashesku DV, Schultz JA, Peng H, Khabashesku VN, Furutani H, Prather KS, Wang HWJ, Jackson SN, Woods AS (2004) Anal Chem 76:6734–6742
Watanabe T, Kawasaki H, Yonezawa T, Arakawa R (2008) J Mass Spectrom 43:1063–1071
Chen CT, Chen YC (2004) Anal Chem 76:1453–1457
Chen CT, Chen YC (2005) Anal Chem. 77:5912–5919
Kawasaki H, Yonezawa T, Watanabe T, Arakawa R (2007) J Phys Chem C 11:16278–16283
McLean JA, Stumpo AA, Russell DH (2005) J Am Chem Soc 127:5304–5305
Huang YF, Chang HT (2006) Anal Chem 78:1485–1493
Wu HP, Su CL, Chang HC, Tseng WL (2007) Anal Chem 79:6215–6221
Kawasaki H, Sugitani T, Watanabe T, Yonezawa T, Moriwaki H, Arakawa R (2008) Anal Chem 80:7524–7533
Wei J, Buriak JM, Siuzdak G (1999) Nature 399:243–246
Shen Z, Thomas JJ, Averbuj C, Broo KM, Engelhard M, Crowell JE, Finn MG, Siuzdak G (2001) Anal Chem 73:612–619
Go EP, Prenni JE, Wei J, Jones A, Hall SC, Witkowska HE, Shen Z, Siuzdak G (2003) Anal Chem 75:2504–2506
Teng CH, Ho KC, Lin YS, Chen YC (2004) Anal Chem 76:4337–4342
Vanderpuije Benjamin NY, Han G, Rotello VM, Vachet RW (2006) Anal Chem 78:5491–5496
Sherrod SD, Diaz AJ, Russell WK, Cremer PS, Russell DH (2008) Anal Chem 80:6796–6799
Chiu TC, Chang LC, Chiang CK, Chang HT (2008) J Am Soc Mass Spectrom 19:1343–1346
Agrawal K, Wu HF (2008) Rapid Commun Mass Spectrom 22:283–290
Chen WY, Chen YC (2006) Anal Bioanal Chem 386:699–704
Li Y, Leng T, Lin H, Deng C, Xu X, Yao N, Yang P, Zhang X (2007) J Proteome Res 6:4498–4510
Chen CT, Chen WY, Tsai PJ, Chien KY, Yu JS, Chen YC (2007) J Proteome Res 6:316–325
Sun S (2006) Adv Mater 18:393–403
Gu H, Zheng R, Zhang X, Xu B (2004) J Am Chem Soc 126:5664–5665
Shukla N, Liu C, Jones PM, Weller D (2003) J Magn Magn Mater 266:178–184
Xu C, Xu K, Gu H, Zhong X, Guo Z, Zheng R, Zhang X, Xu B (2004) J Am Chem Soc 126:3392–3393
Gibot P, Tronc E, Chanéac C, Jolivet JP, Fiorani D, Testa AM (2005) J Magn Magn Mater 290:555–558
Yonezawa T, Kawasaki H, Tarui A, Watanabe T, Arakawa R, Shimada T, Mafune F (2009) Anal Sci 25:339–346
Chen WY, Wang LS, Chiu HT, Chen YC, Lee CY (2004) J Am Soc Mass Spectrom 15:1629–1635
Tarui A, Kawasaki H, Taiko T, Watanabe T, Yonezawa T, Arakawa R (2009) J Nanosci Nanotechnol 9:159–164
Levinson SS, Elin RJ, Yam L (2002) Clin Chem 48:1131–1132
Harrison JF, Lunt GS, Scott P, Blainey JD (1968) Lancet 1:371–375
Klockars M, Reitamo S, Weber T, Kerttula Y (1978) Acta Med Scand 203:71–75
Acknowledgments
We are grateful to Ms. Michiyo Sato for the SEM measurements. This work was partially supported by Grants-in-Aid for Scientific Research (B), for Young Scientists (B) (No. 20710091, 21310072, and 19350045) and for Priority Areas (“Strong Photon-Molecule Coupling Fields for Chemical Reactions (470, No. 21020010) and “Molecular Science for Supra Functional Systems” (477, No. 20050007)) from the Japan Society for the Promotion of Science and the Ministry of Education, Culture, Sports, Science, and Technology-Japan (MEXT). This research was also partly supported by the Core-to-Core Program promoted by the Japan Society for the Promotion of Science (Project No.18004) and by the Strategic Project to Support the Formation of Research Bases at Private Universities: Matching Fund Subsidy from MEXT.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kawasaki, H., Akira, T., Watanabe, T. et al. Sulfonate group-modified FePtCu nanoparticles as a selective probe for LDI-MS analysis of oligopeptides from a peptide mixture and human serum proteins. Anal Bioanal Chem 395, 1423–1431 (2009). https://doi.org/10.1007/s00216-009-3122-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-009-3122-0