Abstract
Polymorphisms of N-acetyltransferase 2 (NAT2) are well known to modify urinary bladder cancer risk as well as efficacy and toxicity of pharmaceuticals via reduction in the enzyme’s acetylation capacity. Nevertheless, the discussion about optimal NAT2 phenotype prediction, particularly differentiation between different degrees of slow acetylation, is still controversial. Therefore, we investigated the impact of single nucleotide polymorphisms and their haplotypes on slow acetylation in vivo and on bladder cancer risk. For this purpose, we used a study cohort of 1,712 bladder cancer cases and 2,020 controls genotyped for NAT2 by RFLP-PCR and for the tagSNP rs1495741 by TaqMan® assay. A subgroup of 344 individuals was phenotyped by the caffeine test in vivo. We identified an ‘ultra-slow’ acetylator phenotype based on combined *6A/*6A, *6A/*7B and *7B/*7B genotypes containing the homozygous minor alleles of C282T (rs1041983, *6A, *7B) and G590A (rs1799930, *6A). ‘Ultra-slow’ acetylators have significantly about 32 and 46 % lower activities of caffeine metabolism compared with other slow acetylators and with the *5B/*5B genotypes, respectively (P < 0.01, both). The ‘ultra-slow’ genotype showed an association with bladder cancer risk in the univariate analysis (OR = 1.31, P = 0.012) and a trend adjusted for age, gender and smoking habits (OR = 1.22, P = 0.082). In contrast, slow acetylators in general were not associated with bladder cancer risk, neither in the univariate (OR = 1.02, P = 0.78) nor in the adjusted (OR = 0.98, P = 0.77) analysis. In conclusion, this study suggests that NAT2 phenotype prediction should be refined by consideration of an ‘ultra-slow’ acetylation genotype.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Large meta-analyses have shown an association between NAT2 slow acetylation genotypes and increased urinary bladder cancer (UBC) risk (García-Closas et al. 2005; Sanderson et al. 2007). Although the association was significant in meta-analyses, the results are controversial at the level of individual studies. For example, 35 of 46 studies included into a meta-analysis did not amount to statistical significance (Moore et al. 2011). Possible explanations for the lack of significance at the level of individual studies are differences in exposure to bladder carcinogens and ethnicity (Golka et al. 2002; Selinski 2012; Bolt 2013a, b), as well as the relatively low case numbers and statistical power limitations.
A further aspect may limit the studies on NAT2 genotypes with bladder cancer risk, namely the technique of NAT2 genotyping and phenotype prediction (Deitz et al. 2004). Conventionally, haplotype pairs of seven NAT2 exon SNPs (rs1801279, rs1041983, rs1801280, rs1799929, rs1799930, rs1208 and rs1799931) have been used to derive slow, intermediate and rapid genotypes (Grant et al. 1997; Hein 2006) according to the consensus nomenclature (Hein 2002; Hein et al. 2008, http://www.louisville.edu/medschool/pharmacology/NAT.html). Recently, a tagging SNP rs1495741 (Rothman et al. 2010, García-Closas et al. 2011) and a 2-SNP genotype (Selinski et al. 2011) have been recommended. This fuelled again the debate on the optimal technique of NAT2 genotyping and phenotype prediction (Selinski et al. 2011; Agúndez 2003; He et al. 2012; Hein and Doll 2012a, b, Suarez-Kurtz et al. 2012a, b). A remaining crucial problem is the understanding of the relevance of different slow haplotypes (Cascorbi et al. 1995; Gross et al. 1999; Bolt et al. 2005; Hein 2006, 2009; Golka et al. 2008; Ruiz et al. 2012).
In the present study, we used the IfADo UBC case–control series, consisting of 1,712 cases and 2,020 controls (Golka et al. 2009; Lehmann et al. 2010; Selinski et al. 2011, 2012, 2013a; Schwender et al. 2012). In this case–control series, neither the 7-SNP-based genotype, nor the tagging SNP nor the 2-SNP genotype was significantly associated with bladder cancer risk. Interestingly, we could identify an ‘ultra-slow’ genotype based on comparison with in vivo caffeine metabolism.
In contrast to solely discriminating between slow and rapid genotypes, the consideration of this ‘ultra-slow’ genotype showed an association with bladder cancer risk. If confirmed in independent cohorts, the ‘ultra-slow’ genotypes should be used for refinement of NAT2 phenotype prediction in future.
Materials and methods
Patients
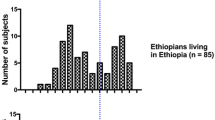
We used eight case–control series, in total 1,712 UBC cases and 2,020 controls from Germany (1,268/1,693), Hungary (226/76), Pakistan (106/61) and Venezuela (112/190) as described earlier (Golka et al. 2009; Selinski et al. 2012, 2013a; for details, see Supplementary Tables S1A-C, Supplementary Figure S1, Supplementary Materials and Methods). NAT2 7-SNP genotypes were determined in 1,692 cases and 1,995 controls, the NAT2 tagSNP genotype was determined in 1,702 cases and 1,957 controls. A subgroup of 344 Caucasians was phenotyped for NAT2 using the caffeine test (for details see Selinski et al. 2011). The sample collection was approved by the local ethics committee and by the IRB (institutional review board). Exclusion criteria were a missing written informed consent, age of less than 20 years and, in case of the German and Hungarian study groups, non-Caucasian descent.
Genotyping and phenotyping
The A/G substitution of the NAT2 tagSNP (rs1495741) on chromosome 8p23, position 18,272,881, was detected via TaqMan® SNP Genotyping Assay (Applied Biosystems, Darmstadt, Germany). Genotyping of the NAT2 polymorphisms G191A (rs1801279), C282T (rs1041983), T341C (rs1801280), C481T (rs1799929), G590A (rs1799930), A803G (rs1208) and G857A (rs1799931) was performed using PCR- and RFLP-based methods (Blaszkewicz et al. 2004). All SNPs were in Hardy–Weinberg equilibrium (HWE; Supplementary Table S2). NAT2 phenotyping was performed using the caffeine test according to standard methods determining the ratio of two caffeine metabolites 5-acetylamino-6-formylamino-3-methyluracil (AFMU) and 1-methylxanthine (1-MX) in urine by liquid–liquid extraction, HPLC separation and UV detection (Blaszkewicz 2004). The person under investigation was given two cups of coffee containing caffeine—if possible after complete evacuation of the bladder. Urine was sampled routinely 2 and 4 h after the administration of caffeine. For quality control, five test persons provided urine samples 2, 4 and 6 h after caffeine intake. The results showed no deviation in the phenotyping, indicating that the time of sampling played a minor role. The influence of a full or empty bladder before the administration of caffeine was investigated on seven persons. Deviations in phenotyping were not observed. During the evaluation of the method, urine samples were taken without regard to food intake or medication. All these variations in the sampling procedures gave no indication of interferences of the results in these different aspects. For the classification, a cut-off of 0.85 was used to differentiate between slow (<0.85) and rapid acetylators (≥0.85). For details, see Supplementary Materials and Methods.
Statistical methods. Deviations from HWE were checked for each of the eight SNPs in each study group separately in cases and controls using chi-square tests. The NAT2 haplotype pairs were determined from the seven NAT2 SNPs using PHASE, v2.1.1 (Stephens et al. 2001; Li and Stephens 2003; Stephens and Donnelly 2003) as described earlier (Agúndez et al. 2008; Selinski et al. 2011, 2013a), and the 7-SNP genotype was derived according to the nomenclature of the N-acetyltransferases (Hein 2002; Hein et al. 2008, http://www.louisville.edu/medschool/pharmacology/NAT.html). The A/A, A/G and G/G genotypes of the NAT2 tagSNP rs1495741 were considered as slow, intermediate and rapid, respectively (García-Closas et al. 2011). The 2-SNP genotype was defined as the sum of variant alleles of the two NAT2 SNPs rs1041983 (C282T) and rs1801280 (T341C) as described earlier (Selinski et al. 2011). Associations of genetic markers with UBC were evaluated using asymptotic chi-square tests, odds ratios (OR) and 95 % confidence intervals (CI) when expected cell frequencies were ≥5; otherwise, we used exact tests and 95 % CIs. For combined study groups, the method of Mantel–Haenszel was used to adjust for the different study regions if homogeneity of the ORs could be assumed according to the Breslow–Day test (P ≥ 0.05). In case of inhomogeneity, random effect models were used (DerSimonian and Laird 1986). Adjusted ORs and 95 % confidence intervals as well as Wald tests adjusted for age, cigarette smoking, gender and study region in case of combined study groups were calculated using logistic regression. This analysis was repeated stratifying for smoking habits. Effects of exposure on NAT2 risks were investigated using exact chi-square tests in a stratified analysis. Genotype differences in the in vivo activity were tested using the Wilcoxon or the Kruskal–Wallis test in case of equal and Welch’s t test in case of unequal variances. Global differences were checked using the F test; pairwise comparisons were made using the Tukey test. Equality of the variances was tested using the Levene test; normality could not be assumed according to the Kolmogorov–Smirnov test. The level of significance was α = 0.05 for all tests and confidence intervals. The software R, version 2.15.0 (R Development Core Team 2008), was used to test for HWE and fit the random effect models. For all further calculations, we used the software package SAS/STAT®, version 9.2 (SAS/STAT® software, version 9.2. Copyright© 2002–2008).
Results
Identification of an ‘ultra-slow’ genotype
In a first step, we studied a possible association of traditionally used NAT2 genotypes with bladder cancer risk in the IfADo UBC case–control series. Neither the 7-SNP genotype, nor the tagging SNP as well as the 2-SNP genotype showed a significant association with bladder cancer risk (Table 1; Supplementary Tables S3-S5). This was the case for the univariate as well as the multivariate analysis adjusted for smoking habits, age, gender and study group. Stratifying for smoking habits yielded a similar result. Next, we studied the seven individual SNPs that are usually applied to derive the 7-SNP genotype. Interestingly, two of them showed a borderline significant association with bladder cancer risk, namely C282T (P = 0.0319) and G590A (P = 0.0763) (Table 2A, B; Supplementary Tables S6-S7).
To understand the relationship between C282T and G590A and NAT2 activity, we studied a group of 344 individuals who have been phenotyped by the caffeine test. The homozygous minor alleles of both, C282T and G590A, showed lower activities than those of all other individual SNPs (at least 37 % reduction, P < 0.0001, Table 3). Note that we can distinguish two groups of SNPs: homozygous minor alleles of C282T, G590A and G857A (mainly present in the *6 and *7 haplotype clusters) usually cannot be observed together with homozygous minor alleles of T341C, C481T and A803G (mainly present in the *5 haplotype cluster; Table 4; Supplementary Table S8).
Next we studied how C282T and G590A are linked to haplotypes. The C282T variant is mainly present in the slow *6A and *7B haplotypes (Table 4; 98.6 %). G590A is mainly present in the *6A haplotypes (99.9 %). The homozygous minor allele of C282T corresponds mainly to *6A/*6A, *6A/*7B and *7B/*7B (95.6 %), whereas the homozygous minor allele of G590A corresponds to *6A/*6A (100 %). The *6A/*6A genotypes showed significantly lower activities than *5B/*5B (0.17 vs. 0.28; P = 0.0001; Table 5A, B). Also, the combination of *6A/*6A and *6A/*7B (for *7B/*7B no in vivo data were available) resulted in lower activities than *5B/*5B (0.15 vs. 0.28; P < 0.0001; Table 5A, C). The concept that *6A and *7B define ‘ultra-slow’ haplotypes is further supported by the observation that an increasing number of *6A and *7B haplotypes is associated with a decreasing activity of caffeine metabolism (Table 5A). Pairwise comparison showed significantly lower activities of the *6A/*6A genotypes compared to *5B/*6A (P = 0.0186) and *5B/*5B (P = 0.0001). Also, the combined *6A/*6A and *6A/*7B resulted in lower activities than the combined *5B/*6A and *5B/*7B genotypes (P = 0.0081) as well as the *5B/*5B genotypes (P < 0.0001). In conclusion, in vivo the combined *6A/*6A and *6A/*7B define a genotype that is associated with a particularly low activity of caffeine metabolism.
Association of the ‘ultra-slow’ genotype with bladder cancer risk
The aforementioned ‘ultra-slow’ genotype (combined *6A/*6A, *6A/*7B, *7B/*7B) was associated with bladder cancer risk in the combined study cohort of 1,692 cases and 1,995 controls (OR = 1.31, P = 0.0123) (Table 6A, B). After adjusting for age, gender and smoking habits, a trend was obtained (OR = 1.22; P = 0.0816). Analysis of individuals with reported high levels of occupational exposure to bladder carcinogens did not reveal an increased relevance of the ‘ultra-slow’ genotype in this subgroup (Supplementary Table S12A, B).
Discussion
Genotyping NAT2 is of high relevance, since many drugs but also urinary bladder carcinogens are substrates of this enzyme (Golka et al. 1996, 2002; Hengstler et al. 1998; Vineis et al. 2001; Hung et al. 2004; García-Closas et al. 2005; Hein 2006, 2009; Sanderson et al. 2007; Agúndez 2008; Moore et al. 2011; Cai et al. 2012). In this study, we further refined NAT2 phenotype prediction by identification of an ‘ultra-slow’ genotype. This ‘ultra-slow’ genotype is defined by a combination of *6A/*6A, *6A/*7B, *7B/*7B haplotype pairs and corresponds mainly to the homozygous minor alleles of C282T (rs1041983 T/T) and G590A (rs1799930 A/A). Individuals with the ‘ultra-slow’ genotype show much lower activities of caffeine in vivo metabolism compared to all other slow genotypes (0.15 vs. 0.22; P < 0.0001).
Interestingly, the ‘ultra-slow’ genotype but not the common slow NAT2 genotype was associated with increased urinary bladder cancer risk in the IfADo UBC case–control series. This suggests that individuals with extremely low NAT2 activity may be particularly susceptible to urinary bladder carcinogens. Discrimination between these ‘ultra-slow’ and other slow metabolizers may be superior over conventional pooling of all slow acetylators. Regarding the reduction in genotyping effort, the C282T-T341C 2-SNP genotype suggested by Cascorbi et al. (1995) and Selinski et al. (2011) also yields this discrimination between slow and ‘ultra-slow’ haplotypes. In Caucasians and similar populations, C282T may serve as a tagSNP for the *6A and *7B haplotypes and T341C indicates the *5 haplotype cluster. Our results correspond well to previous studies of Cascorbi et al. (1995) and Ruiz et al. (2012), demonstrating a notably decreased NAT2 activity in vivo in *6/*6 compared to *5/*5 genotypes. Similarly large and recent studies on UBC indicate similar effects of *5B, *6A and *7B haplotypes (García-Closas et al. 2005) or no effects of the homozygous variant alleles of the NAT2 SNPs that are present in these haplotypes (Moore et al. 2011). Interestingly, the well-known enhanced risk of anti-tuberculosis drug-induced hepatotoxicity in slow acetylators (Cai et al. 2012) seems to be more pronounced for *6/*6 genotypes in European (Leiro-Fernandez et al. 2011) and Asian (Huang et al. 2002; Lee et al. 2010; An et al. 2012) study groups, though the slow *5 haplotypes are less common in East Asian populations (Sabbagh et al. 2011), as well as in several large Brazilian studies (Possuelo et al. 2008; Teixeira et al. 2011) but not all (Santos et al. 2013). A meta-analysis of 14 studies indicates that tuberculosis patients with NAT2*6/*6 genotypes may have an even higher risk of anti-tuberculosis drug-induced hepatotoxicity than *5/*5 tuberculosis patients (all slow vs. rapid genotypes OR = 3.05, *6/*6 vs. rapid OR = 3.61, *5/*5 vs. rapid OR = 2.45; Selinski et al. 2013b).
A limitation of the current study is that the ‘ultra-slow’ genotype was significantly associated with bladder cancer risk in the univariate analysis, whereas only a trend was obtained in the multivariate regression adjusted for age, gender and smoking habits. It should also be noted that the current literature concerning the identification of differences between slow haplotypes regarding velocity and substrates remains controversial (Cascorbi et al. 1995; Gross et al. 1999; Bolt et al. 2005; Hein 2006, 2009; Golka et al. 2008; Ruiz et al. 2012). In particular, Hein (2006) showed the lowest acetylation speed in vitro for the *5B haplotypes containing the slow T341C, C481T and A803G SNPs. Generally, G191A (*14 haplotypes), T341C (*5 haplotypes) and G590A (*6 haplotypes) are associated with reductions in N- and O-acetylation capacity in a number of different bacterial and eukaryotic expression systems (Zang et al. 2007; Walraven et al. 2008). The effect of G857A (*7 haplotypes) seems to be substrate dependent (Zang et al. 2007; Walraven et al. 2008). These four missense SNPs result in a reduction in protein. In case of G191A, G590A and G857A but not T341C, this seems to be related to the NAT2 protein stability. The silent SNPs C282T and C481T as well as the missense SNP A803G seem to have no effect on acetylation capacity, protein stability or mRNA levels in vitro (Zang et al. 2007; Walraven et al. 2008). In summary, in vitro studies suggest that *5 haplotypes are particularly slow, whereas in vivo *6 seems to be more important. Therefore, it will be of high interest whether the ‘ultra-slow’ NAT2 genotype and its association with particularly low caffeine metabolism and increased bladder cancer risk will be confirmed in independent cohorts.
In conclusion, we suggest to further refine NAT2 phenotype prediction by consideration of an ‘ultra-slow’ genotype based on combined *6A/*6A, *6A/*7B and *7B/*7B genotypes.
References
Agúndez JA (2003) NAT2 genotyping: equilibrium between accuracy and feasibility in routine analyses. J Appl Res 3:1–6
Agúndez JA (2008) Polymorphisms of human N-acetyltransferases and cancer risk. Curr Drug Metab 9:520–531
Agúndez JA, Golka K, Martínez C, Selinski S, Blaszkewicz M, García-Martín E (2008) Unraveling ambiguous NAT2 genotyping data. Clin Chem 54:1390–1394
An HR, Wu XQ, Wang ZY, Zhang JX, Liang Y (2012) NAT2 and CYP2E1 polymorphisms associated with antituberculosis drug-induced hepatotoxicity in Chinese patients. Clin Exp Pharmacol Physiol 39:535–543
Arylamine N-Acetyltransferase Nomenclature Committee. Update July 22, 2011. http://www.louisville.edu/medschool/pharmacology/NAT.html. Accessed Feb 7, 2013
Blaszkewicz M (2004) N-acetyltransferase 2 (phenotyping: caffeine test). In: Angerer J, Müller M, Weiss T et al. (eds) Analyses of hazardous substances in biological materials, vol 9. Special issue: Markers of susceptibility. Wiley-VCH, Weinheim, pp 165–182
Blaszkewicz M, Dannappel D, Thier R, Lewalter J (2004) N-acetyltransferase 2 (genotyping). In: Angerer J, Müller M, Weiss T et al. (eds) Analyses of hazardous substances in biological materials, vol 9. Special issue: markers of susceptibility. Wiley-VCH, Weinheim, pp 135–163
Bolt HM (2013a) Human bladder cancer risk calculation based on genome-wide analysis of genetic variants. Arch Toxicol 87:397–399
Bolt HM (2013b) Relevance of genetic disposition versus environmental exposure for cancer risk: an old controversy revisited with novel methods. EXCLI J 12:79–80
Bolt HM, Selinski S, Dannappel D, Blaszkewicz M, Golka K (2005) Re-investigation of the concordance of human NAT2 phenotypes and genotypes. Arch Toxicol 79:196–200
Cai Y, Yi J, Zhou C, Shen X (2012) Pharmacogenetic study of drug-metabolising enzyme polymorphisms on the risk of anti-tuberculosis drug-induced liver injury: a meta-analysis. PLoS ONE 7:e47769
Cascorbi I, Drakoulis N, Brockmöller J, Maurer A, Sperling K, Roots I (1995) Arylamine N-acetyltransferase (NAT2) mutations and their allelic linkage in unrelated Caucasian individuals: correlation with phenotypic activity. Am J Hum Genet 57:581–592
Deitz AC, Rothman N, Rebbeck TR, Hayes RB, Chow WH, Zheng W et al (2004) Impact of misclassification in genotype-exposure interaction studies: example of N-acetyltransferase 2 (NAT2), smoking, and bladder cancer. Cancer Epidemiol Biomarkers Prev 13:1543–1546
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
García-Closas M, Malats N, Silverman D, Dosemeci M, Kogevinas M, Hein DW et al (2005) NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet 366:649–659
García-Closas M, Hein DW, Silverman D, Malats N, Yeager M, Jacobs K et al (2011) A single nucleotide polymorphism tags variation in the arylamine N-acetyltransferase 2 phenotype in populations of European background. Pharmacogenet Genomics 21:231–236
Golka K, Prior V, Blaszkewicz M, Cascorbi I, Schöps W, Kierfeld G et al (1996) Occupational history and genetic N-acetyltransferase polymorphism in urothelial cancer patients of Leverkusen, Germany. Scand J Work Environ Health 22:332–338
Golka K, Prior V, Blaszkewicz M, Bolt HM (2002) The enhanced bladder cancer susceptibility of NAT2 slow acetylators towards aromatic amines: a review considering ethnic differences. Toxicol Lett 128:229–241
Golka K, Blaszkewicz M, Samimi M, Bolt HM, Selinski S (2008) Reconstruction of N-acetyltransferase 2 haplotypes using PHASE. Arch Toxicol 82:265–270
Golka K, Hermes M, Selinski S, Blaszkewicz M, Bolt HM, Roth G et al (2009) Susceptibility to urinary bladder cancer: relevance of rs9642880[T], GSTM1 0/0 and occupational exposure. Pharmacogenet Genomics 19:903–906
Grant DM, Hughes NC, Janezic SA, Goodfellow GH, Chen HJ, Gaedigk A et al (1997) Human acetyltransferase polymorphisms. Mutat Res 376:61–70
Gross M, Kruisselbrink T, Anderson K, Lang N, McGovern P, Delongchamp R et al (1999) Distribution and concordance of N-acetyltransferase genotype and phenotype in an American population. Cancer Epidemiol Biomarkers Prev 8:683–692
He YJ, Shapero MH, McLeod HL (2012) Novel tagging SNP rs1495741 and 2-SNPs (rs1041983 and rs1801280) yield a high prediction of the NAT2 genotype in HapMap samples. Pharmacogenet Genomics 22:322–324
Hein DW (2002) Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat Res 506–507:65–77
Hein DW (2006) N-acetyltransferase 2 genetic polymorphism: effects of carcinogen and haplotype on urinary bladder cancer risk. Oncogene 25:1649–1658
Hein DW (2009) N-acetyltransferase SNPs: emerging concepts serve as a paradigm for understanding complexities of personalized medicine. Expert Opin Drug Metab Toxicol 5:353–366
Hein DW, Doll MA (2012a) A four-SNP NAT2 genotyping panel recommended to infer human acetylator phenotype. Pharmacogenomics 13:855
Hein DW, Doll MA (2012b) Accuracy of various human NAT2 SNP genotyping panels to infer rapid, intermediate and slow acetylator phenotypes. Pharmacogenomics 13:31–41
Hein DW, Boukouvala S, Grant DM, Minchin RF, Sim E (2008) Changes in consensus arylamine N-acetyltransferase gene nomenclature. Pharmacogenet Genomics 18:367–368
Hengstler JG, Arand M, Herrero ME, Oesch F (1998) Polymorphisms of N-acetyltransferases, glutathione S-transferases, microsomal epoxide hydrolase and sulfotransferases: influence on cancer susceptibility. Recent Results Cancer Res 154:47–85
Huang YS, Chern HD, Su WJ, Wu JC, Lai SL, Yang SY et al (2002) Polymorphism of the N-acetyltransferase 2 gene as a susceptibility risk factor for antituberculosis drug-induced hepatitis. Hepatology 35:883–889
Hung RJ, Boffetta P, Brennan P, Malaveille C, Hautefeuille A, Donato F et al (2004) GST, NAT, SULT1A1, CYP1B1 genetic polymorphisms, interactions with environmental exposures and bladder cancer risk in a high-risk population. Int J Cancer 110:598–604
Lee SW, Chung LS, Huang HH, Chuang TY, Liou YH, Wu LS (2010) NAT2 and CYP2E1 polymorphisms and susceptibility to first-line anti-tuberculosis drug-induced hepatitis. Int J Tuberc Lung Dis 14:622–626
Lehmann ML, Selinski S, Blaszkewicz M, Orlich M, Ovsiannikov D, Moormann O et al (2010) Rs710521[A] on chromosome 3q28 close to TP63 is associated with increased urinary bladder cancer risk. Arch Toxicol 84:967–978
Leiro-Fernandez V, Valverde D, Vázquez-Gallardo R, Botana-Rial M, Constenla L, Agúndez JA et al (2011) N-acetyltransferase 2 polymorphisms and risk of anti-tuberculosis drug-induced hepatotoxicity in Caucasians. Int J Tuberc Lung Dis 15:1403–1408
Li N, Stephens M (2003) Modelling linkage disequilibrium, and identifying recombination hotspots using SNP data. Genetics 165:2213–2233
Moore LE, Baris DR, Figueroa JD, Garcia-Closas M, Karagas MR, Schwenn MR et al (2011) GSTM1 null and NAT2 slow acetylation genotypes, smoking intensity and bladder cancer risk: results from the New England bladder cancer study and NAT2 meta-analysis. Carcinogenesis 32:182–189
Possuelo LG, Castelan JA, de Brito TC, Ribeiro AW, Cafrune PI, Picon PD et al (2008) Association of slow N-acetyltransferase 2 profile and anti-TB drug-induced hepatotoxicity in patients from Southern Brazil. Eur J Clin Pharmacol 64:673–681
R Development Core Team (2008) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Rothman N, Garcia-Closas M, Chatterjee N, Malats N, Wu X, Figueroa JD et al (2010) A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet 42:978–984
Ruiz JD, Martínez C, Anderson K, Gross M, Lang NP, García-Martín E et al (2012) The differential effect of NAT2 variant alleles permits refinement in phenotype inference and identifies a very slow acetylation genotype. PLoS ONE 7:e44629
Sabbagh A, Darlu P, Crouau-Roy B, Poloni ES (2011) Arylamine N-acetyltransferase 2 (NAT2) genetic diversity and traditional subsistence: a worldwide population survey. PLoS ONE 6:e18507
Sanderson S, Salanti G, Higgins J (2007) Joint effects of the N-acetyltransferase 1 and 2 (NAT1 and NAT2) genes and smoking on bladder carcinogenesis: a literature-based systematic HuGE review and evidence synthesis. Am J Epidemiol 166:741–751
Santos NP, Callegari-Jacques SM, Santos AK, Silva CA, Vallinoto AC, Fernandes DC et al (2013) N-acetyltransferase 2 and cytochrome P450 2E1 genes and isoniazid-induced hepatotoxicity in Brazilian patients. Int J Tuberc Lung Dis 17:499–504
SAS/STAT® software, version 9.2. Copyright © 2002–2008, SAS Institute Inc., Cary
Schwender H, Selinski S, Blaszkewicz M, Marchan R, Ickstadt K, Golka K et al (2012) Distinct SNP combinations confer susceptibility to urinary bladder cancer in smokers and non-smokers. PLoS ONE 7:e51880
Selinski S (2012) Genetic variants confer susceptibility to urinary bladder cancer: an updated list of confirmed polymorphisms. EXCLI J 11:743–747
Selinski S, Blaszkewicz M, Lehmann ML, Ovsiannikov D, Moormann O, Guballa C et al (2011) Genotyping NAT2 with only two SNPs (rs1041983 and rs1801280) outperforms the tagging SNP rs1495741 and is equivalent to the conventional 7-SNP NAT2 genotype. Pharmacogenet Genomics 21:673–678
Selinski S, Lehmann ML, Blaszkewicz M, Ovsiannikov D, Moorman O, Guballa C et al (2012) Rs11892031[A] on chromosome 2q37 in an intronic region of the UGT1A locus is associated with urinary bladder cancer risk. Arch Toxicol 86:1369–1378
Selinski S, Blaszkewicz M, Agúndez JA, Martínez C, García-Martín E, Hengstler JG et al (2013a) Clarifying haplotype ambiguity of NAT2 in multi-national cohorts. Front Biosci (Schol Ed) 5:672–684
Selinski S, Blaszkewicz M, Ickstadt K, Hengstler JG, Golka K (2013b) Improvements in algorithms for phenotype inference: The NAT2 example. Curr Drug Metab (in press)
Stephens M, Donnelly P (2003) A comparison of Bayesian methods for haplotype reconstruction. Am J Hum Genet 73:1162–1169
Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989
Suarez-Kurtz G, Sortica VA, Vargens DD, Bruxel EM, Petzl-Erler ML, Tsuneto LT et al (2012a) Impact of population diversity on the prediction of 7-SNP NAT2 phenotypes using the tagSNP rs1495741 or paired SNPs. Pharmacogenet Genomics 22:305–309
Suarez-Kurtz G, Vargens DD, Sortica VA, Hutz MH (2012b) Accuracy of NAT2 SNP genotyping panels to infer acetylator phenotypes in African, Asian, Amerindian and admixed populations. Pharmacogenomics 13:851–854
Teixeira RL, Morato RG, Cabello PH, Muniz LM, Moreira Ada S, Kritski AL et al (2011) Genetic polymorphisms of NAT2, CYP2E1 and GST enzymes and the occurrence of antituberculosis drug-induced hepatitis in Brazilian TB patients. Mem Inst Oswaldo Cruz 106:716–724
Vineis P, Marinelli D, Autrup H, Brockmöller J, Cascorbi I, Daly AK et al (2001) Current smoking, occupation, N-acetyltransferase-2 and bladder cancer: a pooled analysis of genotype-based studies. Cancer Epidemiol Biomarkers Prev 10:1249–1252
Walraven JM, Zang Y, Trent JO, Hein DW (2008) Structure/function evaluations of single nucleotide polymorphisms in human N-acetyltransferase 2. Curr Drug Metab 9:471–486
Zang Y, Doll MA, Zhao S, States JC, Hein DW (2007) Functional characterization of single-nucleotide polymorphisms and haplotypes of human N-acetyltransferase 2. Carcinogenesis 28:1665–1671
Acknowledgments
The authors thank Ms. Doris Dannappel, Ms. Kirsten Liesenhoff-Henze, Ms. Marion Page, Ms. Claudia Schulte-Dahmann and Mr. Roland Wycislok for excellent technical support.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jan G. Hengstler and Klaus Golka have shared senior authorship.
Electronic supplementary material
Below is the link to the electronic supplementary material.
204_2013_1157_MOESM1_ESM.docx
Supplementary Materials and Methods.doc Detailed description of the study groups and the genotyping and phenotyping methods. Supplementary material 1 (DOCX 46 kb)
204_2013_1157_MOESM2_ESM.doc
Supplementary Tables S1-S12 and Figure S1.doc Study group characteristics and a more detailed analysis on individual study group level. Supplementary material 2 (DOC 1855 kb)
Rights and permissions
About this article
Cite this article
Selinski, S., Blaszkewicz, M., Ickstadt, K. et al. Refinement of the prediction of N-acetyltransferase 2 (NAT2) phenotypes with respect to enzyme activity and urinary bladder cancer risk. Arch Toxicol 87, 2129–2139 (2013). https://doi.org/10.1007/s00204-013-1157-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-013-1157-7