Abstract
Near infrared spectroscopy (NIR) has been used to determine important indicators of the quality of beers, for example original and real extract and alcohol content, using a partial least squares (PLS) calibration approach. A population of 43 samples, obtained commercially in Spain and including different types of beer, was used. Cluster hierarchical analysis was used to select calibration and validation data sets. Absorbance sample spectra, in transmission mode, were obtained in triplicate by using a 1-mm pathlength quartz flow cell and glass chromatography vials of 6.5 mm internal diameter. The two methods of sample introduction were compared critically, on the basis of spectral reproducibility for triplicate measurements and after careful selection of the best spectral pre-processing and the spectral range for building the PLS model, to obtain the best predictive capability. For each mode of sample introduction two calibration sets were assayed, one based on the use of 15 samples and a second extended based on use of 30 samples, thus leaving 28 and 13 samples, respectively, for validation. The best results were obtained for 1 mm flow cell measurements. For this method original zero-order spectra data in the ranges 2220–2221 and 2250–2350 nm were chosen. For the real extract, original extract, and alcohol dx-y and sx-y values of −0.04 and 0.07% w/w, −0.01 and 0.13% w/w, and −0.01 and 0.1% v/v, respectively, were obtained. The maximum errors in the prediction of any of these three indicators for a new sample were 2.2, 1.2, and 1.9%, respectively. This method compares favorably with the automatic reference method in terms of speed, reagent consumed, and waste generated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Beer is an alcoholic beverage obtained by fermentation, in the presence of yeast, of cereals germinated in water [1]. The main components of beers are water, carbohydrates and ethanol. These three components are commonly used in the brewing industry for quality control of the final product, based on the determination of real extract, original extract, and ethanol content. Original and real extract correspond to the amount of sugars in the beers before and after the fermentation process, respectively, and are normally expressed in % w/w. Ethanol content is a key economic and organoleptic property affecting both beer classification (in term of taxes) and taste [1, 2]. It is usually expressed in % v/v. The real extract is important because it is a measure of the amount of sugars that did not undergo fermentation and remains in the beer. This property tells consumers the sweetness of the product and its energy value as source of carbohydrates. Determination of the original extract and the ethanol content together with the real extract are important for knowing the fermentation grade of the beer to determination the efficiency factor based on the fermentation yield.
The official methods of the analytical division of the European Brewery Convention for the determination of the aforementioned quantities are based on distillation of the beer and measurement of the density of the distillate and the remaining solution after diluting to standard final volumes [3]. The densities of both solutions are compared with data in semi-empirical tables to obtain the percentage of extracts and the ethanol content. These methods involve much sample handling and are time-consuming and therefore inefficient in terms of time and cost. There is therefore a need to develop rapid alternative methods for routine quality control programs.
Use of autoanalyzers in the brewing industry reduces sample handling but involves an analysis time at 30 min to obtain triplicate values of each quantity. There are precedents for determination of ethanol in beers by use of direct FTIR [4] or near infrared spectroscopy (NIR) measurements [5, 6, 7] at selected wavelengths using external calibration and based on classical least square monoparametric treatments. It is, however, clear that for monitoring the brewing process and for rapid quality control, simultaneous determination of real and original extracts and alcohol content are needed and for that the use of multivariate calibration models is required.
To the best of our knowledge the papers reported in Table 1 report the most relevant work performed to develop alternative methods based on NIR measurements and chemometrics. This table summarizes the most relevant aspects of the papers indicating the number of samples used for both calibration and validation, the type of samples considered, the number of factors selected for each determination, the best spectral range selected, and the prediction error found for each.
In addition to these papers, Mendes et al. [8] studied the determination of ethanol in fuel ethanol and some beverages based upon NIR-PLS and Raman-PLS. This work shows that this determination can be achieved successfully, although assessment of predictive capability for beers must be regarded as far from exhaustive because it was proved for a single sample only. Schropp et al. [9] evaluated an Anton Paar NIR system for evaluation of beer quality.
In the study of Maudoux et al. [10], it was reported that alcohol, real extract, original gravity (original extract), nitrogen, and polyphenols could be estimated from the transmission NIR spectra of samples by use of multiple linear regression and partial least squares (PLS) analysis by using actual beer samples for calibration. Papers by Norgaard et al. [11] and Westad and Martens [12] focus solely on original extract determination.
Despite the few references found for determination of real and/or original extract and ethanol in beers by NIR it is clear that this method combines the high sample throughput achievable by spectroscopic techniques with the capability of multi-property determination of autoanalyzers.
All previously published work on these determinations used rectangular quartz cuvettes of 1 [10], 10 [12], or 30 mm [11] pathlength, thus requiring a washing step to avoid cross-contamination. The washing steps slow down the speed of the analysis and may be avoided by using individual vials for each sample. These could be used both to obtain the transmission spectra and as containers for keeping the samples for the period of time required by good laboratory practice. To the best of our knowledge, there is no report on the effect of using individual glass vials for PLS–NIR determination of beer quality.
This work was therefore devoted to the development of a method based upon NIR measurements and chemometry for evaluation of beer quality. To increase the range of application of the multivariate model, a heterogeneous sample population was chosen for selecting the calibration and the validation datasets. The process of selection of each sample set was carried out using hierarchical cluster analysis [13], which was based on the classification of the beers obtained from their NIR spectra. Different stages in the development of a PLS–NIR method for prediction of ethanol and real and original extracts are discussed. Analytical figures of merit, based on net analyte signal calculations [14], were obtained for the sensitivity and selectivity of the determination of original and real extracts and the ethanol content in beers.
These studies focused on two different sample-measurement modes, one based on the use of a flow cell and the second based on the use of glass chromatography vials. The data obtained from both were critically compared.
Experimental
A Fourier transform NIR Brucker MPA spectrometer controlled by Opus for Windows software from Brucker Optik (Bremen, Germany) and circular cell holder was used for acquisition of spectra. A 1.00 mm pathlength (50 μL volume) quartz flow cell from Hellma (Müllheim, Germany) was mounted on a homemade adaptor. For vial experiments an MPA heatable vial cell holder (Bruker) for vials of 8.2 mm o.d. was used. The vial fits tightly inside the holder and no lateral movement can be observed (only spinning of the vial is allowed). The vial temperature was set to 32°C. Room temperature was monitored using a mercury thermometer with a precision of ±0.5°C and it did not vary significantly during acquisition of the spectra of all the samples. For sample preparation, a magnetic stirrer (IKA, Staufen, Germany) and a thermostatic bath (Grant, Cambridge, UK) were used. A Gilson Minipuls 2 peristaltic pump (Gilson, Villiers-le-Bel, France) was used to fill the flow cell through Tygon pump tubes and 0.5 mm i.d. Teflon connection tube.
Samples
A total of 44 beer samples contained in sealed aluminium cans were obtained commercially in Spain. Duplicate cans of the same batch were always collected; one was used for NIR measurements and the other was used to measure the properties of interest by reference procedures.
The sample population contained different types of beer:
-
1
Normal beers (34 samples): These beers are made from barley and another cereal (normally the most abundant in the production area). For these beers, alcohol content is higher than 3% v/v and the original extract is lower than 12.5% w/w.
-
2
100% malt beers (three samples): for these beers the unique source of carbohydrates is malt and the beer does not contain any other cereal.
-
3
Special brewed beers (two samples): the original extract in these beers is higher than 12.5% w/w.
-
4
Alcohol-free beer (one sample): although its name may lead to confusion, the alcohol content of this beer is below 0.9–1% v/v. This sample was obtained from normal beer after undergoing thermal evaporation of most of the alcohol.
-
5
Beer with soda (two samples): produced as a mix of normal beer with soda, which is done after the fermentation, just before the canning. This mixture results in dilution of the beer, changing its flavor and reducing the extract and alcohol content.
-
6
Beer with lemon (one sample): similar to the previous type, but using a lemon-based soda.
-
7
German beer (one sample): apart from its certificate of origin and its flavor, its composition is not significantly different. It has a higher alcohol and extract content and a darker color than the normal beers considered.
Table 2 summarizes, for each sample, the main characteristics of the beer and reference values obtained for the density, original extract, real extract, and alcohol content (expressed both in % v/v and % w/w). Standard procedures for the Anton Paar autoanalyzer provided by the manufacturer were followed in analysis of the samples. The standard deviation of reference concentrations (sc) for real and original extract and alcohol was estimated as 0.05% w/w, 0.05% w/w, and 0.05% v/v, respectively. Results obtained by use of the autoanalyzer for sample 16 were unsuccessful; for this sample, therefore, analysis is qualitative, not quantitative.
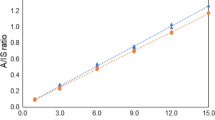
Table 3 shows the correlation for the whole sample population of the quantities measured in this study. As expected, a high linear correlation was found between alcohol content expressed as % w/w and % v/v—% w/w=0.7852 % v/v, R2=0.9957. There is also a good correlation between the real extract and the original extract and between the real extract or original extract and the alcohol content. It should also be noticed that the correlation between the experimentally obtained data has a similar trend when it was considered each data set separately or the whole sample population.
NIR analysis
Samples were placed in the same temperature-controlled room where the spectrometer was located before performing the analysis. The sample compartment temperature was monitored and it remained stable at 26±1°C during acquisition of NIR spectra using a flow cell. For measurements made in glass vials the samples were placed in a water bath at 32°C and measured under thermostatted conditions.
Samples were degassed by stirring for 5 min and filtered before filling the measurement cell. In the vial experiments, one vial for each sample was used. Triplicate measurements were performed by rotating the vial. During the acquisition of triplicate spectra using the flow cell the sample was continuously pumped through the cell using a peristaltic pump (flow rate≅1.5 mL min−1) and refilled after each measurement.
For both flow cell and vials sample spectra were scanned between 800 and 2857 nm, by averaging 25 scans per spectrum with a nominal resolution of 2 cm−1 and using a zero filling factor of 2. The acquisition of each averaged spectrum required 35 s. The maximum standard deviation of the response (sR) was estimated to be 0.0005 a.u.
The background and blank spectra were acquired by filling the cells with Milli-Q purified water (Millipore, Bedford, MA, USA) and using the same instrumental conditions than those employed for samples. Background spectra were scanned at intervals of seven samples and blank spectra were acquired after measurement of each sample, to ensure cross-contamination of NIR spectra was minimized by cleaning the flow cell.
For flow-cell measurements, after finishing data acquisition for each sample, warm water (45–50°C) was flushed through the cell and the tubing manifold for 20 s to clean the apparatus by removing any beer residue. Then, to reduce the cell temperature, water at room temperature was pumped for another 20 s. Before measuring the next sample two spectra of the blank were acquired to assess the success of the cleaning procedure. For vial measurements, because a different glass vial was used for each sample, no cleaning stage was necessary. To assess vial variability, ten vials were filled with the same sample and the spectrum for each was acquired in triplicate.
Both sample and blank spectra were collected in absorbance mode. For vial measurements two analytical windows were available—the 800–1370 and 1590–1817-nm regions. The other regions were eliminated before the calculations, because it was observed that spectral variations in these regions could not be ascribed to variations in sample composition. For measurements obtained using the 1-mm flow cell the spectral analytical windows were wider—800–1850 and 2050–2400 nm.
Data analysis
Spectra obtained from the Opus software were automatically converted from cm−1 to nm and exported in text format (data spacing 0.098 nm). Data were analyzed using Matlab 6.0 (The Mathworks, South Natick, MA, USA). First, internal and laboratory-written Matlab functions were used for hierarchical cluster analysis to evaluate the similarity of samples in terms of their NIR spectra and to assess the number of characteristic subsets into which the available samples could be divided. Similar criteria to those already published for milk classification were used [15]. Multivariate calibration calculations were made with the MVC1 toolbox [16].
The following figures for the model’s fit to the data and the predictive power were used throughout the text. In all cases the scope was to evaluate the average deviation of the model from the actual data.
PRESS is the sum of squares prediction error (quadratic sum term in Eq. 1) for the model, which includes A factors. The root-mean-square error of calibration (RMSEC) is a measure of how well the model fits the calibration data, and is defined as:
where \(\hat C_i \) means the values of the predicted data (in our case extracts and alcohol content) when all samples are included in the model building and d.f. is the number of degrees of freedom calculated as the number of calibration samples with known concentration (Ci) minus A+1, the number of factors kept in the model plus one.
The root-mean-square error of cross-validation (RMSECV) is a measure of predictive ability of the model formed on part of a dataset to predict the rest of the data. The RMSECV is defined as the previous equation, except that \(\hat C_i \) are predictions for samples not included in the model formulation and d.f. is the number of times in which the cross-validation is repeated (i.e. in the leave-one-out cross-validation d.f. is equal to the number of calibration samples).
As the ability of the model to fit the calibration data is not a direct measurement of its prediction capabilities, it is mandatory to compare the values predicted for well-known new samples not used to build the model. This can be performed by calculating the root-mean-square error of prediction (RMSEP) when the model is applied to new data for which the reference values are available. The RMSEP is calculated exactly as in Eq. (1) except that the estimates for Ci are based on a previously developed model in which the sample concentrations of the validation set are excluded in the model-building step and the degree of freedom is the number of samples used for validation.
To validate the NIR methodology against the autoanalyzer data found, different quality indicators are also given. These are the absolute mean difference (dx-y) between NIR predicted values \({\left({\ifmmode\expandafter\hat\else\expandafter\^\fi{C}_{i}} \right)}\) and reference data (Ci), the standard deviation of mean differences (sx-y), the quality coefficient (QC), and the pooled standard error of prediction for validation samples (sreg) [17]. As stated by Massart et al., the QC is to be preferred over correlation coefficient (of \({\ifmmode\expandafter\hat\else\expandafter\^\fi{C}_{\rm i}}\) versus Ci) “not only because it gives a better idea of the spread of the data points around the fitted straight line but also because it gives some indication on the percentage error to be expected for the estimated concentration”.
To build and select PLS models, the following iterative procedure was carried out. To build the best calibration model a selection of the optimum number of factors which minimize the root-square-of-cross-validation was based on the criterion of Haaland and Thomas [18]. To improve the predictive performance of the regression method, a search for suitable sensors was considered. In this sense, one subroutine from the MVC1 toolbox was used to find the minimum PRESS, as a function of the number of factors, based on a moving spectral window strategy [16]. Several spectral windows were tested to evaluate their prediction capabilities for the validation set. Only most significant results will be shown here.
Cluster analysis
In hierarchical cluster analysis the similarity between samples is calculated by comparison of their NIR spectra using the distance concept, calculated using a mathematical relationship (i.e. Euclidian norm) of numerical properties of the samples (i.e. absorbance at different wavelengths). In successive procedures, each sample is linked to the closest sample or group of samples and a characteristic distance is used to describe this union. This distance between groups of samples can be evaluated in different ways and is the main difference among common linkage methods (Ward, complete, average, etc.). In other words, by this procedure, each sample is replaced by a group comprising the sample and its neighboring samples located within the given similarity distance between NIR spectra. The results are represented in a dendrogram, which shows at which normalized or re-scaled distance (i.e. each distance rationed to the maximum distance, multiplied by a factor) of a group of samples is differentiated from others, when it is read from right to left. At the far-left end each replicate of each sample comprises a group of one member, i.e. each spectrum is unique. Thus, for a given rescale distance, a different number of groups are kept. At this stage we proposed [13] to use the similarity distance between triplicate spectra from the same sample as a minimum cut-off criterion. Actually, taking into account the concepts of limit of detection and quantification, we chose ten times the average distance between replicates as cut-off value.
Results and discussion
Beer NIR spectra
Because beers are aqueous solutions, only the NIR spectral region in which water does not absorb strongly can be used for analytical purposes. For vial cells there was strong absorption of the incident light by the components of the system, mainly water, in the regions between 1370 and 1590 nm and between 1817 and 2857 nm, so these regions were ignored in this study. Because the pathlength of the flow cell is smaller than that of the vials, a wider spectral window can be used, i.e. from 800–1850 and from 2050–2400 nm.
Figure 1 shows the spectra of three different types of beers obtained from the 1 mm pathlength cell. Characteristics of NIR spectra of beers are mainly related to the absorption of water in this spectral region, with some features of the other two main constituents, ethanol and carbohydrates. Because the background signal was acquired using water, the beer spectra shown arise from absorption of some beer constituents plus reduction of the amount of water. For the former, positive bands are observed, whereas for the latter kind of bands, negative peaks are found.
The main water absorption bands in the NIR spectra are located at approximately 970, 1200, 1450 and 1930 nm [19, 20]. These correspond to different combination or overtone bands of the normal vibration modes of water—symmetric stretching, bending, and asymmetric stretching modes. The most intense of these bands (that located at 1930 nm) means that the 1850–2050 spectral range cannot be useful for measurements made either in 6.5 mm vial or 1 mm flow cell experiments. Apart from these water bands, in the spectrum of all the beers a series of bands between 1560–1760 and 2050–2360 was observed. From the bibliography, the following general assignments can be made [21, 22]. The 1660–1760 nm region is ascribed to the C–H first overtone stretch vibration modes (in CH3 and CH2 groups), whereas the bands located between 2200 and 2400 nm are the first set of CH combination bands. For the OH, the first overtone vibration modes occurs between 1520 and 1640 and combination bands between 2050 and 2200 nm, a region in which a broad band is observed. When comparing the spectrum of a beer without alcohol and that of a normal beer, it can be observed that the differences correspond to the bands ascribed to all three main constituents.
Correction of NIR data
For all vial and flow-cell experiments, when comparing all NIR spectra collected for all samples a shift of the baseline was observed, sometimes even within replicates of the same sample. For vials this problem was more severe, because we rotated the vial position between replicate measurements. To preserve signal/noise ratio we tried to correct the original absorbance spectra without applying any derivative procedures.
Different ways of evaluating the efficiency of correction can be used, for example evaluating the root square error of prediction. In our opinion this efficiency can be tested before conducting any calibration procedure. In fact, it was tested by classification of the samples by hierarchical cluster analysis (HCA, see below). In the ideal case replicates of the same sample should be grouped together, so any deviation from this ideal case can be interpreted in terms that the intra-sample variance (for triplicates not clustered together) is higher than the inter-sample variance. Obviously, in such cases performing the calibration model is precluded.
Our previous experience with mid-infrared analysis of milk and oil samples [13, 15] showed that good results were achieved when the average absorbance in a fairly flat region was subtracted from each spectrum before data treatment for correcting additive artefacts. In the NIR region, however, there is no flat region in the spectrum useful for correction of the whole spectrum. For example, for flow cell data, if the region between 800 and 900 nm is used, for many triplicates significant variations in the 2000–2400 nm region were observed. Therefore, it was decided to use two different wavelengths for zeroing the region after and before the main water absorption band (~1850–2050 nm):
-
1
the average absorbance between 1060 and 1065 nm, for the bands from 800 to 1850 nm, and
-
2
the average absorbance between 2220 and 2222 nm, for the bands from 2050 to 2400 nm.
When using vials, subtraction of the baseline in a given region did not provide a good correction results. Therefore, other well-known algorithms were tested, for example multiplicative scatter correction (MSC) and standard normal variate (SNV), either considering together or separately the two spectral windows found for these cells. In all the cases unsuccessful results were obtained. Therefore, derivative spectra were tested. Good signal-to-noise derivative spectra were obtained by fitting a third-degree polynomial function to a 5 nm moving window (51 data points). Figure 2 shows the derivative spectra for selected samples. It is apparent there are many non-informative regions in the first-order derivative spectra (Figure 2A). When checking the replicate spectra for each sample, it is possible to see that in region 1 (800–1350 nm) the variability between 1160 and 1175 and between 980 and 1020 are much higher than at other wavelengths. The absorption bands in these regions are related to water and are highly temperature-dependent, so this may be explained by small thermal differences among triplicates. The signal-to-noise ratio for the spectral range between 1600 and 1800 (region 2) is better than that found for region 1, and is even better when only the region 1640–1760 nm is considered.
Derivative NIR spectra of main types of beer sample. A and B were calculated from vial experiments, for C flow cell data were used. a, b, and c describe the type of beer as indicated in the footnote of Fig. 1
When regions 1 and 2 were studied in more depth, some high variability sub-regions were detected. Consequently, a wavelength-elimination algorithm was developed to calculate, at each wavelength, the pooled variance of triplicate analysis and to compare this with the variance between samples (similar to ANOVA) [17]. Only those wavelengths in which the inter-sample variance is significantly higher than the intra-sample variance were kept. For region 1 the intra samples variability is still of the order of the inter-sample variability, especially for normal beers, resulting in poor classification of samples. In contrast, successful results were obtained for region 2 after applying this algorithm. Good classification of samples and triplicates were obtained, as will be discussed below. Therefore, only region 2 will be used for processing vial experiments; this is shown in Fig. 2B.
For comparison purposes only, the derivative spectra for all samples were also calculated for data obtained using the flow cell. Figure 2C shows the derivative spectra in the region not accessible by use of vial experiments.
Clustering of beer samples from their NIR spectra
A clustering method was used before PLS data treatment to evaluate possible classes among samples considered in this study and to enable us to select properly a representative calibration set, thus improving the prediction of unknown samples. Furthermore, differences among sample composition can be used to evaluate the classification of samples based on their NIR spectra.
In previous work the calibration set for different complex groups of samples has been successfully selected by the use of hierarchical cluster analysis [13, 15]. In this work this strategy was followed using the best combination of distance measurement and linkage methods previously found, that is, Euclidian distance and Ward linkage. To calculate the spectral distance the scores of the most significant principal component (PC) were used. The PC was selected after application of principal component analysis (PCA) to the corrected flow cell zero-order absorbance data in the region from 2220 to 2370 nm. By use of only one factor 99.9% of the variance in the X-block (spectral data) was explained. Figure 3 shows the dendrographic classification of samples obtained using zero-order spectra obtained using the flow cell (Fig. 3A) and those found by any first-order derivative spectra obtained with the vials (Fig. 3B) and flow cell (Fig. 3C).
Dendrographic classification of samples using the Euclidean distance after PCA analysis of NIR spectra. A Flow cell data, 2 PC obtained in the zero order spectral range 2224–2350 nm (one point baseline established at 2220–2221 nm), B vial data, 2 PC obtained from first order spectra, C flow cell, 2 PC obtained from first order spectra
In all cases the three replicates of each sample were grouped together. Next, it is interesting to elucidate if the agglomeration level has a clear interpretation. The main clusters formed (from right to left) are directly correlated with the mean intensity of the NIR spectra of the samples in these groups. Thus, samples with high absorbance levels are grouped together. As an increase in the absorbance is mainly related to an increase in the total amount of carbohydrates, alcohol, and proteins, clusters are basically related with the similar content of these analytes between the samples. Table 4 shows, for clusters obtained using zero-order spectra from the flow cell measurements, the mean and the standard deviation for each analyte in each cluster.
Basically, clustering criteria seem to be based first on the original extract and second on the alcohol content, which separates samples into two main groups. This classification can distinguish (1) the beer sample with very low original extract and without alcohol (sample 19) from the rest. In the second group, another clear distinction can be made: (2a) the beer with lime (sample 16) and the beers with lemonade (samples 11 and 20) and (2b) the German type beer (sample 14), together with the 100% malt beers (samples 2 and 8). The other samples (clusters 6–10) are normal beers with different alcohol and extract content. The agglomeration in these clusters seems to follow the alcohol content of the samples, with a content around 5.0% v/v for cluster 8, around 4.7% v/v for clusters 9 and 10, and around 4.5% v/v for clusters 6 and 7. The separation between clusters 6 and 7, and between 9 and 10, cannot be explained by the available data, and could indicate high similarity between samples classified in a same cluster.
To compare the different data treatment, the cluster analysis was repeated on the derivative spectra obtained using both vial and flow cell. For vials the region between 1640 and 1760 nm, before elimination of non-informative wavelengths, was used. By use of three factors 99.5% of the variance in the X-block (spectral data) was explained. Figure 3B shows the dendrographic classification of samples obtained using the glass vials. For flow cell data, when the same region is used plus the region from 2220 to 2360 nm, the dendrogram shown in Fig. 3C was obtained. It is apparent that for most samples their neighbouring samples are the same in both figures. When Figs. 3B and 3C are compared with the dendrogram in Fig. 3A it can be seen that discrimination among samples was better for the latter. Nevertheless, sample agglomeration in all three figures is closely similar and could be useful for calibration set selection.
Selection of the calibration set
Determination of the number and the nature of samples to be used for calibration is always a critical factor in multivariate analysis and in this study it was based on the results from hierarchical cluster analysis. The calibration and validation datasets were selected using the dendrogram in Fig. 3A (without sample 16). Selection was based on the following criteria:
-
(i)
At least one sample from each cluster was selected for calibration.
-
(ii)
If the cluster comprised more than one sample, the number of samples selected for calibration was approximately the root square of the total number of samples included in the cluster. The remaining samples were assigned to the validation data set. So, the number of samples assigned to the validation was equal to or higher than the number used for calibration.
-
(iii)
Samples within a given cluster were selected randomly.
By following these rules calibration and a validation sets comprising 15 and 28 samples, respectively, were established.
To evaluate the representativeness of the aforementioned calibration set, an extended calibration model was also used. This extended calibration set included the samples of all individual clusters but now the root square of samples from clusters with more than one was reserved for validation and the rest included in the calibration set. Therefore, in this case there were more calibration samples than validation samples (30 and 13, respectively). Table 5 summarizes the descriptive statistics (mean and standard deviation) of the calibration and validation sets in the single (cal 1 and val 1) and the extended (cal 2 and val 2) models, considering each quality characteristic to be determined. It is apparent that the calibration data set selected can always be regarded as representative.
Some attempts were made to obtain calibration models for density, but prediction results were poor. There are two reasons for this:
-
1
the small range of this property (for all samples density is between 1.007 and 1.011 g mL−1), and
-
2
the range of errors predicted for triplicates, which was 0.002–0.003 g mL−1 .
No additional comment on this property will therefore be made in this study.
Determination of real extract, original extract, and ethanol using flow cell
Different models were built using calibration sets 1 and 2 and compared in terms of RMSECV and RMSEP values for the validation data sets. It should be noticed that in the second case the prediction errors are not significantly different from those obtained with a reduced number of samples employed for calibration, indicating that the selection strategy used avoids the extensive effort normally required in multivariate calibration. It can, in fact, be seen that the number of samples used as calibration set 1 is far below that used in other published work (Table 1), although it may be pointed out that no outliers (either in calibration or validation) were detected in any of the models presented.
When correlating absorbance data obtained from flow cell measurements the best PLS models were always built using the 2250–2360 nm range only. All models built using the other spectral regions (in combination or not with the aforementioned region) yield models with low predictive capability. The performance of the models built in the 2250–2360 nm spectral region increased by approximately 2–3% when the baseline was based in the 2220–2221 nm wavelength range.
Table 6 shows the calibration and validation results obtained for the three properties analyzed using the optimized models. For real extract determination the optimum PLS method was based on four extracted factors in the spectral range 2249–2303 nm. For original extract the same number of factors was obtained in the optimized model, but the spectral region was shifted to high wavelength numbers (2259–2348 nm).
Figure 4 shows the net sensitivity vector associated to the PLS method for real and original extract determination. It is apparent there is no direct match between both vectors, and thus, despite the correlation observed between reference data for these two properties (Table 3), both extracts are not associated to the same NIR spectral feature.
For ethanol determination the optimum spectral range was between 2298 and 2337 nm, requiring three factors to build the PLS model. So, as can be observed, the optimum spectral range for original extract shares a region with the real extract model and other part with the ethanol model, which seems closely coherent.
Table 6 lists the figures of merit obtained under all the aforementioned conditions. The reproducibility of the determination, established from the mean standard deviation of each triplicate, and the standard error of prediction (that includes the uncertainty from the model [23, 24], considering the standard deviation of the reference concentration (sc) and the standard deviation of the response (sR)) were 0.05 and 0.01% w/w, 0.07 and 0.01% w/w, and 0.02 and 0.01% w/w, for real extract, original extract, and ethanol, respectively. It is apparent that these values are similar to those found using the autoanalyzer method. It can also be seen from Table 6 that there is no significant difference between the figures of merit obtained for the simplest model and those obtained for the extended calibration set. The prediction of the validation samples in set 2 is always of similar quality to that for validation data set 1. This similarity was obtained for all the optimized models obtained from zero-order absorbance data found using the flow cell and the first-order derivative data obtained using the vial and the flow cell. Therefore, in further comparison, only references to Cal/Val 2 will be made
As can be seen from the dx-y, sx-y, and QC values found for the PLS model, there is no significant difference between results provided by the proposed PLS–NIR method and those provided by the reference method. The maximum percentage error to be expected for new determinations (QC) of real extract, original extract, and ethanol are 2.2, 1.2, and 1.9%, respectively.
Sensitivity and selectivity data were obtained from the net analyte signal [14] and show that selectivity and sensitivity for ethanol determination (22% and 0.028 (% v/v)−1) were higher than for the other two properties. The selectivity for real extract is the lowest (3.5%) whereas sensitivity for this (0.011 (% w/w)−1) is slightly higher than for original extract (0.009 (% w/w)−1).
Beer analysis using glass vials
A similar procedure to that described above for building calibration–prediction models was followed for determination of real extract, original extract, and ethanol in beers based on measurements made on samples in glass chromatography vials. It must be pointed out that only the region below 1850 nm is available because of use of a 6.5-mm pathlength and that first-order derivative spectral data were used. For comparison purposes, derivative spectra were also calculated for data obtained using the flow cell. In this case the optimum model was also obtained in the 2250–2360 nm spectral region, but only model characteristics in the same spectral region available for vials will be shown here. The most relevant improvements achievable in the 2250–2360 nm range will be mentioned at the end of this section.
Regarding prediction capabilities, Table 7 summarizes different characteristics of the optimum PLS model built for all the properties using the extended calibration set (figures for calibration/validation set 1 are always similar). It should be also said that in calibration and validation outliers were detected. For determination of real extract the optimum PLS method based on derivative spectra was found for four extracted factors in the spectral range 1662–1684 nm. For original extract, the same number of factors and nearly the same spectral region (1667–1686 nm) were obtained in the optimized model. Analysis of net sensitivity vector (data not shown) associated for each PLS method provided evidence that the spectral features correlated with these two properties are not exactly the same. For ethanol determination, on the other hand, the optimum spectral range was between 1677 and 1742 nm, requiring only one factor for building the PLS model.
Table 7 also includes the figures of merit obtained under all the aforementioned conditions. Prediction capabilities using vials for real and original extract are worse than those presented in Table 6, by a factor of 1.5–2, whereas nearly the same performance was obtained for determination of ethanol. As is also apparent, there is no significant difference between characteristic values obtained for vials compared with those obtained when using the flow cell, for the model built in the same spectral range. This may be explained by the best transparency of the flow cell based on its low optical path length, which offers most possibilities of selecting an appropriate spectral range; also, flow cell measurements reduce the signal-to-noise ratio (see, for example, Fig. 2). This latter fact is evident when comparing the sensitivity obtained for each property when using vials and flow cell—it always drops by nearly one order of magnitude for the flow cell, whereas the selectivity remains fairly constant. For the vial, the reproducibility of the determination, established from the mean standard deviation of each triplicate and the standard error of prediction, were 0.11 and 0.06, 0.14 and 0.06, and 0.01 and 0.01% w/w, for real extract, original extract, and ethanol, respectively, which are higher than those presented in Table 6. The maximum percentage error to be expected for new determinations of real extract, original extract, and ethanol were 3.3, 2.5 and 1.8%, respectively, which are worse than those found for the best spectral range using the flow cell and also worse than those obtained under the same conditions for the flow cell data, except for ethanol determination, which does not depend on the measurement.
For use of derivative spectra obtained from the flow cell and the predictive performance in the optimised spectral range (which is in the region between 2250 and 2350 nm) the following trends were observed (data not shown) in comparison with the data in Table 6. First, sensitivity increases between four and seven times. The RRMSEP values are 3.3, 1.4, and 2% for real extract, original extract, and ethanol, respectively. Thus, the performance for ethanol was not significantly different; this may be because the selectivity in this region decreases sharply (it is approx. 30%) and the noise in this region is higher than in the 1650–1750 nm region.
The reproducibility of vials was analyzed using ten different vials filled with the same sample. Acquisition of spectra for each vial was performed in triplicate. The intra-vial pooled standard deviation for original extract, real extract and ethanol were 0.18% w/w, 0.20% w/w, and 0.03% v/v, respectively. The inter-vial standard deviations were 0.12% w/w, 0.19% w/w, and 0.02% v/v, respectively. It can thus be concluded that the loss of performance observed in this study for determination of beer extracts using glass vials for NIR spectra acquisition is because of lack of transparency of the 8.2 mm o.d. vials in the region from 2249 to 2348 nm and not to the variability of the vials.
Conclusions
Hierarchical cluster analysis, performed after PC analysis of NIR absorbance spectra, has been proved to be an excellent means of selecting representative samples for calibration in PLS–NIR determination of real and original extracts and alcohol content of beers.
From comparison of the two sample-introduction methods employed in this study it can be concluded that use of the 1-mm flow cell enables the use of the 2249–2348 nm region, which results in high sensitivity and excellent predictive performance, especially for extracts determination and using zero-order NIR absorbance spectra. However, the use of 6.5 mm glass vials improves the speed of sample analysis, by avoiding the need for cleaning of the measurement cell, and improves the selectivity of determination of beer properties when first-order derivative NIR spectra in the region from 1662 to 1742 nm were employed; under these conditions predictive performance was similar to that obtained by using the flow cell for ethanol determination and slightly worse than when using flow-cell measurements for real and original extract determination.
Compared with previously published work on chemometric NIR analysis of beer samples, the method developed provides RMSEP values of 0.12, 0.14, and 0.08% for real, original extract and ethanol, respectively, which are better than the prediction error obtained by Norgaard et al. [11] and by Westad and Martens [12] for original extract (0.18 and 0.17% w/w), respectively, and clearly better than the coefficient of validation errors reported by Maudoux et al. [10] for all the properties studied, which varied between 5.94 and 6.19%. In all this work NIR measurements were made using quartz cells of different path length and using spectral ranges different from those selected by us.
References
Varnam AH, Sutherland JP (1996) Bebidas: Tecnología, Química y Microbiología. Editorial Acribia, S.A., Zaragoza
Council Directive 92/84/EEC: OJ L 316, 31.10.1992. Bull 10:1992
(1987) Analytica—European brewery convention, 4th edn. Brauerei—und Getranke-Rundschau, Zurich
Pérez-Ponce A, Garrigues S, de la Guardia M (1996) Analyst 121:923–928
Gallignani M, Garrigues S, de la Guardia M (1993) Analyst 118:1167–1173
Gallignani M, Garrigues S, de la Guardia M (1994) Anal Chim Acta 296:155–161
Engelhard S, Lohmannsroben HG, Schael F (2004) Appl Spectrosc 58:1205–1209
Mendes LS, Oliveira FCC, Suarez PAZ, Rubim JC (2003) Anal Chim Acta 493:219–231
Schropp P, Bruder T, Forstner A (2002) Mon Schr Brauwiss 55:212–216
Maudoux M, Yan SH, Collin S (1998) J Near Infrared Spectrosc 6:A363–A366
Norgaard L, Saudland A, Wagner J, Nielsen JP, Munck L, Engelsen SB (2000) Appl Spectrosc 54:413–419
Westad F, Martens H (2000) J Near Infrared Spectrosc 8:117–124
Inon FA, Garrigues JM, Garrigues S, Molina A, de la Guardia M (2003) Anal Chim Acta 489:59–75
Lorber A, Faber K, Kowalski BR (1997) Anal Chem 69:1620–1626
Inon FA, Garrigues S, de la Guardia M (2004) Anal Chim Acta 513:401–412
Olivieri AC, Goicoechea HC, Inon FA (2004) Chemom Intell Lab Syst 73:189–197
Massart DL, Vandeginste BGM, Buydens LMC, de Jong S, Lewi PJ, Verbeke J (1997) Handbook of chemometrics and qualimetrics (Part A and B). Elsevier Science BV, Amsterdam
Haaland DM, Thomas EV (1988) Anal Chem 60:1193–1202
Buijs K, Choppin GR (1963) J Chem Phys 39:2035–2041
Maeda H, Ozaki Y, Tanaka M, Hayashi N, Kojima T (1995) J Near Infrared Spectrosc 3:191–201
Weyer LG (1985) Appl Spectrosc Rev 21:1–43
Workman JJ (1996) Appl Spectrosc Rev 31:251–320
Faber K, Kowalski BR (1997) J Chemom 11:181–238
Olivieri AC (2002) J Chemom 16:207–217
Acknowledgments
Financial support by the Direcció General d’Investigació i Transferència Tecnològica de la Conselleria d’Empresa, Universitat i Ciència de la Generalitat Valenciana (Project GV 04B-247, Grupos 03-118 and invited professor FAI grant), Universitat de València (Projecte UV-AE-20050203), University of Buenos Aires (X013), and ANPCyT (PICT-2003 17932)) is acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Iñón, F.A., Llario, R., Garrigues, S. et al. Development of a PLS based method for determination of the quality of beers by use of NIR: spectral ranges and sample-introduction considerations. Anal Bioanal Chem 382, 1549–1561 (2005). https://doi.org/10.1007/s00216-005-3343-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-005-3343-9