Abstract
The development of predictive in vitro stem cell-derived hepatic models for toxicological drug screening is an increasingly important topic. Herein, umbilical cord tissue-derived mesenchymal stem cells (hnMSCs) underwent hepatic differentiation using an optimized three-step core protocol of 24 days that mimicked liver embryogenesis with further exposure to epigenetic markers, namely the histone deacetylase inhibitor trichostatin A (TSA), the cytidine analogue 5-azacytidine (5-AZA) and dimethyl sulfoxide (DMSO). FGF-2 and FGF-4 were also tested to improve endoderm commitment and foregut induction during Step 1 of the differentiation protocol, being HHEX expression increased with FGF-2 (4 ng/mL). DMSO (1%, v/v) when added at day 10 enhanced cell morphology, glycogen storage ability, enzymatic activity and induction capacity. Moreover, the stability of the hepatic phenotype under the optimized differentiation conditions was examined up to day 34. Our findings showed that hepatocyte-like cells (HLCs) acquired the ability to metabolize glucose, produce albumin and detoxify ammonia. Global transcriptional analysis of the HLCs showed a partial hepatic differentiation degree. Global analysis of gene expression in the different cells revealed shared expression of gene groups between HLCs and human primary hepatocytes (hpHeps) that were not observed between HepG2 and hpHeps. In addition, bioinformatics analysis of gene expression data placed HLCs between the HepG2 cell line and hpHeps and distant from hnMSCs. The enhanced hepatic differentiation observed was supported by the presence of the hepatic drug transporters OATP-C and MRP-2 and gene expression of the hepatic markers CK18, TAT, AFP, ALB, HNF4A and CEBPA; and by their ability to display stable UGT-, EROD-, ECOD-, CYP1A1-, CYP2C9- and CYP3A4-dependent activities at levels either comparable with or even higher than those observed in primary hepatocytes and HepG2 cells. Overall, an improvement of the hepatocyte-like phenotype was achieved for an extended culture time suggesting a role of the epigenetic modifiers in hepatic differentiation and maturation and presenting hnMSC-HLCs as an advantageous alternative for drug discovery and in vitro toxicology testing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The pharmaceutical industry faces considerable challenges regarding political and social pressure to reduce the cost of medicines. The drug development process is expensive, lasts on average 10–15 years and shows a success rate of only 11% for all therapeutic areas. The majority of drug attrition is due to the lack of efficacy or safety that occurs essentially in the clinical phases IIb and III of drug development (Kola and Landis 2004). Liver damage is also an important cause of new drug failures, post-marketing withdrawals or relabelling with black box warnings, of which 21% accounts for hepatotoxicity (Wilke et al. 2007). This is mostly due to the fact that hepatotoxicity is difficult to predict in early stages of drug development, making the improvement of pre-clinical models, namely human cellular models, particularly relevant concerning human safety. The need of an available source of human hepatocytes is therefore of major importance for in vitro toxicology applications (Gomez-Lechon et al. 2014).
Human primary hepatocytes (hpHeps) are not easily available and they are not much proliferative in healthy conditions or in vitro given their highly differentiated cell type (Godoy et al. 2013). Additionally, these cells quickly lose their normal functions, namely metabolism of xenobiotics, when isolated and maintained in culture. Human cell lines as HepG2, Huh7, and more recently HepaRG, have also been used to study hepatic metabolism of new molecules and toxicants. However, these immortalized cell models still present crucial drawbacks for predictive hepatotoxicity such as an incomplete metabolic profile regarding phase I, phase II and hepatic transporters (Godoy et al. 2013) resulting in distinct toxicity profiles for the same compounds, when compared to primary hepatocytes (Gerets et al. 2012; Sjogren et al. 2014).
Differentiation of stem cells (SCs) has been suggested as an alternative approach for obtaining functional hepatocyte-like cells (HLCs). A study from Sjogren et al. (Sjogren et al. 2014) showed that induced pluripotent stem cell (iPSC)-derived hepatocytes are better toxicity predictors than Huh7 and HepaRG for drugs that cause apoptosis via mitochondrial-driven mechanisms. However, although the scientific community has already achieved impressive results, a completely mature HLC population derived from SCs has not yet been achieved (Godoy et al. 2015; Schwartz et al. 2014). Mesenchymal SCs (MSCs), in particular, are a promising SC type, regarding availability, expandability, ethical and safety concerns (Miranda et al. 2015; Santos et al. 2008; Santos et al. 2013, 2015), when compared to iPSCs or embryonic SCs (ESCs). Despite its mesenchymal nature, MSCs derived from human bone marrow (Snykers et al. 2007), human adipose tissue (Li et al. 2014) or human umbilical cord tissue (Campard et al. 2008; Yoon et al. 2010; Zhang et al. 2009) have been described as able to undergo hepatocyte differentiation. In a comparative study, Lee et al. (Lee et al. 2012) showed that neonatal MSCs, namely placenta-derived MSCs (including chorionic villi MSCs, amnion MSCs, chorionic plate MSCs and Wharton’s jelly MSCs), presented a better hepatocyte-like phenotype after differentiation than adult MSCs, such as human adipose tissue-derived MSCs (AD-MSCs) or bone marrow-derived MSCs (BM-MSCs), reinforcing that each MSC type has a different differentiation potential and the importance of the choice of the primary MSC source (De Kock et al. 2012).
The few papers reporting hepatic differentiation of human neonatal umbilical cord tissue-derived MSCs (hnMSC-UCM) (Campard et al. 2008; Yoon et al. 2010) characterize the obtained HLCs in terms of urea and albumin production, positive glycogen storage, acetylated low-density lipoprotein (Ac-LDL) uptake and the presence of hepatic markers as CK-18, albumin (ALB) and α-fetoprotein (AFP) through immunohistological or gene expression analyses. These studies did not contemplate a deeper evaluation of the metabolic potential of these cells regarding phase I, II and III metabolic activities. Most importantly, the maintenance of cell functionality in culture was not evaluated. Some publications do evaluate CYP and hepatic transporter presence in iPSC or human ESC-derived HLCs (Brolen et al. 2010; Ulvestad et al. 2013), but this analysis using MSCs has still not been thoroughly explored. Considering hnMSC-UCM, Campard et al. (2008) investigated the activity of CYP3A4 only and tested whether hnMSC-UCM-derived HLCs would respond to the phenobarbital induction stimuli, which was not observed. Additionally, essential hepatic markers such as HNF-4α and CYP2B6 were not detected, as well as HepPar1, a highly sensitive marker of hepatocyte’s mitochondria. More recently, Zhang et al. (2009) and Zhou et al. (2014) published on differentiation of hnMSC-UCM into HLCs, using a two-step protocol but the achieved phenotype is less mature than that reported by Campard et al. (2008), presenting positive staining for CK-18, ALB and AFP. Nevertheless, the cells presented a fibroblast-like morphology and did not present glycogen storage and Ac-LDL uptake capacities (Zhang et al. 2009; Zhou et al. 2014).
Considering that the transcription of genes governing cell fate during development critically depends on the chromatin accessibility, epigenetic modifiers (EMs) have also been suggested for improving HLC phenotype. In this study, a total of 8 protocols were used to test the differentiation capacity of a specific population of hnMSC-UCM, UCX® (Miranda et al. 2015; Santos et al. 2013, 2015), isolated using a method which was fully adapted in compliance with advanced therapy medicinal product (ATMP) regulatory requirements (Martins et al. 2014). This differentiation procedure was performed using a combination of different cytokines in order to mimic liver embryogenesis and the addition of EM. The EMs tested in this work are small molecules that have been previously described and applied in vitro as promoters of hepatic differentiation, namely trichostatin A (TSA), dimethyl sulfoxide (DMSO) and 5-azacytidine (5-AZA). TSA is a histone deacetylase inhibitor (HDAC-I) (Snykers et al. 2007), DMSO exerts an epigenetic modulation through less described mechanisms (Pal et al. 2012), and the cytidine analogue 5-AZA is a small molecule that inhibits DNA methylation (Seeliger et al. 2013; Yoshida et al. 2007). hnMSCs showed the ability to commit with the endoderm lineage and to differentiate into mature HLCs. Moreover, the addition of EM and the optimized differentiation protocol resulted in an improved cell phenotype and a remarkable maintenance of HLC hepatic functions in culture was achieved, locating HLCs closer to hpHeps as shown by global gene expression.
Materials and methods
Reagents
Cell culture supplements were purchased from Lonza (Basel, Switzerland), unless otherwise stated. Trypsin–EDTA, foetal bovine serum (FBS), insulin-transferrin-selenium solution (ITS) and non-essential amino acid supplement (NEAA) were purchased from Gibco®/Life Technologies (Madrid, Spain). 7-Ethoxyresorufin and 7-ethoxycoumarin were purchased from Alfa Aesar (Karlsruhe, Germany). Hepatocyte growth factor (HGF), fibroblast growth factors (FGF-2 and FGF-4) and oncostatin M (OSM) were purchased from Peprotech (Rocky Hill, NJ, USA). Finally, Iscove’s modified Dulbecco’s medium (IMDM), alpha-modified Eagle’s medium (α-MEM), epidermal growth factor (EGF), dexamethasone, DMSO, nicotinamide, 5-AZA, trypan blue, periodic acid, Schiff’s reagent, Mayer’s haematoxylin, amylase, Hank’s balanced salt solution (HBSS), 4-methylumbelliferone (4-MU), β-glucuronidase/arylsulfatase, resorufin, 3-methylcholantherene (3-MC), omeprazole and rifampicin were acquired from Sigma-Aldrich (Madrid, Spain).
Cell cultures
hnMSCs were isolated as described in Miranda et al. (2015) and Santos et al. (2008, 2015) and expanded as undifferentiated cells in α-MEM supplemented with 10% of FBS (growing medium). HepG2 cells were cultured in α-MEM supplemented with 10% FBS, 1 mM of sodium pyruvate and 1% NEAA. Cryopreserved human primary hepatocytes (hpHeps; pool of 10 donors) were purchased from Invitrogen™ (Carlsbad, CA, USA; HEP10, A12176), thawed on cryopreserved hepatocyte recovery medium (CHRM; CM7000, Invitrogen™) and manipulated according to manufacturer instructions. Rat primary hepatocytes (rpHeps) were isolated from Wistar rats 3 to 6 months old, weighting 200–400 g, obtained from Charles River Laboratories (Barcelona, Spain) and cultured according to our previous reports (Leite et al. 2011; Miranda et al. 2009, 2010). Prior to each experiment, the animals were kept in a separate cage for at least 24 h, with ad libitum access to food and water. All applicable institutional and governmental regulations concerning the ethical use of animals were followed, according to the European guidelines for the protection of animals used for scientific purposes (European Union Directive 2010/63/EU) and the Portuguese Law nº 113/2013. Cell cultures of hnMSCs, HepG2, hpHeps and freshly isolated rpHeps were maintained at 37 °C in a humidified atmosphere with 5% CO2 in air. Cell viability was assessed through trypan blue exclusion method.
Hepatocyte differentiation of hnMSCs
hnMSCs were seeded in 24-well plates (Nunc™, Wiesbaden, Germany) at a density of 1.5 × 105 cells/cm2 in rat-tail collagen-coated surface. Rat-tail collagen was produced in-house according to Rajan et al. (2006) and used at a concentration of 0.2 mg/mL. Cells were allowed to proliferate for 24 h in hnMSC growing medium. A three-step differentiation protocol was applied using as basal medium (BM) IMDM with 1% penicillin–streptomycin–fungizone (P/S/A). Briefly, in the first step (Step 1), consisting of 48 h, 4 different combination of growth factors were tested: BM supplemented with 2% FBS, 10 ng/mL of EGF and (1) 10 ng/mL of FGF-2; (2) 4 ng/mL of FGF-2; (3) 4 ng/mL of FGF-2 and 10 ng/mL of FGF-4 or (4) 10 ng/mL of FGF-2 and 10 ng/mL of FGF-4. In the second step (Step 2), cells were maintained for 10 days with BM supplemented with 20 ng/mL of HGF, 4 ng/mL of FGF-2, 10 ng/mL of FGF-4, 0.61 g/L of nicotinamide and 1% ITS. In the third step (Step 3), at day 13 of differentiation, cells were supplemented with BM containing 8 ng/mL of OSM, 1 μM of dexamethasone and 1% ITS. This culture medium was maintained up to day 34 of culture. The medium was renewed every 3–4 days in all protocols. DMSO (1%, v/v), 5-AZA (20 µM) and TSA (500 nM) supplementation was performed and a total of 8 protocols evaluated, as summarized in Table 1.
Gene expression
RNA isolation
Total RNA was isolated from samples with 0.5–1.0 million of cells using Trizol® (Life Technologies), and extraction was performed according to manufacturer instructions.
For microarray analysis of gene expression, RNA was further purified using a NZY Total RNA Isolation Kit (NZYTech, Lisbon, Portugal) and quantified by measuring absorbance at 260 nm using LVis Plate mode (SPECTROstar Omega, BMG Labtech, Ortengerg, Germany). RNA integrity and sample purity were also measured using a 2100 Bioanalyzer (Agilent Technologies; Palo Alto, CA), assuring a RIN score higher than 8. For quantitative real-time PCR (qRT-PCR), RNA concentration was determined as above, prior to cDNA synthesis.
Microarray
Microarray analysis of hpHeps and HLC gene expression was performed using Affymetrix Human Genome U133 Plus 2.0 Gene Arrays at the Bioinformatics and Expression Analysis core facility at Karolinska Institutet (Sweden). Microarray data from undifferentiated hnMSCs (UCX®) and HepG2 cells, performed using the same array platform, were obtained from Gene Expression Omnibus (GSE51869 and GSE40117) (PMID: 24438697; PMID: 23393228). Subsequent analysis of differential gene expression and principal component analysis (PCA) were performed using AltAnalyze (PMID: 20513647). Gene expression levels in HLCs, hnMSCs and HepG2 cells were compared to the gene expression levels in hpHeps. Gplots heatmap.2 R package (Warnes et al. 2009) was used to generate a gene expression heatmap of the 2000 genes with highest variance across the samples. Bioinformatic pathway analysis was conducted using the Ingenuity Pathways Analysis (IPA) software for genes with a fold change greater than 2 compared to the expression levels in hpHeps (Benjamini–Hochberg-corrected p < 0.05). The network biology-based computational platform CellNet (PMID: 25126793) was used to assess cell identity based on gene expression profiles. Venn diagrams were generated using Venny (Oliveros 2007–2015).
qRT-PCR
cDNA was synthesized from 1 µg RNA using NZY First-Strand cDNA Synthesis Kit (NZYTech) according to the manufacturer instructions. Quantitative real-time PCR was performed using SensiFAST™ SYBR® Hi-ROX Kit (Bioline®, Luckenwalde, Germany). Master mix was prepared for a final reaction volume of 25 µL, using 1 µL of template cDNA and 0.1 µM of forward and reverse primers. Table A (Supplementary Material 1, Table A) presents the specific human primers used for endoderm markers, progenitor hepatocytes and hepatocytes. The reaction was performed on StepOne™ Real-Time PCR System (Applied Biosystems®/Life Technologies) consisting of a denaturation step at 95 °C for 10 min, 40 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 1 min and extension at 72 °C for 30 s. A dissociation stage was added to determine the melting temperature (Tm) of a single nucleic acid target sequence as a quality and specificity measure. The comparative Ct method (2−ΔΔCt) was used to quantify the amount of target genes which were normalized to a reference gene GADPH and relative to undifferentiated cells, hnMSCs. The efficiency of each reaction was estimated from a serially diluted cDNA from a cell type expressing the target gene in order to construct a standard curve (1, 10−1 and 10−2) for each gene.
Histology
Immunocytochemistry
Cells cultured on collagen-coated glass slides (Nunc™, Lab-Tek™ II Chamber Slide™) were fixed with 4% paraformaldehyde (PFA; Sigma-Aldrich) in PBS with 4% sucrose and analysed for detection of CK-18, CK-19, multidrug resistance protein-2 (MRP-2), organic anion-transporting polypeptide C (OATP-C), CYP1A2 (Santa Cruz Biotechnology®, Inc), HNF-4α (R&D Systems, Minneapolis, MI, USA) and CYP3A4 (Nosan Corporation, Yokohama, Japan). Cryopreserved hpHeps, freshly isolated rpHeps cultured for one day and HepG2 were used as positive controls, while undifferentiated hnMSCs were used as negative control (for details, see Supplementary Material 1).
Periodic acid Schiff’s (PAS) staining
HLCs cultured in 24-well plates were fixed with 4% PFA in PBS and stained with Schiff’s reagent and Mayer’s haematoxylin counterstaining. Positive and negative controls consisted of cryopreserved hpHeps, freshly isolated rpHeps, cultured for 2 days, and HepG2 cells, and undifferentiated hnMSCs when at 70% confluence, respectively (for details, see Supplementary Material 1).
Albumin and urea production
Albumin and urea quantification were determined on cell culture supernatants using an albumin ELISA kit (Bethyl Laboratories, Montgomery, TX, USA) and a quantitative colorimetric urea kit (QuantiChrom™ Urea Assay Kit, BioAssay Systems, Hayward, CA, USA), respectively. Both procedures were performed according to the manufacturer instructions. Data acquisition was done using a microplate reader (SPECTROstar Omega, BMG Labtech). The data refer to the production rate and are expressed as pg/106 cells.h and µg/106 cells.h, respectively.
Biotransformation activity
Phase I metabolism
CYP enzyme activity was measured by means of 7-ethoxyresorufin-O-deethylase (EROD) activity, 7-ethoxycoumarin-O-deethylase (ECOD) activity and P450-Glo™ assays (Promega, Madison, WI, USA). EROD assay mainly reflects the activity of the human isozymes CYP1A2 and CYP1A1 (Wilkening et al. 2003). The procedure consisted of cell incubation with 8 mM of 7-ethoxyresorufin in non-supplemented BM for 90 min. The product concentration, 7-hydroxyresorufin, was measured in supernatants after 2 h of enzymatic digestion with β-glucuronidase/arylsulfatase. The quantification was calculated using a calibration curve of resorufin in culture medium. Fluorescence was measured (Ex 530 nm/Em 590 nm). ECOD assay measures human CYP2B6, CYP1A2 and CYP2E1 activities (Miranda et al. 2010; Wilkening et al. 2003). The procedure consists of 90-min incubation with 7-ethoxycoumarin at 0.8 mM in non-supplemented BM. 7-Hydroxycoumarin concentration was determined after the same digestion procedure as used for EROD assay, followed by a liquid–liquid extraction step with chloroform and back-extraction of the organic phase with a solution of 5.84% of NaCl in 0.1 M of NaOH. Fluorescence was measured (Ex 340 nm/Em 460 nm). Induction of ECOD and EROD activities was performed by 48-h incubation with 2.5 μM of 3-MC or 100 μM of omeprazole. CYP3A4, CYP2C9 and CYP1A1 activities were assessed using the luminescent P450-Glo™ assay with the respective substrates Luciferin-PPXE, Luciferin-H and Luciferin-CEE. Each assay was performed with a final volume of 60 μL and luminescent intensity measured according to the manufacturer instructions. Induction was performed by 48-h incubation with 2.5 μM of 3-MC for CYP1A1 or 25 μM of rifampicin for CYP2C9 and CYP3A4. Luminescence and fluorescence measurements were done using a microplate reader (SPECTROstar Omega, BMG Labtech). The data are expressed as pmol/106 cells.h, except for CYP3A4 activity that is expressed as nmol/106 cells.h.
Phase II metabolism
Uridine 5′-diphosphate glucuronosyltransferases’ (UGTs) activity was determined by quantification of the substrate 4-MU before and after cell incubation. The procedure was performed according to Miranda et al. (2010) with slight modifications. Briefly, a 100 μM of 4-MU in HBSS was incubated with cells for 1 h. The supernatants were freshly transferred into a black 96-well plate, 4 μL of NaOH at 0.1 M added per well in order to achieve pH = 11, and fluorescence was measured (Ex 340 nm/Em 460 nm). The data are expressed as nmol/106 cells.h.
Protein quantification
For protein quantification, cells were lysed with 0.1 M of NaOH and incubated overnight at 37 °C. Protein concentration was determined using the bicinchoninic acid (BCA) protein assay kit (Novagen, Madison, WI, USA) according to manufacturer instructions and also measured by absorbance at 280 nm using LVis Plate mode (SPECTROstar Omega, BMG Labtech). A linear calibration curve to relate total protein with cell number was generated to estimate cell number, from each cell type and for both methods of quantification.
Statistical analysis
The results are given as the mean ± SEM. Statistical data analysis was performed using GraphPad Prism version 6.0 (GraphPad Software, La Jolla, CA, USA). The effects on metabolic activity were evaluated by two-way ANOVA with Holm–Sidak method in order to determine whether the metabolic activity of each compound was significantly altered by the protocol, the induction of specific CYP450 enzyme inducers, over the time of culture, and to evaluate differences between groups and controls. In all cases, p < 0.05 was considered statistically significant.
Results
The endoderm marker HHEX is expressed in hnMSCs and induced by FGF-2
The three-step protocol developed for hepatocyte differentiation was optimized based on previously described methodologies (Campard et al. 2008; Snykers et al. 2006, 2007) and considering the liver development stages: commitment, differentiation and maturation. Since hnMSCs are of mesodermal origin, particular attention was given to the endoderm commitment by testing the effect of FGF-2 at 4 ng/mL and 10 ng/mL, in the presence or absence of FGF-4 on HHEX gene expression (Fig. 1).
Gene expression of HHEX on hnMSCs after exposure to different FGF cocktails in the differentiation Step 1 (D3). qRT-PCR shows HHEX higher expression with 4 ng/mL of FGF-2 when compared to 10 ng/mL of FGF-2, or 4 ng/mL of FGF-2 and 10 ng/mL of FGF-4, or 10 ng/mL of FGF-2 and 10 ng/mL of FGF-4 (black bars). Positive controls (white bars) are cryopreserved hpHeps and HepG2 cell line. Data are normalized to the reference gene GAPDH and expressed in Log10 relative to hnMSCs (n = 3). FGF fibroblast growth factor, HHEX hematopoietically expressed homeobox protein, GAPDH glyceraldehyde-3-phosphate dehydrogenase, hpHep human primary hepatocytes, hnMSC undifferentiated human neonatal mesenchymal stem cells
HHEX is an endoderm marker essential in liver development (Kubo et al. 2010). A basal expression of the HHEX gene was detected on hnMSCs. The addition of FGF-2 at 4 ng/mL produced a 200-fold increase in HHEX expression, whereas it only produced an 85-fold increase when cells were exposed to a combination of FGF-2 at 4 ng/mL and FGF-4 (10 ng/mL). Moreover, HHEX gene expression in the presence of FGF-2 (10 ng/mL) resulted in an 8- and 17-fold increase in the absence or presence of FGF-4, respectively. Thus, FGF-2 only was added to Step 1 of all differentiation protocols to a final concentration of 4 ng/mL.
Epigenetic modifiers improve hepatocyte phenotype on HLCs
The epigenetic modifiers (EMs) tested in this work consist of small molecules that have been previously described and applied in vitro as promoters of hepatic differentiation, namely TSA, DMSO and 5-AZA. Previously, we have evaluated TSA at concentrations ranging from 100 nM to 1000 nM being 500 nM the best-tested condition (data not shown), and therefore, this concentration is used in this work. As for DMSO and 5-AZA, 1% and 20 μM, respectively, were adopted according to the literature (Guillouzo et al. 2007; Seeliger et al. 2013). Furthermore, DMSO was tested alone, from days 10 and 13 onwards, or in combination with TSA or 5-AZA in a total of 8 different differentiation protocols (Table 1).
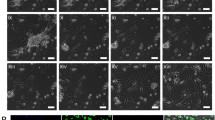
During the differentiation protocol, cells in culture were maintained confluent and in serum-free conditions in order to prompt differentiation rather than proliferation. The fibroblastic morphology of hnMSCs was kept during the first two steps of the differentiation protocol except for protocols 4 and 5, where TSA was added at day 6. This resulted in cell detachment (Fig. 2), and therefore, protocols 4 and 5 were not further pursued. From day 13 onwards, after the addition of OSM and dexamethasone, cell morphology started to change to a more epithelial-like shape with granular cytoplasm in all other protocols, i.e. protocols 1–3 and 6–8 (Fig. 2). At day 24, an epithelial polygonal hepatocyte-like morphology, with some binucleated cells, and glycogen storage ability were observed in HLCs obtained with protocols 1, 2, 6 and 8, where TSA was absent (Fig. 2). Accordingly, ALB and CK-18 were detected by immunocytochemistry. Overexpression of HHEX (data not shown), CK18, TAT, CEBPA, HNF4A and CYP3A4 was observed at day 24 in all protocols (Fig. 3). CK19, a biliary cell marker, showed low expression in all protocols. ALB and AFP overexpression was observed on protocols 1, 6 and 8 (Fig. 3). In parallel, the HLC enzymatic competence, namely phase I and II activities, was analysed (Fig. 4a). Similar EROD activity was obtained with all protocols. However, the protocols including DMSO at day 10 (protocols 6–8) presented significant induction of EROD activity (p < 0.01) whereas protocols with TSA only revealed induction when concomitantly added with DMSO (protocols 3 and 7; p < 0.05). In particular, protocol 6 resulted in higher CYP3A4 activity when compared to the positive controls (p < 0.05). CYP3A4 induction was also observed with this protocol (p < 0.05). Regarding phase II enzymatic activity, an increased UGTs’ activity was observed in differentiation protocols where DMSO was added at day 10 (protocols 6, 7 and 8) when compared to its supplementation at day 13 (protocols 2 and 3), independently of TSA presence (p < 0.001; Fig. 4a). A positive effect of 5-AZA (protocol 8) was also observed with improved UGTs’ activity (p < 0.01) when compared to the protocol where it was absent (protocol 6), as previously observed by Seeliger et al. (Seeliger et al. 2013). Finally, the protocols without TSA, but supplemented with DMSO at day 10 (protocols 6 and 8) presented higher urea production (Fig. 4b), especially protocol 6 (p < 0.05) where the achieved urea production was comparable to HepG2. All protocols also presented a significant increase in albumin production ability when compared to the negative control (Fig. 4c).
Comparative characterization of hnMSC-derived HLCs obtained with the differentiation protocols 1–8 (D24). Phase-contrast, PAS staining and immunocytochemical representative images reveal an epithelial morphology, glycogen storage ability and the presence of CK-18 and ALB, respectively, in HLCs from protocols 1–3, 6 and 8. In immunocytochemical analysis, cell nuclei were counterstained with DAPI. Scale bar 50 μm. PAS periodic acid Schiff, ALB albumin, CK cytokeratin, HLC hepatocyte-like cells, hnMSC undifferentiated human neonatal mesenchymal stem cells
Gene expression analysis of HLC derived from hnMSCs upon exposure to protocols 1–3 and 6–8 (D24). qRT-PCR shows overexpression of the hepatic markers ALB, AFP, CK18, HNF4A, CYP3A4, TAT and CEBPA and underexpression of the biliary marker CK19 (grey bars and black bar for protocol 6). Positive controls (white bars) are HepG2 cell line and cryopreserved hpHeps. Data are normalized to the reference gene GAPDH and expressed in Log10 relative to hnMSCs (n = 3). ALB albumin, AFP alpha-fetoprotein, CK cytokeratin, HNF4A hepatocyte nuclear factor-4α, CYP cytochrome P-450, TAT tyrosine aminotransferase, CEBPA CCAAT/enhancer binding protein, GAPDH glyceraldehyde-3-phosphate dehydrogenase, HLC hepatocyte-like cells, hpHep human primary hepatocytes, hnMSC undifferentiated human neonatal mesenchymal stem cells
Functional capacity of HLC derived from hnMSCs upon exposure to protocols 1–3 and 6–8 (D24). a Phase I (EROD and CYP3A4 activities) and phase II (UGTs’ activity) enzymatic activities. Induction studies (striped bars) were performed for 48 h with 25 μM of rifampicin (CYP3A4) or 2.5 μM of 3-MC (EROD). rpHeps and hpHeps were cultured for 72 and 24 h for phase I and II assays, respectively; b urea secretion; and c albumin production. Grey bars correspond to HLCs differentiated using protocols 1–3 and 7–8 and black bars to protocol 6. Controls (white bars) are hnMSCs (negative control) and HepG2 cell line, rpHeps and hpHeps (positive controls). Data are represented as mean ± SEM (n = 4–6). *, **, ***Significantly differ among the controls with p < 0.05, p < 0.01 and p < 0.001, respectively. $, $$ and $$$Significantly differ from protocol 6 with p < 0.05, p < 0.01 and p < 0.001, respectively. #, ##Significantly induced activity with p < 0.05 and p < 0.01, respectively. CYP cytochrome P-450, EROD 7-ethoxyresorufin-O-deethylase, UGT uridine 5′-diphosphate glucuronosyltransferase, 3-MC 3-methylcholanthrene, HLC hepatocyte-like cells, hpHep human primary hepatocytes, rpHep rat primary hepatocytes and hnMSC undifferentiated human neonatal mesenchymal stem cells
Overall, TSA supplementation resulted in a more heterogeneous cell phenotype considering morphology and expression of hepatocyte-specific proteins (protocols 3 and 7), whereas DMSO supplementation resulted in HLCs expressing hepatocyte markers, epithelial cell morphology, homogeneous glycogen storage and improved HLC biotransformation activity (protocols 2, 6 and 8). The later was more evident when DMSO was added at day 10 (protocol 6) rather than at day 13 (protocol 2).
hnMSC-derived HLCs present a partial hepatic differentiation at the transcriptional and functional levels at day 34
The need for a competent alternative model for drug metabolism studies encouraged us to examine the stability of hepatic phenotype in culture. Overall, protocol 6 provided the best condition where HLCs showed results that are comparable to the positive controls. Thus, hnMSC-derived HLCs from protocol 6 were kept in culture until day 34, revealing an epithelial morphology and the presence of binucleated cells and maintaining confluence throughout the cell culture period (Fig. 5). Differentiation efficiency and phenotype maintenance were further evaluated at the transcriptional and functional levels.
hnMSCs-HLC morphology and glycogen storage ability upon sequential exposure to liver-specific factors and DMSO supplementation from day 10 onwards. a Cell morphology of hnMSCs during the differentiation process at days 4, 13, 17, 24, 27 and 34 and of undifferentiated hnMSCs (negative control), HepG2 cell line and rpHeps (positive controls); b PAS staining of HLCs at days 27 and 34 of undifferentiated hnMSCs (negative control) and rpHeps (positive controls). HLCs and rpHeps stained after digestion with amylase in order to access the specificity of the observed staining are also presented. Controls are hnMSCs (negative control) and rpHeps (positive controls). Representative images were acquired with phase-contrast microscopy. Scale bar 50 μm. DMSO dimethyl sulfoxide, PAS periodic acid Schiff, HLC hepatocyte-like cells, rpHep rat primary hepatocytes, hnMSC undifferentiated human neonatal mesenchymal stem cells
PAS staining demonstrated glycogen storage ability of HLCs up to day 34 in culture (Fig. 5). Albumin and urea secretion was also stable during the full culture period, showing levels higher than hnMSCs (p < 0.05; Fig. 6).
Effect of culture time on albumin and urea production in HLC derived from hnMSCs up to day 34 in culture upon sequential exposure to liver-specific factors and DMSO supplementation from day 10 onwards (black bars). Undifferentiated hnMSCs and HepG2 cell line, rpHeps and hpHeps are negative and positive controls, respectively (white bars). Data are represented as mean ± SEM (n = 4–6). *, **, ***Significantly differ among the controls with p < 0.05, p < 0.01 and p < 0.001, respectively. DMSO dimethyl sulfoxide, HLC hepatocyte-like cells, rpHep rat primary hepatocytes, hpHep human primary hepatocytes, hnMSC undifferentiated human neonatal mesenchymal stem cells
To compare gene expression signatures between our HLCs (D34) and hpHeps, an unbiased whole genome analysis was performed. To get a better understanding on how hnMSC-derived HLCs compared to other cells we then compared our data with previously published gene array data from undifferentiated hnMSCs and HepG2 cell line available at Gene Expression Omnibus. Interestingly, global gene expression analysis (Fig. 7a), comparing differentially expressed genes in HLCs and hpHeps, estimated that ~70% of transcripts remain the same between HLCs and hpHeps (n = 17152), using a combined criterion of p < 0.05 and fold change >2. The heatmap representation and unsupervised clustering of the 2000 genes with highest variance (Fig. 7b) showed that in some clusters the differentiation protocol approximates the HLC’s gene expression profile to that of hpHeps, when compared to the undifferentiated hnMSCs. It also revealed similar gene expression clustering between HLCs and hpHeps, which was not observed between HepG2 and hpHeps. In HLCs, the expression of some genes involved in drug transport, amino acid metabolism and proliferation of hepatocytes was comparable to hpHeps, whereas others involved in xenobiotic metabolism remained poorly expressed (Fig. 7b and 7d). Accordingly, a PCA plot (Fig. 7c) evidenced a partial gain of mature hepatic features of the HLCs revealing that HLCs did not reach the hpHep global gene expression profile, but are closer than the HepG2 cell line and distant from hnMSCs. In fact, a Venn diagram representing the number of differentially expressed genes in HLCs, hnMSCs and HepG2 compared to hpHeps (fold change > 2, p < 0.05) shows that HLCs have less differentially expressed genes than hnMSC or HepG2, when these cells are compared to hpHeps (Figure S1 a). Additionally, CellNet analysis for assessing cell type identity, plotted as gene regulatory networks (GRNs; Figure S1 b) and network influence scores (Figure S1 c), demonstrated that HLCs consist of a mixed cell type with both “fibroblast” and “ovary” features. CellNet is a platform that estimates how closely cell populations resemble their target cell/tissue type (Cahan et al. 2014). The rational is that by measuring the establishment of cell and tissue (C/T)-specific GRNs in a cell population would serve both as a robust metric of cellular identity and as a tool to identify aberrant regulatory nodes. Thus, as expected, undifferentiated hnMSCs and liver cells (hpHeps and HepG2) had a maximum GRN status score for “fibroblast” and “liver”, respectively. Moreover, from the total of 16 cell/tissue types listed, no significant changes in GRN scores were observed between HLCs and hpHeps for “esc”, “neuron”, “skin”, “hspc”, “macrophage”, “bcell”, “tcell”, “heart”, “sk. muscle” and “lung”. Regarding the “fibroblast” GRN status, the HLCs showed a decrease relative to the hnMSCs revealing some degree of differentiation, but still some features of MSCs from where they were derived. The network influence profile, on the other hand, was similar in HLCs and hnMSCs, whereas hpHeps presented high influence on transcription factors such as NR1H4 (FXR), NR1/3 (CAR), NR1/2 (PXR), HNF4A (Figure S1 c). The GRN score is based on the expression levels of a limited number of transcriptional regulators (Figure S1 c). Therefore, to get a better assessment of the activation status of biological pathways and functions in HLCs, we performed a bioinformatics pathway analysis herein plotted within three categories: metabolism, cell death and survival, and cell growth, proliferation and cancer in all groups relative to hpHeps (Fig. 7d). All groups (HLC, HepG2 and hnMSC) presented a negative z-score indicating repression of the expression profile for drug transport, xenobiotic metabolism and amino acid metabolism though HLCs presented a higher activation than hnMSCs and HepG2 cell line for the later two subcategories. In contrast to HepG2 or hnMSCs, cell death and survival and cell growth, proliferation and cancer categories presented the same activation in HLCs as in hpHeps (Fig. 7d), suggesting that HLCs are closer to hpHeps regarding these particular biofunctions. Most importantly, Fig. 7d further indicates that, unlike HepG2, HLCs display a less “cancer cell”-like profile as shown by normalized gene expression signatures associated with cell death, apoptosis, proliferation of hepatoma cell lines, proliferation of tumour cells, colony formation and liver cancer. Finally, canonical pathway representation from the IPA analysis showed an approximation of HLCs to hpHeps in glucose and lipid metabolism and in signalling pathways such as PI3 K/AKT, PKA, oncostatin M and Wnt/β-catenin (Figure S1 d); and major differences in the PXR/RXR and FXR/RXR activation pathways followed by xenobiotic metabolism signalling and glutathione-mediated detoxification.
Genome-wide analyses of HLC derived from hnMSCs at day 34 of differentiation. a Volcano plot of differently expressed genes in HLC vs hpHep (fold change >2, p < 0.05); b Heatmap representation of the 2000 genes with highest variance across samples in hpHeps, HLCs, hnMSCs and HepG2; c principal component analysis (PCA) of gene expression profiles in hpHeps, HLCs, hnMSCs and HepG2; d expression profile-based prediction of biological function activation performed with IPA for HLCs, hnMSCs and HepG2 cells compared to hpHeps. Positive Z-scores indicate activation, whereas negative z-scores indicate repression; e relative expression levels of genes involved in drug metabolism comprehending phase I, phase II and drug transporters in HepG2, hpHeps, HLCs and hnMSCs, compared to hpHeps. HLC hepatocyte-like cells, hpHep human primary hepatocytes, hnMSC undifferentiated human neonatal mesenchymal stem cells
In order to have a deeper insight on gene expression profile, genes were also grouped considering genes coding for key enzymes in phases I, II and III of drug metabolism (Fig. 7e). High expression of the phase I enzymes CYP1B1, CYP3A7 and MAO was observed in HLCs but not in the other groups relative to hpHeps. Moreover, there was a higher expression of CYP3A5 and CYP3A7 relative to HepG2 and hnMSCs, whereas the expression of relevant CYPs such as CYP2D6, CYP2C9 and CYP3A4 was similar in HLCs, HepG2 and hnMSCs, being considerably lower than hpHeps. Remarkably, some aldehyde dehydrogenase isoforms, namely from ALDH1, ALDH3, ALDH7, ALDH9, ALDH18 subfamilies, were induced in HLCs to levels comparable to hpHeps. Phase II metabolism analysis showed a high expression of GST family genes and a low expression of UGT transcripts. In the “drug transporters” group, similar expression of some transcripts was observed in both ABC and SLC transporters, namely ABCA, ABCC, SLC5, SLC7, SLC28, SLC29 and SLCO subfamilies.
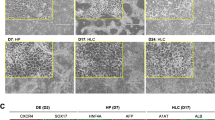
To support global transcriptomic characterization, further gene and protein expression of selected mature hepatocyte markers was evaluated by qRT-PCR and immunofluorescence. Confirming qRT-PCR gene expression data (Fig. 8), and despite low expression relative to hpHeps (Fig. 7e), the hepatic transporters OATP-C and MRP-2 and the biotransformation enzymes CYP1A2 and CYP3A4 were detected by immunofluorescence, along with the presence of CK-18, ALB and HNF-4α and the absence of CK-19 (Fig. 9). In fact, the transcription factors HNF4A and CEBPA, the biotransformation enzymes CYP1A1 and CYP3A4, the genes encoding for ALB, AFP and the enzyme involved on amino acid metabolism TAT, all presented increased expression from day 24 up to day 34, being TAT, CK18 and CYP3A4 higher than HepG2 (Fig. 8), suggesting some level of maturation. The observed CK18 expression level is comparable with hpHeps and higher than HepG2 cell line. CK19 was underexpressed as expected.
Gene expression analyses during hnMSC differentiation into HLCs upon sequential exposure to liver-specific factors and DMSO supplementation from day 10 onwards. qRT-PCR shows increased gene expression over time of the hepatic markers ALB, AFP, CK18, HNF4A, CYP3A4, TAT and CEBPA and decreased expression of the biliary marker CK19 in HLCs (black bars). Positive controls (white bars) are HepG2 cell line and cryopreserved hpHeps. Data are normalized to the reference gene GAPDH and expressed in Log10 relative to hnMSCs (n = 3). DMSO dimethyl sulfoxide, ALB albumin, AFP α-fetoprotein, CK cytokeratin, HNF4A hepatocyte nuclear factor-4α, CYP cytochrome P-450, TAT tyrosine aminotransferase, CEBPA CCAAT/enhancer binding protein, GAPDH glyceraldehyde-3-phosphate dehydrogenase, HLC hepatocyte-like cells, hpHep human primary hepatocytes, hnMSC undifferentiated human neonatal mesenchymal stem cells
Immunocytochemical analysis of HLC derived from hnMSCs upon sequential exposure to liver-specific factors and DMSO supplementation from day 10 onwards (D27). Representative images show the presence of the hepatic markers ALB, CK-18, HNF-4α, MRP-2, OATP-C, CYP1A2, CYP3A4 and the absence of the biliary marker CK-19. hnMSCs and HepG2 cell lines were used as negative and positive controls, respectively. Cell nuclei are stained with DAPI. Scale bar 50 μm. DMSO dimethyl sulfoxide, ALB albumin, CK cytokeratin, HNF-4α hepatocyte nuclear factor-4α, MRP-2 multidrug resistance protein-2, OATP-C organic anion-transporting polypeptide C, CYP cytochrome P-450, HLC hepatocyte-like cells, hnMSC undifferentiated human neonatal mesenchymal stem cells
The functional characterization of HLCs further supported the gain of hepatic features, namely by examining the effect of culture time on CYP and UGTs’ activities (Fig. 10). ECOD and CYP1A1 activities were maintained with time in culture. Conversely, EROD, CYP2C9 and CYP3A4 activities and induction abilities were greatly improved by day 34, reaching levels higher than HepG2 for CYP3A4 and CYP2C9. At day 34, CYP3A4, CYP1A1 and EROD activities are also enhanced when compared to rpHeps. A 2.5-fold and ~3-fold activity increase was even observed for CYP3A4 when compared to hpHep basal and induced activities, respectively. Accordingly, UGTs’ activity was maintained during 13 days counting from the last day of the differentiation protocol, revealing a 2.2-fold and 2.7-fold increase in UGTs’ activity by day 27, when compared to HepG2 (p < 0.01) and rpHeps (p < 0.001), respectively.
Effect of culture time on phase I and II activities in HLC derived from hnMSCs up to day 34 in culture upon sequential exposure to liver-specific factors and DMSO supplementation from day 10 onwards. Phase I and II activity was assessed by measuring EROD, ECOD, CYP1A1, CYP2C9 and CYP3A4 and UGTs’ activities, respectively, at days 21, 27 and 34 (black bars). Induction assays (stripped bars) were performed with 48-h incubation with 2.5 µM of 3-MC (CYP1A1 activity), 100 μM of omeprazole (ECOD and EROD activities) and 25 µM of rifampicin (CYP3A4 and CYP2C9 activities). Controls (white bars) are hnMSCs (negative control) and HepG2 cell line, hpHeps and rpHeps (positive controls). rpHeps and hpHeps were cultured for 72 and 24 h for phase I and II assays, respectively. Data are represented as mean ± SEM (n = 3). *, **, *** Significantly differ among the controls with p < 0.05, p < 0.01 and p < 0.001, respectively. #, ##, ###Significantly induced activity with p < 0.05, p < 0.01 and p < 0.001, respectively. CYP cytochrome P-450, DMSO dimethyl sulfoxide, EROD 7-ethoxyresorufin-O-deethylase, ECOD 7-ethoxycoumarin-O-deethylase, UGT uridine 5′-diphosphate glucuronosyltransferase, 3-MC 3-methylcholanthrene, HLC hepatocyte-like cells, hpHep human primary hepatocytes, rpHep rat primary hepatocytes, hnMSC undifferentiated human neonatal mesenchymal stem cells
In summary, the differentiation process herein developed evidenced a partial hepatic differentiation of the HLCs with relevant improvements regarding the HepG2 cell line.
Discussion
Hepatotoxicity studies are usually limited in duration, and results may not provide a complete understanding of drug’s cumulative and long-term in vivo effects. The inability to culture, characterize and challenge cells over longer periods of time is a relevant drawback of primary hepatocyte cell culture. Maintaining metabolic competence for longer periods of time is thus of foremost importance to investigate enzyme induction and inhibition, drug metabolism and drug chronic toxicity, and it is the main reason why alternative in vitro models have not yet been fully achieved (Hewitt et al. 2007). The differentiation of stem cells, MSCs in particular, into HLCs has been attempted to provide qualified model systems to study hepatic drug metabolism in vitro (Campard et al. 2008; Li et al. 2014; Snykers et al. 2007; Yoon et al. 2010; Zhang et al. 2009). Many reports have shown distinct hepatic differentiation abilities among different MSCs. Although the differentiation efficiency has been greatly improved by modifying the culture conditions, the obtained HLCs are still insufficient for either toxicological or clinical applications (Snykers et al. 2007). In this work, we have demonstrated that a specific population of human neonatal MSCs isolated from the umbilical cord tissue, UCX® (Santos et al. 2008, 2013), is actually a promising cell source for deriving functional HLCs. The use of epigenetic modifiers on hepatic differentiation, especially DMSO, enabled the achievement of highly efficient hnMSC-derived HLCs, and most importantly, metabolic competence could be enhanced up to day 34 in culture. Whole genome analysis further confirmed the gain of hepatic features of HLCs and improvement regarding HepG2 cell line.
We firstly investigated the in vitro endoderm commitment potential of hnMSCs. Different MSCs have been reported to present important differences in their transcriptomes depending on their tissue source. De Kock et al. (2012) suggested that Wharton’s jelly-derived MSCs presented a more favourable transcriptome regarding hepatocyte differentiation when compared to other types of stem cells, namely AD-MSCs, BM-MSCs and human skin-derived precursor cells. In fact, hnMSC-UCM have been described as expressing early liver-specific markers, endoderm genes such as GATA4 (La Rocca et al. 2009), GATA5, GATA6, SOX9 and SOX17 and liver progenitor markers as DKK1, DPP4, DSG2, CX43 and CK19 (Buyl et al. 2014; De Kock et al. 2012). GATA4 has been reported to be a transactivator of HHEX expression (Denson et al. 2000) required for definitive endoderm tissues for normal liver development. Conversely, GATA6 gene deletion impairs HHEX upregulation needed for liver bud formation and a normal hepatic developmental program, showing that GATA6 is essential for the earliest stages of hepatic development (Zhao et al. 2005). In liver development, the FGF family of cytokines is secreted by cardiogenic mesoderm and is required for the hepatic commitment of endoderm. Kang et al. (2013), using mouse embryos, showed that FGF-4 is important for cell distribution in primitive endoderm but not for its formation, being later required for lineage restriction. On the other hand, FGF-2 is a powerful inducer of foregut-derived cell lineages (Zhang et al. 2004). FGF-2 concentration determines cell differentiation fate as Ameri et al. (2010) observed in studies of embryo development, showing that low concentrations of FGF-2 (4 ng/mL) triggers hepatic fate, whereas higher concentrations promote pancreatic and pulmonary differentiation. Thus, the effect of FGF-2 at 4 ng/mL and 10 ng/mL, alone or in combination with FGF-4, on endoderm commitment/foregut induction was evaluated by means of HHEX gene expression. Our data showed a basal HHEX expression in undifferentiated hnMSCs that was further induced by FGF-2 in a concentration-specific manner, reinforcing the differentiation potential of these cells into endoderm-derived lineages. The observed upregulation of HHEX gene expression by FGF-2 at 4 ng/mL on differentiating cells is in accordance with Zhang et al. (2004) that showed that 5 ng/mL of FGF-2 increased HHEX expression. Ameri et al. (2010) also showed that higher concentrations of FGF-2 would reduce HHEX expression and inhibit further ALB expression, determining 4 ng/mL as the optimal concentration to direct hepatic cell fate. On the other hand, FGF-4 was reported to downregulate HHEX expression (Dessimoz et al. 2006), which is also in agreement with our findings. The importance of HHEX for hepatic lineage commitment was also shown in vitro by differentiating human ESCs (Kubo et al. 2010).
In this work, the differentiation strategy adopted consisted in a three-step sequential induction of hnMSC differentiation rather than through cell exposure to a cocktail. As previously reported, a sequential differentiation procedure better mimics the in vivo liver development (Snykers et al. 2006). In spite of the advantages of hnMSCs, few studies on hepatic differentiation are available on this cell type (Campard et al. 2008; Yoon et al. 2010; Zhang et al. 2009). As aforementioned, one example is the report by Campard et al. (2008), where the potential of hnMSC-UCM to derive HLCs was evaluated although not reaching a mature phenotype. Later, Zhang et al. (2009) and Zhou et al. (2014) published the differentiation of hnMSC-UCM into HLCs, using two-step protocols but the achieved phenotype is less mature than that reported by Campard et al. (2008). Therefore, the later procedure was applied to hnMSCs as our starting point (protocol 1) for the differentiation protocols developed herein.
The effect of the epigenetic modifiers TSA, 5-AZA or DMSO, with different exposure times, was tested in hepatic differentiation or maturation. DMSO is routinely used for HepaRG cell line maturation (Guillouzo et al. 2007) and more recently for progenitor cells (Liu et al. 2012) and SC differentiation (Banas et al. 2009; Czysz et al. 2015; Hay et al. 2008; Pal et al. 2012). HepaRG cell line is an important reference in cell-based assays due to its relevant phase I, II and III activities that are not usually observed on other cell lines such as HepG2 (Andersson 2010). Herein, DMSO was used at 1% as reported in the literature for the maturation of foetal hepatocytes (Liu et al. 2012) and was tested alone, from days 10 and 13 onwards, and in combination with TSA or 5-AZA. TSA is an HDAC-I that presented promising differentiation-inducing properties in human and rodent primary cultures of adult hepatocytes, hepatoma cell lines and that was also tested for hepatocyte differentiation of BM-MSCs (Snykers et al. 2007). In contrast, to our knowledge, 5-AZA effect on hepatocyte differentiation protocols in vitro has only been attempted on AD-MSCs by Seeliger et al. (2013), and on umbilical blood-derived MSCs by Yoshida et al. (2007), being applied 24 h prior to the differentiation induction since proliferation is needed for the incorporation of 5-AZA on cellular DNA.
Our findings indicate that a sequential differentiation protocol gradually induced cellular modifications from spindle fibroblast-like to hexagonal epithelial-like binucleated cells with glycogen storage ability. These morphological changes were particularly evident upon DMSO treatment without TSA cotreatment. Accordingly, by the end of Step 3, HLCs exposed to DMSO from day 10 onwards also had increased expression of the liver-specific markers CK18, ALB, CEBPA, TAT, HHEX, AFP, HNF4A, CYP1A1 and CYP3A4. The mechanisms through which DMSO induces hepatic differentiation are still poorly understood. However, studies by Chetty and Slavin (1994) showed an activation of the retinoblastoma protein in DMSO-induced cells. Furthermore, a high proportion of cells in early G1 phase was also observed. G1 is of high importance for cell differentiation and a prolonged G1 phase has been reported as an indication of a differentiated status by reflecting loss of pluripotency and gain of lineage-specific markers (Calder et al. 2013). Thus, DMSO treatment increases the responsiveness of SC differentiation signals, enhancing differentiation across all germ layers, and improving terminal differentiation into functional derivatives (Chetty and Slavin 1994). DMSO has also been associated to hypo- and hipper-phosphorylation of several cellular proteins (Earp et al. 1983; Yen and Varvayanis 1995) and on phosphatidylinositol signalling (Rubbini et al. 1997). Finally, DMSO at 2% on rpHeps resulted in more relevant CYP enzyme activity by restoring HNF-4α and CEBPA levels that rapidly decreased in culture without DMSO (Su and Waxman 2004), corroborating the observed gene expression data that showed increased expression of these factors up to day 34 in culture.
Nevertheless, the presence and expression of hepatic markers by SC-derived hepatocytes do not make them mature hepatocytes (Hengstler et al. 2005). Moreover, besides obtaining a mature HLC population, a major aim of this study was to obtain functional HLCs that do not dedifferentiate with time in culture. Therefore, we further tested the functional capacity of the HLCs cultured up to day 34. During this period, the HLCs kept on exhibiting a hepatocyte phenotype by possessing liver-specific markers at the mRNA and protein levels. Additionally, functional aspects such as albumin secretion, ammonia detoxification, glycogen storage, inducible CYP activity and phase II activity were also validated against undifferentiated hnMSCs. The increase in CK18, CEBPA, HNF4A, TAT, AFP and ALB expression indicates that cells developed into more mature hepatocytes with time in culture.
Biotransformation capacity was also of particular interest in these extended cultures being the use of hepatocyte cultures hampered mostly by the rapid loss of CYP enzyme expression and activity in vitro. Therefore, regarding phase I metabolism, CYP3A4, CYP2C9 and CYP1A1 were evaluated due to their relevance in drug metabolism (Gerets et al. 2012). EROD and ECOD activities were also investigated covering CYP1A1/2 and CYP2B6/1A2/2E1, respectively. HLCs showed CYP activity, which was either maintained or increased with time, and responded to rifampicin and omeprazole/3-MC induction in CYP3A4/CYP2C9 and ECOD/EROD/CYP1A1, respectively. Not only CYP3A4 and CYP1A1 were overexpressed in HLCs (qRT-PCR analysis), but also they could convert substrates more efficiently than rpHeps, as shown by the P450-Glo™ and EROD activity assays. In addition, the P450-Glo™ substrate (PPXE) for CYP3A4 activity may also detect some activity of other CYP3A enzymes such as CYP3A5 and CYP3A7, being the later overexpressed in our transcriptomic analysis. CYP3A7 is present in liver, though it is more expressed in foetal liver, which may indicate that HLCs are following the liver fate despite presenting a less mature phenotype. Moreover, evidence supporting the shift between CYP3A7 and CYP3A4 immediately after birth has been reported (Vyhlidal et al. 2006). This shift is apparently triggered by HNF-4α (Vyhlidal et al. 2006), herein detected by qRT-PCR at the mRNA expression and protein levels, although at a much lower extent than in hpHeps and showing a network influence negative score. Most importantly, although transcriptomic analysis points out to CYP3A4 and CYP2C9 underexpression on HLCs, their enzymatic activities reached higher levels than positive controls (D34), hence with positive implications for the use of these HLCs as an in vitro model for drug development. To note that primary hepatocytes, used as controls in functional assays, were obtained under their optimal culture conditions, i.e. up to day 3 of culture, otherwise cell dedifferentiation would have been observed; and consisted of a pool of 10 donors to avoid inter-individual differences. Moreover, since cultivation of primary hepatocytes is known to induce cell dedifferentiation the whole genome analyses of hpHeps were performed in non-cultivated cells, which may justify part of the transcriptome profile distance observed between HLCs and hpHeps. In fact, previous results described that overall gene expression profiles of hpHeps approached SC-derived HLCs after an extended cultivation period (Godoy et al. 2015).
Phase II enzymes and the hepatic transporters are also important determinants of pharmacological and toxicological activity of xenobiotics (Gomez-Lechon et al. 2014). Glucuronidation is a major metabolic pathway by which the parent compound is converted in water-soluble derivatives and therefore more readily excreted (Gerets et al. 2012; Gomez-Lechon et al. 2014; Sjogren et al. 2014). Despite the low expression of UGTs observed in the hpHep transcriptome profile, the HLC UGTs’ activity was observed up to day 34 in culture, being 1.7-fold and 2.1-fold higher than HepG2 or rpHeps, respectively, and similar to hpHeps, further suggesting a relevant HLC maturation level.
The hepatic transporters are essential for xenobiotic metabolism and excretion through blood or bile. SLC transporters are involved in pathways as transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds as well as transport of vitamins, nucleosides and related molecules (He et al. 2009). In particular, SLCO1A and SLCO3A, which have drug and organic anion transporter activity, presented an expression level similar to hpHeps in transcriptomic analysis. Moreover, transporters involved in amino acid transport such as SLC7 subfamily were also detected. OATP-C (SLC21A6) is a liver-specific influx transporter, located in the sinusoidal membrane of hepatocytes, and MRP-2 (ABCC2) is an efflux transporter, also highly expressed in liver, and located on the basolateral membrane (Godoy et al. 2013). The presence of both transporters at the protein level was detected suggesting phase III cell phenotype. Gene expression and protein presence of OATP-C have been reported on human ESC-derived HLCs (Brolen et al. 2010). MRP-2 activity has been previously published by Li et al. (2014) on human AD-derived HLCs and gene expression of sinusoidal and basolateral drug transporters, but none of those two were reported on HLCs derived from hnMSCs. To our knowledge, the presence of MRP-2 and of OATP-C was herein shown for the first time on hnMSC-derived HLCs. Moreover, an overexpression of the ABCA1 hepatic transporter was detected in the microarray analysis, relative to hpHeps, hnMSCs and HepG2 cell line despite the low activation, the nuclear receptor canonical pathways LXR/RXR FXR/RXR and PXR/RXR. LXR is activated by cholesterol and its metabolites and consequently activates ABCA1 expression. Bile acids activate FXR and PXR responds to fatty acids by increasing its uptake in hepatocytes (Rui 2014). This may suggest that the focus of HLC differentiation on phase I and II metabolism may be neglecting other essential features of the hepatocyte functions such as bile acids and lipid metabolism. Thus, this observation suggests that a further optimization of differentiation medium composition, namely by increasing its lipid content, should be taken into consideration in future studies, together with a broader characterization of HLCs at lipid and glucose metabolism levels. In fact, the bile salts supplementation, not commonly used in primary hepatocyte cultures, was recently reported to induce mature liver functions, namely BSEP, MRP2 and CYP7A1 (Godoy et al. 2016). In the present report, the results of a total of 8 conditions that differed in the combination and time of supplementation of three EMs are presented. Our data show that hnMSCs are capable of differentiating into HLCs upon mimicking the in vivo development microenvironment of hepatocytes by the addition of the epigenetic modifier DMSO from day 10 onwards. Furthermore, a genome-wide analysis increased the perception of the level of HLC maturation, indicating a partial hepatic phenotype. Remarkably, some features were enhanced along the differentiation protocol, namely in terms of drug transporter and phase I and II metabolism expression and function, taking the HLCs in the direction of a mature phenotype. The acquisition of such a stable and active biotransformation capacity will be determinant for the potential applicability of liver-like in vitro models for longer-term toxicity testing.
Abbreviations
- Ac-LDL:
-
Acetylated low-density lipoprotein
- AFP:
-
α-Fetoprotein
- ALB:
-
Albumin
- 5-AZA:
-
5-Azacytidine
- CK-18:
-
Cytokeratin 18
- CK-19:
-
Cytokeratin 19
- CYP:
-
Cytochrome P-450
- DMSO:
-
Dimethyl sulfoxide
- c/EBPα:
-
CCAAT/enhancer binding protein
- ECOD:
-
7-Ethoxycoumarin-O-deethylase
- EROD:
-
7-Ethoxyresorufin-O-deethylase
- FGF-2:
-
Fibroblast growth factor-2
- FGF-4:
-
Fibroblast growth factor-4
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- HHEX:
-
Hematopoietically expressed homeobox protein
- HLC:
-
Hepatocyte-like cell
- HNF-4α:
-
Hepatocyte nuclear factor-4α
- hnMSC:
-
Human neonatal mesenchymal stromal cell
- hpHep:
-
Human primary hepatocyte
- hrHep:
-
Human rat hepatocyte
- IPA:
-
Ingenuity pathway analysis
- 3-MC:
-
3-Methylcholantherene
- MRP-2:
-
Multidrug resistance protein-2
- 4-MU:
-
4-Methylumbelliferone
- OATP-C:
-
Organic anion-transporting polypeptide C
- TAT:
-
Tyrosine aminotransferase
- PCA:
-
Principal component analysis
- TSA:
-
Trichostatin A
- UCX® :
-
hnMSCs isolated from the umbilical cord matrix
- UGT:
-
Uridine 5′-diphosphate glucuronosyltransferase
References
Ameri J, Stahlberg A, Pedersen J et al (2010) FGF2 specifies hESC-derived definitive endoderm into foregut/midgut cell lineages in a concentration-dependent manner. Stem Cells 28(1):45–56. doi:10.1002/stem.249
Andersson TB (2010) The application of HepRG cells in evaluation of cytochrome P450 induction properties of drug compounds. Methods Mol Biol 640:375–387. doi:10.1007/978-1-60761-688-7_20
Banas A, Teratani T, Yamamoto Y et al (2009) Rapid hepatic fate specification of adipose-derived stem cells and their therapeutic potential for liver failure. J Gastroenterol Hepatol 24(1):70–77. doi:10.1111/j.1440-1746.2008.05496.x
Brolen G, Sivertsson L, Bjorquist P et al (2010) Hepatocyte-like cells derived from human embryonic stem cells specifically via definitive endoderm and a progenitor stage. J Biotechnol 145(3):284–294. doi:10.1016/j.jbiotec.2009.11.007
Buyl K, De Kock J, Najar M et al (2014) Characterization of hepatic markers in human Wharton’s jelly-derived mesenchymal stem cells. Toxicol In Vitro 28(1):113–119. doi:10.1016/j.tiv.2013.06.014
Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, Collins JJ (2014) Cell Net: network biology applied to stem cell engineering. Cell 158(4):903–915. doi:10.1016/j.cell.2014.07.020
Calder A, Roth-Albin I, Bhatia S et al (2013) Lengthened G1 phase indicates differentiation status in human embryonic stem cells. Stem Cells Dev 22(2):279–295. doi:10.1089/Scd.2012.0168
Campard D, Lysy PA, Najimi M, Sokal EM (2008) Native umbilical cord matrix stem cells express hepatic markers and differentiate into hepatocyte-like cells. Gastroenterology 134(3):833–848. doi:10.1053/j.gastro.2007.12.024
Chetty R, Slavin JL (1994) Epithelioid sarcoma with extensive chondroid differentiation. Histopathology 24(4):400–401. doi:10.1111/j.1365-2559.1994.tb00547.x
Czysz K, Minger S, Thomas N (2015) DMSO efficiently down regulates pluripotency genes in human embryonic stem cells during definitive endoderm derivation and increases the proficiency of hepatic differentiation. PLoS ONE 10(2):e0117689. doi:10.1371/journal.pone.0117689
De Kock J, Najar M, Bolleyn J et al (2012) Mesoderm-derived stem cells: the link between the transcriptome and their differentiation potential. Stem Cells Dev 21(18):3309–3323. doi:10.1089/scd.2011.0723
Denson LA, McClure MH, Bogue CW, Karpen SJ, Jacobs HC (2000) HNF3beta and GATA-4 transactivate the liver-enriched homeobox gene, Hex. Gene 246(1–2):311–320. doi:10.1016/S0378-1119(00)00082-2
Dessimoz J, Opoka R, Kordich JJ, Grapin-Botton A, Wells JM (2006) FGF signaling is necessary for establishing gut tube domains along the anterior-posterior axis in vivo. Mech Dev 123(1):42–55. doi:10.1016/j.mod.2005.10.001
Earp HS, Rubin RA, Austin KS, Dy RC (1983) DMSO increases tyrosine residue phosphorylation in membranes from murine erythroleukemia cells. Biochem Biophys Res Commun 112(2):413–418. doi:10.1016/0006-291X(83)91479-1
Gerets HH, Tilmant K, Gerin B et al (2012) Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell Biol Toxicol 28(2):69–87. doi:10.1007/s10565-011-9208-4
Godoy P, Hewitt NJ, Albrecht U et al (2013) Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 87(8):1315–1530. doi:10.1007/s00204-013-1078-5
Godoy P, Schmidt-Heck W, Natarajan K et al (2015) Gene networks and transcription factor motifs defining the differentiation of stem cells into hepatocyte-like cells. J Hepatol 63(4):934–942. doi:10.1016/j.jhep.2015.05.013
Godoy P, Widera A, Schmidt-Heck W et al (2016) Gene network activity in cultivated primary hepatocytes is highly similar to diseased mammalian liver tissue. Arch Toxicol. doi:10.1007/s00204-016-1761-4 [Epub ahead of print]
Gomez-Lechon MJ, Tolosa L, Conde I, Donato MT (2014) Competency of different cell models to predict human hepatotoxic drugs. Expert Opin Drug Metab Toxicol 10(11):1553–1568. doi:10.1517/17425255.2014.967680
Guillouzo A, Corlu A, Aninat C, Glaise D, Morel F, Guguen-Guillouzo C (2007) The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem Biol Interact 168(1):66–73. doi:10.1016/j.cbi.2006.12.003
Hay DC, Zhao D, Fletcher J et al (2008) Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells 26(4):894–902. doi:10.1634/stemcells.2007-0718
He L, Vasiliou K, Nebert DW (2009) Analysis and update of the human solute carrier (SLC) gene superfamily. Hum Genom 3(2):195–206. doi:10.1186/1479-7364-3-2-195
Hengstler JG, Brulport M, Schormann W et al (2005) Generation of human hepatocytes by stem cell technology: definition of the hepatocyte. Expert Opin Drug Metab Toxicol 1(1):61–74. doi:10.1517/17425255.1.1.61
Hewitt NJ, Lechon MJ, Houston JB et al (2007) Primary hepatocytes: current understanding of the regulation of metabolic enzymes and transporter proteins, and pharmaceutical practice for the use of hepatocytes in metabolism, enzyme induction, transporter, clearance, and hepatotoxicity studies. Drug Metab Rev 39(1):159–234. doi:10.1080/03602530601093489
Kang M, Piliszek A, Artus J, Hadjantonakis AK (2013) FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development 140(2):267–279. doi:10.1242/dev.084996
Kola I, Landis J (2004) Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov 3(8):711–715. doi:10.1038/nrd1470
Kubo A, Kim YH, Irion S et al (2010) The homeobox gene Hex regulates hepatocyte differentiation from embryonic stem cell-derived endoderm. Hepatology 51(2):633–641. doi:10.1002/hep.23293
La Rocca G, Anzalone R, Corrao S et al (2009) Isolation and characterization of Oct-4+/HLA-G+ mesenchymal stem cells from human umbilical cord matrix: differentiation potential and detection of new markers. Histochem Cell Biol 131(2):267–282. doi:10.1007/s00418-008-0519-3
Lee HJ, Jung J, Cho KJ, Lee CK, Hwang SG, Kim GJ (2012) Comparison of in vitro hepatogenic differentiation potential between various placenta-derived stem cells and other adult stem cells as an alternative source of functional hepatocytes. Differentiation 84(3):223–231. doi:10.1016/j.diff.2012.05.007
Leite SB, Teixeira AP, Miranda JP et al (2011) Merging bioreactor technology with 3D hepatocyte-fibroblast culturing approaches: improved in vitro models for toxicological applications. Toxicol In Vitro 25(4):825–832. doi:10.1016/j.tiv.2011.02.002
Li X, Yuan J, Li W et al (2014) Direct differentiation of homogeneous human adipose stem cells into functional hepatocytes by mimicking liver embryogenesis. J Cell Physiol 229(6):801–812. doi:10.1002/jcp.24501
Liu WH, Liu ZC, You N et al (2012) Several important in vitro improvements in the amplification, differentiation and tracing of fetal liver stem/progenitor cells. PLoS ONE 7(10):e47346. doi:10.1371/journal.pone.0047346
Martins JP, Santos JM, Almeida JM et al (2014) Towards an advanced therapy medicinal product based on mesenchymal stromal cells isolated from the umbilical cord tissue: quality and safety data. Stem Cell Res Ther 5:9. doi:10.1186/scrt398
Miranda JP, Leite SB, Muller-Vieira U, Rodrigues A, Carrondo MJ, Alves PM (2009) Towards an extended functional hepatocyte in vitro culture. Tissue Eng Part C Methods 15(2):157–167. doi:10.1089/ten.tec.2008.0352
Miranda J, Rodrigues A, Tostoes R et al (2010) Extending hepatocyte functionality for drug-testing applications using high-viscosity alginate-encapsulated three-dimensional cultures in bioreactors. Tissue Eng Part C Methods 16(6):1223–1232. doi:10.1089/ten.TEC.2009.0784
Miranda JP, Filipe E, Fernandes AS et al (2015) The human umbilical cord tissue-derived MSC population UCX® promotes early motogenic effects on keratinocytes and fibroblasts and G-CSF-mediated mobilization of BM-MSCs when transplanted in vivo. Cell Transplant 24(5):865–877. doi:10.3727/096368913X676231
Oliveros JC (2007–2015) Venny. An interactive tool for comparing lists with Venn's diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html
Pal R, Mamidi MK, Das AK, Bhonde R (2012) Diverse effects of dimethyl sulfoxide (DMSO) on the differentiation potential of human embryonic stem cells. Arch Toxicol 86(4):651–661. doi:10.1007/s00204-011-0782-2
Rajan N, Habermehl J, Cote MF, Doillon CJ, Mantovani D (2006) Preparation of ready-to-use, storable and reconstituted type I collagen from rat tail tendon for tissue engineering applications. Nat Protoc 1(6):2753–2758. doi:10.1038/nprot.2006.430
Rubbini S, Cocco L, Manzoli L et al (1997) Phosphoinositide signalling in nuclei of Friend cells: DMSO-induced differentiation reduces the association of phosphatidylinositol-transfer protein with the nucleus. Biochem Biophys Res Commun 230(2):302–305. doi:10.1006/bbrc.1996.5950
Rui L (2014) Energy metabolism in the liver. Compr Physiol 4(1):177–197. doi:10.1002/cphy.c130024
Santos JMS, Soares R, Martins JP, et al (2008) Optimised and defined method for isolation and preservation of precursor cells from human umbilical cord, INPI PAT20081000083882; PCT/IB2008/054067; WO2009044379. Medinfar, ECBio. https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2009044379
Santos JM, Barcia RN, Simões SI et al (2013) The role of human umbilical cord tissue-derived mesenchymal stromal cells (UCX®) in the treatment of inflammatory arthritis. J Transl Med 11:18. doi:10.1186/1479-5876-11-18
Santos JM, Camões SP, Filipe E et al (2015) Three-dimensional spheroid cell culture of umbilical cord tissue-derived mesenchymal stromal cells leads to enhanced paracrine induction of wound healing. Stem Cell Res Ther 6:90. doi:10.1186/s13287-015-0082-5
Schwartz RE, Fleming HE, Khetani SR, Bhatia SN (2014) Pluripotent stem cell-derived hepatocyte-like cells. Biotechnol Adv 32(2):504–513. doi:10.1016/j.biotechadv.2014.01.003
Seeliger C, Culmes M, Schyschka L et al (2013) Decrease of global methylation improves significantly hepatic differentiation of Ad-MSCs: possible future application for urea detoxification. Cell Transplant 22(1):119–131. doi:10.3727/096368912X638946
Sjogren AK, Liljevald M, Glinghammar B et al (2014) Critical differences in toxicity mechanisms in induced pluripotent stem cell-derived hepatocytes, hepatic cell lines and primary hepatocytes. Arch Toxicol 88(7):1427–1437. doi:10.1007/s00204-014-1265-z
Snykers S, Vanhaecke T, Papeleu P et al (2006) Sequential exposure to cytokines reflecting embryogenesis: the key for in vitro differentiation of adult bone marrow stem cells into functional hepatocyte-like cells. Toxicol Sci 94(2):330–341. doi:10.1093/toxsci/kfl058
Snykers S, Vanhaecke T, De Becker A et al (2007) Chromatin remodeling agent trichostatin A: a key-factor in the hepatic differentiation of human mesenchymal stem cells derived of adult bone marrow. BMC Dev Biol 7:24. doi:10.1186/1471-213X-7-24
Su T, Waxman DJ (2004) Impact of dimethyl sulfoxide on expression of nuclear receptors and drug-inducible cytochromes P450 in primary rat hepatocytes. Arch Biochem Biophys 424(2):226–234. doi:10.1016/j.abb.2004.02.008
Ulvestad M, Nordell P, Asplund A et al (2013) Drug metabolizing enzyme and transporter protein profiles of hepatocytes derived from human embryonic and induced pluripotent stem cells. Biochem Pharmacol 86(5):691–702. doi:10.1016/j.bcp.2013.06.029
Vyhlidal CA, Gaedigk R, Leeder JS (2006) Nuclear receptor expression in fetal and pediatric liver: correlation with CYP3A expression. Drug Metab Dispos 34(1):131–137. doi:10.1124/dmd.105.005967
Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M (2009) gplots: Various R programming tools for plotting data. R Package Version 2(4)
Wilke RA, Lin DW, Roden DM et al (2007) Identifying genetic risk factors for serious adverse drug reactions: current progress and challenges. Nat Rev Drug Discov 6(11):904–916. doi:10.1038/nrd2423
Wilkening S, Stahl F, Bader A (2003) Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties. Drug Metab Dispos 31(8):1035–1042. doi:10.1124/dmd.31.8.1035
Yen A, Varvayanis S (1995) DMSO, sodium butyrate, and TPA induce hypophosphorylation of RB with HL-60 cell differentiation. In Vitro Cell Dev Biol Anim 31(3):164–167. doi:10.1007/BF02639427
Yoon HH, Jung BY, Seo YK, Song KY, Park JK (2010) In vitro hepatic differentiation of umbilical cord-derived mesenchymal stem cell. Process Biochem 45(12):1857–1864. doi:10.1016/J.Procbio.2010.06.009
Yoshida Y, Shimomura T, Sakabe T et al (2007) A role of Wnt/beta-catenin signals in hepatic fate specification of human umbilical cord blood-derived mesenchymal stem cells. Am J Physiol Gastrointest Liver Physiol 293(5):G1089–G1098. doi:10.1152/ajpgi.00187.2007
Zhang W, Yatskievych TA, Baker RK, Antin PB (2004) Regulation of Hex gene expression and initial stages of avian hepatogenesis by Bmp and Fgf signaling. Dev Biol 268(2):312–326. doi:10.1016/j.ydbio.2004.01.019
Zhang YN, Lie PC, Wei X (2009) Differentiation of mesenchymal stromal cells derived from umbilical cord Wharton’s jelly into hepatocyte-like cells. Cytotherapy 11(5):548–558. doi:10.1080/14653240903051533
Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, Duncan SA (2005) GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol Cell Biol 25(7):2622–2631. doi:10.1128/MCB.25.7.2622-2631.2005
Zhou R, Li Z, He C et al (2014) Human umbilical cord mesenchymal stem cells and derived hepatocyte-like cells exhibit similar therapeutic effects on an acute liver failure mouse model. PLoS ONE 9(8):e104392. doi:10.1371/journal.pone.0104392
Acknowledgements
The authors thank Alexandra Medeiros for technical support on immunochemistry assays. This work was supported by Fundação para a Ciência e a Tecnologia (FCT, Portugal) through research grants (PTDC/SAU-TOX/110457/2009, PEst-OE/SAU/UI4013/2011, UID/DTP/04138/2013, SFRH/BD/87508/2012 to M.C., PD/BD/114280/2016 to S.P.C. and SFRH/BPD/96719/2013 and IF/00846/2015 to J.P.M.) and by Human Frontier Science Program (Young Investigator grant to J.L.R). We also acknowledge the support of the COST action BM1305 (A FACTT: Action to focus and accelerate cell-based tolerance-inducing therapies). The work herein presented was performed at iMed.ULisboa and Karolinska Institutet.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
H.C. and P.C. are shareholders of ECBio; J.M.S. is employee of ECBio. The other authors declare that they have no conflict of interests.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This study was approved by the Ethics Committee of the Hospital Dr. José de Almeida (Cascais, Portugal), in scope of a research protocol between ECBio (Research & Development in Biotechnology, S.A.) and HPP Saúde (Parcerias Cascais, S.A.). Umbilical cord donations, with written informed consents, as well as umbilical cord procurement, were made according to Directive 2004/23/EC of the European Parliament and of the Council of 31 March 2004 on setting standards of quality and safety for the donation, procurements, testing, processing, preservation, storage and distribution of human tissues and cells. All applicable international, national and/or institutional guidelines for the care and use of animals were followed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cipriano, M., Correia, J.C., Camões, S.P. et al. The role of epigenetic modifiers in extended cultures of functional hepatocyte-like cells derived from human neonatal mesenchymal stem cells. Arch Toxicol 91, 2469–2489 (2017). https://doi.org/10.1007/s00204-016-1901-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-016-1901-x