Abstract
In recent years, various chemiluminescent clinical immunoassay kits have been widely applied to the detection of hormones. However, a kit for chloramphenicol (CAP) is often absent from most commercial product lists, even though it is important to control the levels of CAP residues in foodstuffs too. Therefore, we describe a simple, solid-phase chemiluminescence immunoassay (CLIA) for the measurement of CAP in foodstuffs. A rabbit anti-CAP IgG is passively adsorbed onto the walls of polypropylene plates. The labeled antigen is horseradish peroxidase (HRP) conjugate of CAP. Luminol solution is used as the substrate of HRP. The light yield is inversely proportional to the concentration of CAP. The method has a similar sensitivity (0.05 ng/ml), specificity, precision, and accuracy to a conventional enzyme immunoassay (EIA). The intra-assay and inter-assay CVs of ten samples were <8% and <20%, respectively, and the analytical recovery of the method was 87–100%. The experimental correlation coefficient of dilution was found to be 0.999 using milk supernatant as buffer. The detection limit for the method was 0.1–10 ng/ml, and it displayed good linearity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chemiluminescence (CL) is a highly sensitive method that enables non-isotopic detection in immunoassays [1, 2]. CL detection of molecules of synthetic or natural origin, such as proteins and nucleic acids (DNA and RNA), as well as other biological molecules, is increasingly replacing radioactive detection as the method of choice where sensitivity is critical. In many hospitals, CL immunoassay (CLIA) kits have become popular [3, 4, 5, 6]. They can detect most hormones and antibiotics with high sensitivity, specificity, precision, and accuracy. Horseradish peroxidase (HRP) is detected using one of the most popular CL substrates (H2O2 and luminol). A variety of substituted phenols have been known to act as enhancers for this system, such as 4-iodophenol [7], 4-phenylboronic acid [8] and 4-iodophenylboronic acid [9]. Toxicological problems associated with the administration of the drug chloramphenicol in humans are well documented [10]. The use of chloramphenicol is prohibited when breeding cattle and birds [11], because chloramphenicol (ingested via foodstuffs) is highly toxic to humans. In many countries, including China, a zero tolerance policy has been established for the presence of CAP residues in meat and animal products. CAP can be detected using various techniques, such as radioimmunoassay, enzyme immunoassay (EIA), high-performance liquid chromatography, and so on. Although EIA is one of the most widely used of these methods, many believe that it will be superceded by CLIA due to its superior performance. We have therefore investigated a CAP detection system based on CL immunoassay, and using HRP as the enzyme label.
Materials and methods
Apparatus and reagents
A luminometer (Victor2 1420 Multilabel Counter) with software was purchased from Wallac (Gaithersburg, MD, USA). EIA-Micro and CLIA-Micro 96-well microtiter plates were purchased from NUNC (Denmark). Chloramphenicol (CAP) and rabbit anti-CAP IgG were supplied by Beijing Atom Hightech Co. Ltd. Horseradish peroxidase (HRP), dicyclohexyl carbodiimide (DCC), N-hydroxysuccinimide ester (NHS), bovine serum albumin (BSA) and N-ethyl-N′-[3-(dimethyl)amino-propyl] carbodiimide hydrochloride (EDC) were all purchased from Sigma (St. Louis, USA).
Assay protocol
EIA protocol (used to check the stabilities of the standards, the CAP-labeled, coated plates, and other prophase work): Rabbit anti-CAP IgG were diluted in 0.05 M, pH 9.5 carbonate buffer (CB) to particular concentrations, and then used to coat the EIA-microtiter plates and incubated overnight at 4 °C. Before the experiment, the plate was washed with 0.02 M, pH 7.4 phosphate buffer (PB), and unoccupied sites were blocked with the blocking solution (1% BSA) for 1 h at 37 °C. The plate was washed, and then labeled conjugates and standards diluted to appropriate concentrations in diluent were added. Upon every dilution, the absorbance was measured at 450 nm (OD450).
CLIA protocol (used to review assay performance and results): immunoreactions were performed as described for the EIA protocol, except that the luminescence of each well was checked after adding CL substrate too. These luminescence results are given in counts per second (cps).
Purification of rabbit anti-CAP IgG
About 1 ml of rabbit anti-CAP serum was purified with Hitrap Protein G HP. The concentration of rabbit anti-CAP IgG was 0.815 mg/ml, shown by the UV absorption at 280 nm.
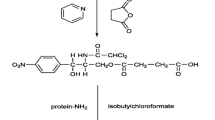
Synthesis of HRP-conjugated CAP
The CAP (1 mg), NHS (1 mg) and DCC (1 mg) were mixed and stirred in 0.1 ml dimethylformamide (DMF) for 4 h at room temperature. The horseradish peroxidase (HRP, 1.0 mg) in 0.5 ml of 0.2 M NaHCO3 was then added to the reaction mixture with stirring. After the addition of EDC (1 mg), the reaction mixture was stirred at room temperature overnight. The reaction solutions were then dialyzed against PBS for two days, then mixed with 2 ml glycerol and stored at −20 °C [12, 13].
Results
Selecting the dilution level of labeled CAP
The EIA-microtiter plates were coated with rabbit anti-CAP IgG (1.5 μg/ml). Labeled conjugates diluted to appropriate concentrations in diluent were added. Upon every dilution we monitored the OD450 counts for standard S0 (the CAP concentration was 0 ng/ml) and NSB (nonspecific binding). 100 μl of standard CAP and 100 μl of labeled CAP were added to each well. The results are shown in Table 1. Based on these results, we chose a dilution level of 1:10000 for the labeled CAP.
Selecting the concentration of the antibody used for the coating
The antibody was diluted to 2, 3, 4, 5 μl in order to coat the EIA-microtiter plates for immunoreaction with the labeled CAP diluted 1:10000, and the results (in terms of OD450 counts) are shown in Table 2.
From the results shown in Table 2, we considered that 3 μg/ml was the appropriate concentration of antibody to use in the following reaction.
Selecting the incubation period
We diluted the standards to 0, 0.1, 30 and 100 ng/ml for immunoreaction. The plates with the standards and the label were incubated for 0.5, 1, 2, and 4 h, then we reviewed the changes in the OD450 counts. The results are shown in Fig. 1 (count in OD450).
According to the results shown and Fig. 1, it takes nearly 3 h to reach reaction equilibrium, so an incubation time of 1 h is acceptable.
The stability of labeled CAP
The labeled CAP was diluted to 1:100, and its stability (in terms of OD450 counts) was monitored for eight days at 37 °C. The results are shown in Fig. 2.
Stabilities of the standards
The standards were diluted to 1 and 10 ng/ml (low and high concentrations, respectively), and their stabilities were monitored at various temperatures over 14 days. The results are shown in Fig. 3.
The stabilities of the standards over 14 days. A1 standard at low concentration at 4 °C, A2 standard at high concentration at 4 °C, B1 standard at low concentration at room temperature, B2 standard at high concentration at room temperatures, C1 standard at low concentration at 37 °C, C2 standard at high concentration at 37 °C
According to Fig. 3, the standards were most stable at 4 °C.
Stability of the coated plates
Plates were coated in the antibody at a concentration of 3 μg/ml, and the stabilities (in terms of OD450 counts) of the coated plates were monitored for ten days at 37 °C. The results are shown in Fig. 4.
The luminescence persistence of the CL substrate
After immunoreaction, 180 μl of CLIA substrate was added to each well, and then the luminescence was monitored every 15 min. The luminescence results are shown in Table 3.
We can obtain the concentrations of the samples by constructing a luminescence vs concentration calibration curve. Now, it takes a significant time, 52 s, for all of the samples to be measured during one detection cycle, and since the luminescence is time-dependent, we must account for the fact that the samples are not actually sampled simultaneously during each cylce. Therefore, in Table 4, each row represents the results for a particular detection cycle, “I” shows the concentrations of the samples obtained via the standard calibration curve, “II” shows the concentrations of the samples obtained via the standard calibration curve at the end of the detection cycle, and “III” shows the relative error: III = (II − I)/I×100.
According to the upper data we consider that offset error is overlook, and we can get perfect result after adding our CLIA substrate in 3–15 min.
The linearity of the calibration curve and the limit of quantitation
We diluted the standards to 0, 0.1, 0.3, 1, 3 and 10 ng/ml, then 100 μl of standard and 50 μl of label was added to wells coated in antibody for immunoreaction. We monitored the counts per second from each well, and constructed a luminescence vs concentration calibration curve (see Fig. 5). According to the curve we can reliably quantitate the levels of CAP in foodstuff samples from 10 ng/ml to 0.1 ng/ml with good linearity. This method can be used to detect CAP concentrations as low as 0.1 ng/ml.
The sensitivity of the method
100 μl of standard and 50 μl of labeled CAP were added to wells coated in antibody (200 μl at 3 μg/ml) and left overnight. After immunoreaction and washing-up, we added 180 μl of CLIA substrate to each well and then detected the luminescence after five minutes. Ten samples were monitored in this experiment. After reviewing the data, we calculated the sensitivity of the method to be 0.046 ng/ml.
The precision of the method
The luminescence of three different samples were monitored over 30 days, and the intra-assay and inter-assay CVs were derived. These are shown in Table 5.
From the results in Table 5, we can see that the intra-assay CVs were <8%, and the inter-assay CVs were <20%.
The correlation coefficient of dilution experiment
We pretreated skim milk with Carrez reagents [14], and diluted the supernatant with standard diluent 1:1. We regarded the upper solution as the sample diluent, and diluted the sample to 1/2, 1/4, 1/8. The results are shown in Fig. 6.
The analytical recovery of CAP from the skim milk sample
We added 1, 4 and 10 ng of CAP to samples of 5 ml of skim milk, and then pretreated the milk as in the dilution experiment. We then derived the concentration of CAP in the skim milk using our developed method. The results are shown in Table 6.
From the data in the table, we can see that the analytical recovery of CAP can be acceptable.
Discussion
The label is not stable at room temperature or at 37 °C, but at 4 °C it is very stable, for at least a few months. From these facts, we conclude that HRP-conjugated CAP is different to HRP-conjugated protein. CAP is a small molecule and it conjugates with HRP via a single chemical bond. As the temperature rises, this bond has a greater tendency to break.
The concentrations of standards used ranged from 0.1 ng/ml to 10 ng/ml. However, this does not mean that the technique is only linear over this concentration range; in fact, if the results for a 0.05 ng/ml standard are added to the calibration curve, the range 0.05 ng/ml to 10 ng/ml still has good linearity.
Because the sensitivity [15] (0.05 ng/ml) with 3μg/ml antibody well coating is acceptable, We recommend an antibody concentration of 3 mg/ml in order to improve the sensitivity and the linearity range for special users. If necessary, however, we can make the sensitivity almost 10 ppt, or lower.
Although the immunoreaction reaches equilibrium after 3 h of incubation, the experiment shows that acceptable results can be achieved after 1 h incubation. In most cases, especially in hospitals, it is imperative to obtain experiment data as quickly as possible, so 1 h incubation is reasonable.
Compared to the results from ELISA, CLIA has a more cragged standard curve, which is why it has a higher sensitivity and a wider detection limit. Otherwise, the CLIA has more effective numeric than ELISA, so CLIA has a higher precision. According to the intra-and inter-assay CVs, we can know about the above conclusion.
It is important to obtain an idea of the persistence of the substrate luminescence over time [16, 17]. It will result in a larger error if the persistence is not as good, because the apparatus detects the luminescence well by well, so the luminescence offset error (caused by the finite time, 52 s, taken to measure all of the wells in sequence) becomes more important. We have proved that, after 3–15 min of the onset of luminescence, the relative luminescence offset error within each set of well measurements is <5%. If the samples are monitored every 5 min, we can reduce the luminescence offset error to <3%.
Aside from the skim milk sample, we determined the CAP in some other foodstuff samples [18, 19]. The recoveries are also satisfactory, and the correlation coefficients in the dilution experiments are >0.99. Our CAP-CLIA technique is therefore suitable for detecting CAP in various foodstuff samples.
References
Ohno K-I, Arakawa H (1999) Development of novel high-sensitivity chemiluminescence assay for luminol using thiourea derivatives. Luminescence 14:355–360
Whitehead TP, Kricka LJ, Carter TJN, Thorpe GHG (1979) Analytical luminescence: its potential in the clinical laboratory. Clin Chem 25(9):1531–1546
Kim JB, Barnard GJ, Collins WP, Kohen F, Lindner HR, Eshhar Z (1982) Measurement of plasma estradiol-17β by solid-phase chemiluminescence immunoassay. Clin Chem 28(5):1120–1124
Javier Bagazgoitia F, Luis Garcia J, Ibarra JM, Patino R (1989) Chemiluminescent immunoassay (CLIA) for urinary albumin. J Biolumin Chemilum 3:169–174
Hubl W, Thorpe GHG, Hofmann F, Meissner D, Thiele HJ (1990) Enhanced cheniluminescent immunoassay for aldosterone. J Biolumin Chemilumin 5:49–52
Aoyagi S, Iwata T, Miyasaka T, Sakai K (2001) Determination of human serum albumin by chemiluminescence immunoassay with luminol using a platinum-immobilized flow-cell. Analytica Chimica Acta 436:103–108
Thorpe GHG, Kricka LJ (1986) Enhanced chemiluminescent reactions catalyzed by horseradish peroxidase. Methods Enzymol 133:331–353
Kricka LJ, Ji X (1995) 4-Phenylylboronic acid: a new type of enhancer for the horseradish peroxidase catalyzed chemiluminescent oxidation of luminol. J Biolumin Chemilumin 10:49–54
Kricka LJ, Cooper M, Ji X (1996) Synthesis and characterization of 4-iodophenylboronic acid: a new enhancer for the horseradish peroxidase-catalyzed chemiluminescent oxidation of luminol. Anal Biochem 240:119–125
Lynas L (1998) Screening for chloramphenicol residues in the tissues and fluids of treated cattle by the four-plate test, Charm II radioimmunoassay and Ridascreen CAP-Glucuronid enzyme immunoassay. Analyst 123:2773–2777
Liu Z-h (1995) Enzyme immunoassay for assaying chloramphenicol in animal foodstuff. Chin J Vet Drug 29(2):47–48
Dzgoev AB (1999) High-sensitivity assay for pesticide using a peroxidase as chemiluminescent label. Anal Chem 71:5258–5261
Wang C-J (1993) A sensitive enzyme immunoassay (EIA) for quantitation of the topical anti-inflammatory agent SCH 40120 in unectracted human plasma. J Pharm Biomed Anal 11(10):871–879
Xu CL (2003) A determination of chloramphenicol in animal foodstuff with enzyme immunoassay[P]. CN1389729A, 2003-1–8
Zhou DG, Fang H (2003) The evaluation of three kinds of chloramphenicol-ELISA kits. Chin J Vet Drug 37(9):28–31
Wang J (2002) A high stability chemiluminescence substrate and its determination[P]. CN1333464A, 2002-1–30
Iwata R, Ito H, Hayashi T, Sekine Y, Koyama N, Yamaki M (1995) Stable and general-purpose chemiluminescent detection system for horseradish peroxidase employing a thiazole compound enhancer and some additives. Anal Biochem 231:170–174
De-shi S, Bin Z, Ya-li Q, Gui-zhi W (2002) Establishment of indirect competitive enzyme-linked immunosorbent assay for detecting the residues of CAP. Chin J Vet Sci 22(1):77–79
Meifang S, Wenya Z, Zhiliang F, Guanghong W, Xuebing G (2003) Enzyme linked immunosorbent assay (ELISA) for quantitative analysis of chloramphenicol residues in aquatic products. J Nanjing Normal Univ (Eng Technol) 3(2):1–4
Conflict of interest:
No information supplied
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lin, S., Han, Sq., Liu, Yb. et al. Chemiluminescence immunoassay for chloramphenicol. Anal Bioanal Chem 382, 1250–1255 (2005). https://doi.org/10.1007/s00216-005-3273-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-005-3273-6