Abstract
Objectives
The aim of the study is to generate a spherical three-dimensional (3D) aggregate of hepatocyte-like cells (HLCs) differentiated from human embryonic stem cells and to investigate the effect of the 3D environment on hepatic maturation and drug metabolism.
Results
Quantitative real-time PCR analysis indicated that gene expression of mature hepatocyte markers, drug-metabolizing enzymes, and hepatic transporters was significantly higher in HLCs cultured in the 3D system than in those cultured in a two-dimensional system (p < 0.001). Moreover, hepatocyte-specific functions, including albumin secretion and bile canaliculi formation, were increased in HLCs cultured in the 3D system. In particular, 3D spheroidal culture increased expression of CES1 and BCHE, which encode hepatic esterases (p < 0.001). The enhanced activities of these hepatic esterases were confirmed by the cholinesterase activity assay and the increased susceptibility of HLCs to oseltamivir, which is metabolized by CES1.
Conclusions
3D spheroidal culture enhances the maturation and drug metabolism of stem cell-derived HLCs, and this may help to optimize hepatic differentiation protocols for hepatotoxicity testing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocyte-like cells (HLCs) derived from human pluripotent stem cells (hPSCs) are an attractive alternative to immortalized/primary hepatocytes and experimental animals for hepatotoxicity and pharmaceutical studies (Hook 2012). Many research groups have developed and optimized hepatic differentiation protocols to generate functional HLCs that mimic embryonic liver development (Cai et al. 2007; Shiraki et al. 2008; Sancho-Bru et al. 2011; Takayama et al. 2012). However, the phenotype and functions of hPSC-derived HLCs differ from those of mature hepatocytes and are similar to those of fetal hepatocytes (Davila et al. 2004). In particular, the expression and activities of drug-metabolizing enzymes (DMEs) and drug transporters are much lower in hPSC-derived HLCs than in primary human hepatocytes (PHHs).
Various three-dimensional (3D) systems, including hanging drop, bioreactors, and synthetic matrixes, have been developed to improve the efficiency of cell culture (Pampaloni et al. 2007; Justice et al. 2009). The physiologic functions of cells in vivo are better mimicked by culture in a 3D system than by culture in a two-dimensional (2D) monolayer. Loss of the hepatic phenotype of rat primary hepatocytes upon culture in a 2D system can be attenuated or even reversed by culture in a 3D system (Bierwolf et al. 2011). In addition, hepatic differentiation in various 3D culture systems promotes the functional maturation of hPSC-derived HLCs (Baharvand et al. 2006; Nagamoto et al. 2012; Ogawa et al. 2013; Takayama et al. 2013; Gieseck et al. 2014).
This study describes an efficient hepatic differentiation protocol employing a simple 3D cell culture method. Human embryonic stem cell (hESC)-derived HLCs cultured in a 3D system were more mature than those cultured in a 2D monolayer. Gene expression of hepatocyte markers, DMEs, and transporters was significantly higher in the former cells than in the latter cells. Moreover, expression of hepatic esterases was increased in HLCs in 3D spheroids. A cholinesterase activity assay and oseltamivir treatment confirmed that the activities of hepatic esterases were increased in these cells.
Materials and methods
Culture and hepatic differentiation of hESCs
hESCs (CHA-hES15) were obtained from CHA University. These cells were maintained and differentiated into HLCs as previously described (Lee et al. 2010; Park et al. 2015). In brief, the cells were cultured in DMEM/F12 medium (Invitrogen) containing 4 ng/ml basic fibroblast growth factor (bFGF; R&D Systems), sodium bicarbonate (Sigma), GlutaMAX (Invitrogen), non-essential amino acids (Invitrogen), β-mercaptoethanol (Sigma), and penicillin–streptomycin (Sigma) on a feeder layer of mouse embryonic fibroblasts treated with mitomycin C (Sigma). To induce differentiation into HLCs, hESCs were first passaged onto Matrigel-coated plates (Becton, Dickinson & Company) and cultured in mTeSR1 medium (Stem cell Technologies) containing 4 ng/ml bFGF. To induce definitive endoderm differentiation, hESCs were cultured in RPMI-1640 medium (Lonza) containing 50 ng/ml Activin A (PeproTech), 0.5 mg/ml bovine serum albumin (BSA, Sigma-Aldrich), 2% vol/vol B27 supplement (Gibco), and 0.5 mM sodium butylate (SB, Sigma) for 1 day and then in this medium containing 0.1 mM instead of 0.5 mM SB for 4 days. To induce differentiation into hepatoblasts, cells were cultured in RPMI-1640 medium (Lonza) containing 10 ng/ml hepatocyte growth factor (HGF, PeproTech), 10 ng/ml fibroblast growth factor 4 (FGF4, PeproTech), 0.5 mg/ml BSA, and 2% vol/vol B27 supplement for 5 days. Thereafter, to induce differentiation into HLCs, cells were cultured in hepatocyte culture medium (HCM, Lonza) containing 10 ng/ml HGF, 10 ng/ml FGF4, and 10 ng/ml oncostatin M (OSM, PeproTech) for 7 days. All cells were cultured at 37 °C in 5% CO2. The culture medium was changed every day.
3D spheroid culture
To induce spheroid formation, hepatoblasts were dissociated with Accutase (Stem Cell Technologies). Dissociated cells were stained with Trypan blue to monitor their viability and counted using a hemocytometer. Cells were seeded into a 96-well round-bottom ultra-low attachment (ULA) plate (Corning) at a density of 1 × 104 cells per well in HCM supplemented with 10 ng/ml HGF, 10 ng/ml FGF4, and 10 ng/ml OSM before being centrifuged at 450 g for 3 min to induce their aggregation. The plates were incubated at 37 °C in 5% CO2 for 10 days. During the 3D culture period, half of the culture medium was changed every day.
Live/Dead and cytotoxicity assays
The Live/Dead assay kit (Invitrogen) was used to evaluate cell viability as described in the manufacturer’s instructions. Briefly, cells and spheroids were incubated with 2 μM calcein acetoxymethyl ester and 4 μM ethidium homodimer at 37 °C for 30 min and then imaged by fluorescence microscopy (Olympus). The fluorescence of each well was measured using a fluorescence microplate reader (Promega). The 50% toxic concentration (TC50) values of oseltamivir were determined after 48 h of treatment.
Quantitative real-time PCR
Total RNA was isolated from the HLCs using TRIzol (Invitrogen) in accordance with the manufacturer’s instructions. cDNA was synthesized from RNA (1–5 μg) with SuperScript II Reverse Transcriptase (Invitrogen). Quantitative PCR was performed using a StepOnePlus Real-Time PCR system (Applied Biosystems) and Power SYBR Green PCR Master Mix (Applied Biosystems). GAPDH was used to normalize the gene expression data.
Immunofluorescence staining
After fixation in 4% paraformaldehyde, cells and spheroids were washed with Dulbecco’s phosphate-buffered saline (DPBS) three times, permeabilized in 0.1% Triton X-100 (Sigma), and then washed again with DPBS. Thereafter, cells and spheroids were blocked in 5% goat serum for 1 h at room temperature, labeled with primary antibodies overnight at 4 °C, and washed with DPBS for three times. Finally, samples were incubated with appropriate secondary antibodies conjugated to Alexa-488 or Alexa-594 (Invitrogen) at RT for 1 h, washed three times with DPBS, and counterstained with DAPI for 10 min.
Albumin secretion assay
The medium of HLCs was collected after 24 h of culture. The concentration of albumin was determined using a Human Albumin ELISA Quantitation Kit (Bethyl Laboratories) in accordance with the manufacturer’s instructions and a Model 680 microplate reader (Bio-Rad). Data were normalized by the cell number.
Urea assay
After differentiation, HLCs were incubated in HCM for 24 h. The medium was collected and assayed using a QuantiChrom Urea Assay Kit (BioAssay Systems) according to the manufacturer’s protocol. Absorbance was measured using a Model 680 microplate reader (Bio-Rad). Data were normalized by the cell number.
Acetylated low-density lipoprotein (Ac-LDL) uptake assay
Cells were incubated with 10 μg/ml acetylated low-density lipoprotein (Ac-LDL) for 5 h at 37 °C and then imaged by fluorescence microscopy (Olympus).
Uptake and release of indocyanine green (ICG)
HLCs were treated with 1 mg/ml indocyanine green (ICG; Sigma) for 1 h at 37 °C in 5% CO2, washed with phosphate-buffered saline, and imaged using an inverted microscope (Olympus).
Bile canaliculi formation assay
Cells and spheroids were incubated with HCM containing 10 μM 5 (and 6)-carboxy-2,7-dichlorofluorescein diacetate (CDFDA, Life Technologies) for 15 min at 37 °C, and then washed with HCM. To inhibit multidrug resistance-associated proteins (MRPs), samples were incubated with HCM containing 2 mM probenecid (Sigma-Aldrich) for 1 h prior to CDFDA treatment. Green fluorescence was visualized using fluorescence microscopy (Olympus).
Cholinesterase activity assay
Cholinesterase activity was analyzed using a commercial ELISA kit (Abcam) according to the manufacturer’s instructions. The absorbance at a wavelength of 415 nm was measured with a microplate reader (Model 680, Bio-Rad).
Statistical analysis
The data are expressed as mean ± standard deviation (SD). Statistical analyses were performed using the t test and Mann–Whitney test. p values less than 0.05 were considered to be statistically significant.
Results
Differentiation of HLCs from hESCs in a 3D culture system
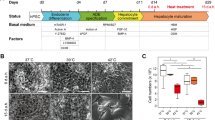
hESCs were treated with growth factors and cytokines to stimulate their differentiation into HLCs (Fig. 1a). At day 10, hepatoblasts were detached from the cell culture plate, seeded into a 96-well ULA plate at a density of 1.0 × 104 cells/well, and centrifuged to induce the formation of spheroids. Spheroids had formed in all wells at 24 h after centrifugation, and this spherical shape was maintained until the end of the differentiation protocol (day 17) (Fig. 1b). The Live/Dead assay confirmed that most HLCs cultured in the 3D system (3D HLCs) were alive (Fig. 1b).
Differentiation of HLCs in a 3D system. a Protocol used to differentiate hESCs into HLCs. Cells were cultured in the 3D system from day 10 to day 17. b Experimental scheme of 3D cell culture method using an ULA plate. A fluorescence image of 3D HLCs stained with a Live/Dead kit (day 17) is shown. Scale bars represent 200 μm
Culture in the 3D system promotes the maturation of HLCs
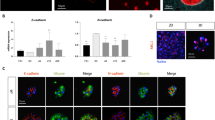
We performed qPCR and immunofluorescence staining to investigate whether this 3D cell culture method induced the maturation of HLCs. qPCR data demonstrated that culture in the 3D system increased the expression of some (constitutive androstane receptor (CAR) and pregnane-X receptor (PXR)), but not all (aryl hydrocarbon receptor (AhR)), hepatic nuclear receptor genes. Expression of the hepatic marker genes hepatic nuclear factor 4 alpha (HNF4α), albumin (ALB), alpha-1 antitrypsin (AAT), transthyretin (TTR), tryptophan 2,3-dioxygenase (TDO2), and glucose 6-phosphatase (G6P) was increased in 3D HLCs. Furthermore, expression of some cytochrome P450 (CYP) genes (CYP1A1, CYP2C9, CYP2D6, and CYP7A1), phase II DMEs (UGT1A1 and UGT2B7), and phase III transporters (MRP2 and MRP3) was higher in 3D HLCs than in HLCs cultured in a 2D system (2D HLCs) (Fig. 2a). Immunofluorescence staining revealed that AAT, ALB, CK8, CK18, and CYP1A2 were expressed in both types of HLCs. More 3D HLCs than 2D HLCs were positive for UGT2B7, MDR1, MRP2, and MRP3 (Fig. 2b). Collectively, these results demonstrate that our 3D cell culture system promoted the maturation of HLCs.
Expression in 2D and 3D HLCs. a Relative gene expression levels of hepatic receptors, hepatic markers, DMEs, and transporters in 3D HLCs at the end of the differentiation protocol (day 17). Results are mean ± SD (n = 3), *p < 0.05, **p < 0.01, ***p < 0.001 versus 2D HLCs (t test followed by the Mann–Whitney test). b Immunocytochemical analysis of 2D and 3D HLCs. Expression of hepatic markers, DMEs, and transporters was detected by immunocytochemistry at day 17. Scale bars represent 200 μm
Culture in the 3D system enhances the hepatic functions of HLCs
To determine whether culture in the 3D system enhances hepatic functions, we investigated some hepatic characteristics of HLCs. 3D HLCs secreted a slightly higher level of albumin than 2D HLCs (Fig. 3a). The amount of urea produced by these two types of HLCs did not significantly differ (Fig. 3a). The Ac-LDL uptake assay demonstrated that both types of HLCs could take up LDL (Fig. 3b). Moreover, 3D HLCs took up and released ICG (Fig. 3c). To investigate bile canaliculi formation, we treated HLCs with CDFDA. Bile canaliculi were observed in 3D HLCs, and the fluorescence intensity gradually decreased over time (Fig. 4a). Taken together, these results show that 3D spheroidal culture enhances some hepatic functions of HLCs.
Hepatic functions of 2D and 3D HLCs. a Levels of albumin secretion and urea production were determined in 2D and 3D HLCs. Results are mean ± SD (n = 3), *p < 0.05 versus 2D HLCs (t test followed by the Mann–Whitney test). b The Ac-LDL uptake assay was conducted at the end of the differentiation protocol. Scale bars represent 200 μm. c ICG uptake and release. Scale bars represent 200 μm
Activities of hepatic esterases and transporters in 3D HLCs. a Bile canaliculi formation assay using CDFDA. MRPs were inhibited by treatment with 2 mM probenecid. Scale bars represent 200 μm. b Gene expression analysis of hepatic esterases. c Cholinesterase activity was analyzed by an ELISA. d, e Cytotoxicity curves and Live/Dead staining of oseltamivir-treated 2D and 3D HLCs. Scale bars represent 200 μm. Results are mean ± SD (n = 3), *p < 0.05, **p < 0.01, ***p < 0.001 versus 2D HLCs (t test followed by the Mann–Whitney test)
Hepatic esterases and transporters are active in 3D HLCs
CDFDA diffuses through the plasma membranes and is secreted by MRP2 and MRP3 after being hydrolyzed to CDF by intracellular esterases (Zamek-Gliszczynski et al. 2003). To investigate the activities of MRP2 and MRP3, we treated cells with 2 μM probenecid, an inhibitor of MRPs, prior to CDFDA (Laupèze et al. 2001; Ramboer et al. 2013). At each time point, the fluorescence intensity was higher in cells pretreated with probenecid than in cells that were not (Fig. 4a). Gene expression of hepatic esterases, such as BCHE, CES1, and CES2, which hydrolyze CDFDA and are phase I DMEs (Zamek-Gliszczynski et al. 2003), was up-regulated in 3D HLCs (Fig. 4b). The activities of these esterases were measured to determine the effect of 3D culture. The cholinesterase activity assay confirmed that the activities of cholinesterases (AchE and BCHE) were significantly increased in 3D HLCs (Fig. 4c). To demonstrate the metabolic function of CES1, we treated HLCs with various concentrations of oseltamivir, which was reported to cause hepatotoxicity after being metabolized by CES1 (Shi et al. 2006). After 48 h of treatment, the Live/Dead assay demonstrated that the TC50 values were about 2645 and 1295 μM in 2D and 3D HLCs, respectively. Moreover, more 3D HLCs than 2D HLCs died following treatment with the same concentration of oseltamivir (Fig. 4d, e). These data indicate that CES1 activity was increased in 3D HLCs. Overall, these findings show that culture in the 3D system does not only simply up-regulate the expression of DMEs in HLCs but also increases their activities.
Discussion
Preclinical evaluation of hepatotoxicity is critical for successful drug development. Cell models of hepatic drug metabolism are essential to accurately predict hepatotoxicity. Culture in a 3D system better replicates the in vivo environment, and such cells are expected to be more similar to those in vivo than cells cultured in a 2D system. HLCs differentiated from hPSCs in a 3D system display intrinsic functions of hepatocytes and increased expression of DMEs compared with those differentiated in a 2D system (Takayama et al. 2013). PHHs cultured in a 3D system can be used to evaluate the toxicity of acetaminophen, a drug that typically causes hepatotoxicity (Schyschka et al. 2013). Acetaminophen is converted to the toxic metabolite N-acetyl-p-benzoquinone imine by CYP2E1 and is excreted from hepatocytes by acetaminophen-specific transporters such as MRP1 (Zaher et al. 1998; Aleksunes et al. 2007). In PHHs cultured in a 3D system, acetaminophen treatment increases the activities of CYP2E1 and MRP1 (Schyschka et al. 2013). These reports indicate that 3D culture methods can generate in vitro models applicable for hepatotoxicity testing and mechanistic studies.
However, despite the advantages of 3D hepatic models generated using various technologies, each approach has drawbacks. For example, when cells are cultured using extracellular matrix (ECM) components, such as matrigel, variation may arise between batches. Other problems associated with in vitro hepatotoxicity models include binding of drugs to scaffolds, handling difficulties, and high cost (Godoy et al. 2013; Bell et al. 2016). In this study, we developed a 3D hepatic model using an ULA microplate. This method does not use ECM components or scaffolds, and is easy to perform and relatively inexpensive. In addition, spheroids formed almost simultaneously in all wells, and the number of cells in each spheroid could be controlled. This spheroidal culture did not induce cell death (Fig. 1b). Moreover, expression of DMEs and hepatocyte markers and hepatocyte-specific functions were enhanced in 3D HLCs (Figs. 2, 3). These results indicate that 3D HLCs are more similar to mature hepatocytes than 2D HLCs.
Esterases hydrolyze prodrugs containing ester, amide, and thioester bonds, and this is involved in the metabolism of about 10% of therapeutic drugs (Williams et al. 2004; Fukami and Yokoi 2012). Carboxylesterases play an important role in the hydrolysis of drugs, and recent studies reported genetic polymorphisms of carboxylesterases (CES1 and CES2) and drug–drug and drug–disease interactions of drugs hydrolyzed by these enzymes (Casey Laizure et al. 2013). Yamada and colleagues reported that patients with latent tuberculosis infections treated with isoniazid can develop severe hepatotoxicity depending on the genetic characteristics of CES1 (Yamada et al. 2010). The anti-influenza agent oseltamivir, commonly known as Tamiflu, is also hydrolyzed by CES1 (Shi et al. 2006). Oseltamivir phosphate (OP), an oral prodrug, is rapidly metabolized into oseltamivir carboxylate (OC), an active form in the liver (Davies 2010). Although it is unclear how these compounds cause hepatotoxicity, OC is more toxic than OP (Shi et al. 2006). OC accumulates in CES1-overexpressing cells, causing a high level of cytotoxicity (Shi et al. 2006). In this study, expression of CES1 and CES2 was increased in 3D HLCs (Fig. 4b). In addition, oseltamivir treatment was more cytotoxic to 3D HLCs than to 2D HLCs (Fig. 4d, e). These data demonstrate that CES1 has hydrolysis activity in 3D HLCs.
BCHE, another hepatic esterase, is synthesized in the liver and released into plasma, and many drugs are reportedly substrates of this cholinesterase (Fukami and Yokoi 2012). Moreover, BCHE has various polymorphisms, and sensitivity to nerve agents such as soman is reportedly related to BCHE variants (Dimov et al. 2012). The expression and activities of cholinesterases were higher in 3D HLCs than in 2D HLCs (Fig. 4b, c). Overall, 3D HLCs are a valuable model for predicting and studying hepatotoxicity related to esterases. In particular, these cells have great potential to elucidate the relationship between toxicity and esterase polymorphisms.
Conclusions
We produced hepatic spheroids using a 3D cell culture method. Hepatocyte marker expression and hepatic functions were enhanced in these cells compared with 2D HLCs. Moreover, expression of DMEs and hepatic transporters was significantly up-regulated in 3D HLCs. In particular, we demonstrated that hepatic esterase activity was higher in 3D HLCs than in 2D HLCs. Therefore, these 3D HLCs can be a useful tool for hepatotoxicity testing and esterase-related drug studies.
References
Aleksunes LM, Augustine LM, Cherrington NJ, Manautou JE (2007) Influence of acetaminophen vehicle on regulation of transporter gene expression during hepatotoxicity. J Toxicol Environ Health A 70:1870–1872
Baharvand H, Hashemi SM, Ashtiani SK, Farrokhi A (2006) Differentiation of human embryonic stem cells into hepatocytes in 2D and 3D culture systems in vitro. Int J Dev Biol 50:645–652
Bell CC, Hendriks DFG, Moro SML, Elis E et al (2016) Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci Rep 6:25187
Bierwolf J, Lutgehetmann M, Feng K, Erbes J, Deichmann S, Toronyi E, Stieglitz C, Nashan B, Ma PX, Pollok JM et al (2011) Primary rat hepatocyte culture on 3D nanofibrous polymer scaffolds for toxicology and pharmaceutical research. Biotechnol Bioeng 108:141–150
Cai J, Zhao Y, Liu Y, Ye F et al (2007) Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology 45:1229–1239
Casey Laizure S, Herring V, Hu Z, Witbrodt K, Parker RB (2013) The role of human carboxylesterases in drug metabolism: have we overlooked their importance? Pharmacotherapy 33:210–222
Davies BE (2010) Pharmacokinetics of oseltamivir: an oral antiviral for the treatment and prophylaxis of influenza in diverse populations. J Antimicrob Chemother 65(Suppl 2):5–10
Davila JC, Cezar GG, Thiede M, Strom S, Miki T, Trosko J (2004) Use and application of stem cells in toxicology. Toxicol Sci 79:214–223
Dimov D, Kanev K, Dimova I (2012) Correlation between butyrylcholinesterase variants and sensitivity to soman toxicity. Acta Biochim Pol 59:313–316
Fukami T, Yokoi T (2012) The emerging role of human esterases. Drug Metab Pharmacokinet 27:466–477
Gieseck RL, Hannan NRF, Bort R, Hanley NA, Drake RAL, Cameron GWW, Wynn TA, Vallier L (2014) Maturation of induced pluripotent stem cell derived hepatocytes by 3D-culture. PLoS ONE 9:e86372
Godoy P, Hewitt NJ, Albrecht U, Andersen ME et al (2013) Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 87:1315–1530
Hook LA (2012) Stem cell technology for drug discovery and development. Drug Discov Today 17:336–342
Justice BA, Badr NA, Felder RA (2009) 3D cell culture opens new dimensions in cell-based assays. Drug Discov Today 14:102–107
Laupèze B, Amiot L, Payen L, Drenou B, Grosset JM, Lehne G, Fauchet R, Fardel O (2001) Multidrug resistance protein (MRP) activity in normal mature leukocytes and CD34-positive hematopoietic cells from peripheral blood. Life Sci 68:1323–1331
Lee JE, Kang MS, Park MH, Shim SH, Yonn TK, Chung HM, Lee DR (2010) Evaluation of 28 human embryonic stem cell lines for use as unrelated donors in stem cell therapy: implications of HLA and ABO genotypes. Cell Transplant 19:1383–1395
Nagamoto Y, Tashiro K, Takayama K, Ohashi K, Kawabata K, Sakurai F, Tachibana M, Hayakawa T, Furue MK, Mizuguchi H (2012) The promotion of hepatic maturation of human pluripotent stem cells in 3D co-culture using type I collagen and Swiss 3T3 cell sheets. Biomaterials 33:4526–4534
Ogawa S, Surapisitchat J, Virtanen C, Ogawa M et al (2013) Three-dimensional culture and cAMP signaling promote the maturation of human pluripotent stem cell-derived hepatocytes. Development 140:3285–3296
Pampaloni F, Reynaud EG, Stelzer EHK (2007) The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol 8:839–845
Park H-J, Choi Y-J, Kim JW, Chun HS, Im I, Yoo S, Han Y-M, Song C-W, Kim H (2015) Differences in the epigenetic regulation of cytochrome P450 genes between human embryonic stem cell-derived hepatocytes and primary hepatocytes. PLoS ONE 10:e0132992
Ramboer E, Vanhaecke T, Rogiers V, Vinken M (2013) Primary hepatocyte cultures as prominent in vitro tools to study hepatic drug transporters. Drug Metab Rev 45:196–217
Sancho-Bru P, Roelandt P, Narain N, Pauwelyn K, Notelaers T, Shimizu T, Ott M, Verfaillie C (2011) Directed differentiation of murine-induced pluripotent stem cells to functional hepatocyte-like cells. J Hepatol 54:98–107
Schyschka L, Sánchez JJM, Wang Z, Burkhardt B, Müller-Vieira U, Zeilinger K, Bachmann A, Nadalin S, Damm G, Nussler AK (2013) Hepatic 3D cultures but not 2D cultures preserve specific transporter activity for acetaminophen-induced hepatotoxicity. Arch Toxicol 87:1581–1593
Shi D, Yang J, Yang D, LeCluyse EL, Black C, You L, Akhlaghi F, Yan B (2006) Anti-influenza prodrug oseltamivir is activated by carboxylesterase human carboxylesterase 1, and the activation is inhibited by antiplatelet agent clopidogrel. J Pharmacol Exp Ther 319:1477–1484
Shiraki N, Umeda K, Sakashita N, Takeya M, Kume K, Kume S (2008) Differentiation of mouse and human embryonic stem cells into hepatic lineages. Genes Cells 13:731–746
Takayama K, Inamura M, Kawabata K et al (2012) Efficient generation of functional hepatocytes from human embryonic stem cells and induced pluripotent stem cells by HNF4α transduction. Mol Ther 20:127–137
Takayama K, Kawabata K, Nagamoto Y et al (2013) 3D spheroid culture of hESC/hiPSC-derived hepatocyte-like cells for drug toxicity testing. Biomaterials 34:1781–1789
Williams JA, Hyland R, Jones BC et al (2004) Drug–drug interactions for UDP-glucoronosyltransferase substrates: a pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug Metab Dispos 32:1201–1208
Yamada S, Richardson K, Tang M et al (2010) Genetic variation in carboxylesterase genes and susceptibility to isoniazid-induced hepatotoxicity. Pharmacogenomics J 10:524–536
Zaher H, Buters JT, Ward JM et al (1998) Protection against acetaminophen toxicity in CYP1A2 and CYP2E1 double-null mice. Toxicol Appl Pharmacol 152:193–199
Zamek-Gliszczynski MJ, Xiong H, Patel NJ et al (2003) Pharmacokinetics of 5 (and 6)-carboxy-2′,7′-dichlorofluorescein and its diacetate promoiety in the liver. J Pharmacol Exp Ther 304:801–809
Acknowledgements
The authors would like to thank the CHA Stem Cell Institute (CHA University, Korea) for providing the CHA-hES15 cell line. This work was funded by the Basic Science Research Program through National Research Foundation (NRF) funding from the Ministry of Science and ICT, Republic of Korea (No. NRF-2017R1C1B2010444).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Choi, YJ., Kim, H., Kim, JW. et al. Hepatic esterase activity is increased in hepatocyte-like cells derived from human embryonic stem cells using a 3D culture system. Biotechnol Lett 40, 755–763 (2018). https://doi.org/10.1007/s10529-018-2528-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-018-2528-1