Abstract
Cytochrome P4502E1 (CYP2E1) gene shows genetic polymorphisms that vary markedly in frequency among different ethnic and racial groups. We studied the genotype distributions and allele frequencies of three CYP2E1 polymorphisms: CYP2E1*5B (RsaI/PstI RFLP, C-1053T/G-1293C SNP, rs2031920 /rs3813867), CYP2E1*6 (DraI RFLP, T7632A SNP, rs6413432), and CYP2E1*7B (DdeI RFLP, G-71T SNP, rs6413420) by PCR/RFLP technique in a sample of 206 healthy subjects representing Turkish population. CYP2E1*5B polymorphism analysis yielded the genotype distribution as 96.12% for *1A/*1A (c1/c1), and 3.88% for *1A/*5B (c1/c2). The genotype frequencies for CYP2E1*6 polymorphism were found as 83.98% for *1A/*1A (T/T), 15.53% for *1A/*6 (T/A) and 0.49% for *6/*6 (A/A). For CYP2E1*7B (G-71T) polymorphism, the genotype frequencies were determined to be 86.89% for *1A/*1A (G/G), 12.62% for *1A/*7B (G/T) and 0.49% for *7B/*7B (T/T). Accordingly, the allele frequencies for *5B, *6 and *7B were 1.94, 8.25, and 6.80%, respectively. The genotype distributions of CYP2E1*5B and *6 in Turkish population were similar to those in other Caucasian populations, while differed significantly from East Asian populations. Recently, a novel and functionally important CYP2E1*7B polymorphism was identified in the promoter region. There have been few studies and limited data on CYP2E1*7B polymorphism frequency in the world and, so far, no information has been available for Turkish population. The genotype frequencies of CYP2E1*7B in Turkish population were found to be similar to those of other Caucasian populations. Population studies like this could be useful in assessing the susceptibility of different populations to chemical-induced diseases, including several types of cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cytochrome P450-dependent monooxygenases exist as a large superfamily of proteins and are the principal enzymes involved in the phase I metabolism of foreign compounds including drugs, food additives, industrial solvents, and pollutants (Lu and Levin 1974; Arinç and Philpot 1976; Arinç and Adali 1983; Lieber et al. 1997). Besides detoxification, they often catalyze the metabolic activation of procarcinogens to their ultimate carcinogenic forms. As is the case for most of the xenobiotic-metabolizing enzymes, cytochrome P450s also show genetic polymorphisms which may alter the expression levels or activities.
Among the various P450s, CYP2E1 is of great interest due to its role in the metabolism and bioactivation of many low molecular weight compounds, including ethanol, acetone, drugs like acetaminophen, isoniazid, chlorzoxazone and fluorinated anaesthetics, and many procarcinogens like benzene, N-nitrosodimethylamine (NDMA) and styrene (Peter et al. 1990; Guengerich et al. 1991; Kharasch and Thummel 1993). Besides, CYP2E1 isoform is induced by ethanol, benzene, pyridine and isoniazid (Koop and Casazza 1985; Johansson and Ingelman-Sundberg 1988; Arinç et al. 2000a, b) as well as by some pathophysiological conditions like diabetes, obesity and starvation (Koop and Casazza 1985; Hong et al. 1987; Song et al. 1987; Arinç et al. 2005, 2007). CYP2E1 gene (Accession No. AL161645) shows genetic polymorphisms which are thought to play a major role in interindividual variability in drug response, drug–drug, drug–xenobiotic interactions and in susceptibility to chemical-induced diseases (Lieber 1997; Bolt et al. 2003). Several reports demonstrated that CYP2E1 polymorphisms differ markedly in frequency among different ethnic and racial groups, like most other xenobiotic-metabolizing enzymes do (Garte et al. 2001).
The human CYP2E1 gene is located in 10q24.3-qter region of chromosome 10, and spans 11,413 base pairs with nine exons and a typical TATA box (Umeno et al. 1988). The gene contains several polymorphisms, some of which seem to effect the expression of the protein. Among CYP2E1 polymorphisms, the most frequently studied ones are the CYP2E1*5B (RsaI/PstI RFLP; position:C-1053T/G-1293C, rs3813867/rs2031920) polymorphism located in the 5′-flanking region of the gene, which was related to altered enzyme expression in vitro (Hayashi et al. 1991); and CYP2E1*6 (DraI RFLP; position T7632A, rs6413432) polymorphism located in intron 6 (Uematsu et al. 1991), which was shown to lower ‘chlorzoxazone metabolic ratios’ (Haufroid et al. 2002), and was correlated with single strand breaks in DNA (Vodicka et al. 2001). Several case-control studies have described the influence of these polymorphisms with increased risk for various cancer types in different populations (El Zein et al. 1997; Wu et al. 1998; Farker et al. 1998; Liu et al. 2001).
In order to clarify the role of CYP2E1 polymorphisms in disease/cancer susceptibility, the allele frequencies of these polymorphisms in control populations should be verified by several studies, since, in general, a sample size of only 100–300 subjects has been used in these polymorphism studies. Thus, each study on control populations would add data to the pool, which makes it possible to more precisely define the true population-frequency of polymorphisms in control populations. Garte et al. (2001) analyzed the allele and genotype frequencies of eight metabolic genes from the data obtained for a total of 73 separate studies gathered after 1996 covering nearly 16,000 control subjects. It has also been observed that different groups of investigators reported different frequencies for the same gene in the same population. For example, GSTT1 null frequency was reported to be 30% (Peter et al. 1989) and 19.5% (Garte et al. 2001) for German population. Another example is related to GSTM1 null frequency in Turkish population which was reported to be 16% by Pinarbasi et al. (2001), 37.4% by Aktas et al. (2001), 54.6% by Balta et al. (2003), and 51.9% by Ada et al. (2004). Therefore, it is of crucial importance to verify the genotype frequencies even in the same population.
While the frequencies of mutant alleles of drug metabolizing enzymes like GSTT1, GSTM1, CYP1A1 and CYP2C9 (Aynacioglu et al. 1999; Pinarbasi et al. 2001; Aktas et al. 2001; Balta et al. 2003; Ada et al. 2004) have been studied relatively extensively in Turkish population, only limited information is available for CYP2E1 polymorphisms (Omer et al. 2001).
Furthermore, Fairbrother and co-workers have detected another novel polymorphism of CYP2E1 gene -the DdeI (mutated allele: CYP2E1*7B, G-71T, rs6413420) polymorphism located in the promoter region. Hence, it is suspected to be associated with the expression or regulation of the gene (Fairbrother et al. 1998). So far, studies and data regarding this polymorphism are very limited in the world. No information is available for the frequency of this CYP2E1*7B (G-71T) allele for the Turkish population.
We report here the genotype and allele frequencies of three CYP2E1 polymorphisms, namely CYP2E1*5B, *6 and *7B, in a sample of 206 healthy subjects representing the Turkish population.
Materials and methods
Subjects
The study sample included a total of 206 healthy and unrelated Turkish volunteers between the ages of 12 and 65 years and mean age of 31.5 ± 12.0 years, including 125 females (mean age 30.4 ± 11.6 years; range 12–63 years) and 81 males (mean age 33.1 ± 12.7 years; range 14–65 years), without known history of cancer and other chronic diseases. Venous blood from the participants was collected with the collaboration of Middle East Technical University Health Center, Biochemistry Laboratory. Written informed consents were taken from all blood donors. A small questionnaire for gathering the demographic information was also given to the volunteers.
Only Turkish subjects were included in the study, therefore study sample totally comprised of Caucasians (Turkish), without any other ethnicities (Africans or Asians). When the geographical distributions of subjects were concerned, most of the subjects were from the Central Anatolia region, and other regions; southern, northern and eastern Anatolia as well as Aegean region was also represented in the study sample. Therefore, the study sample used represented the Turkish population from all regions of the country.
DNA isolation
Four to five milliliter of blood samples from subjects were taken in EDTA-containing vacuumed tubes and stored at −20°C until use. Blood samples were kept at 4°C while they were in active use. DNA was isolated from leukocytes manually by standard phenol: chloroform extraction method.
Genotyping analyses
The genetic polymorphisms of CYP2E1 gene were determined by using the polymerase chain reaction/restriction fragment length polymorphism (PCR/RFLP) technique. Three regions of human CYP2E1 gene were amplified with PCR. These were CYP2E1*5B, *6 and *7B. The sequence of primer pairs, the PCR product sizes and restriction enzymes used for genotyping were given elsewhere (Hayashi et al. 1991; Wu et al. 1998; Yang et al. 2001). The amplified PCR products were visualized on 2.0% agarose gel for CYP2E1*5B and *7B regions and on 1.5% gel for CYP2E1*6 region.
Genotyping for CYP2E1*5B was done by digesting 10 μl of PCR product separately with 10 U of RsaI and PstI at 37°C for 18 h, and visualized on 2.5% agarose gel. CYP2E1*6 and *7B genotypes were determined by digesting 20 μl of corresponding PCR products with 6 and 5 U of DraI and DdeI, respectively, at 37°C for 24 h. Results of DraI and DdeI digestions were analyzed on 1.8 and 2.5% agarose gels, respectively.
Statistical analysis
In this study, non-parametric Chi-Square Test was used to check if the calculated frequencies for CYP2E1*5B, CYP2E1*6 and CYP2E1*7B single nucleotide polymorphisms fit to the Hardy–Weinberg equilibrium. The same test, with Yates’ correction for continuity where necessary, was also used for comparison of the CYP2E1 genotype distributions of Turkish population determined in this study with other populations.
Results
In the current study, three CYP2E1 polymorphisms; *5B located in 5′-flanking region, *6 located in intron 6 and *7B located in promoter region of the gene have been investigated in a sample of 206 healthy subjects representing Turkish population.
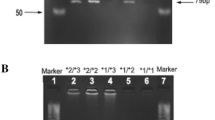
CYP2E1*5B (C-1053T) polymorphism when digested with RsaI produced the wild type genotype *1A/*1A (c1/c1) and heterozygote *1A/*5B (c1/c2). No mutated homozygote (variant) *5B/*5B (c2/c2) was observed in the sample population (Fig. 1a, Table 1). The results were confirmed with digestion with PstI, since this polymorphic site is located on the same fragment and complete linkage disequilibrium was observed between RsaI and PstI positions (Watanabe et al. 1990; Hayashi et al. 1991). The genotype distribution and allele frequencies of CYP2E1*5B polymorphism in Turkish population is given in Table 1. In the total 206 subjects studied, 96.12% were found to be homozygous wild type *1A/*1A (c1/c1), 3.88% were heterozygote *1A/*5B (c1/c2) and none showed homozygote mutated variant genotype. Accordingly, the corresponding frequency for the mutated allele *5B (c2) was 1.94% and for the wild type allele *1A (c1) was 98.06%, as indicated in Table 1.
Genotyping of CYP2E1 gene for *5B, *6 and *7B polymorphisms. a Detection of *5B genotype by digestion with PstI (top panel, A1) and RsaI (bottom panel, A2). In Panel A1, Lane 1, DNA ladder; Lanes 3–9, homozygous wild types *1A/*1A (c1/c1) (412 bp); Lane 2, heterozygote *1A/*5B (c1/c2) (412, 294,118); Lane 10, undigested PCR product (412 bp). In Panel A2, Lane 1, DNA ladder; Lanes 3–9, homozygous wild type *1A/*1A (c1/c1) (352, 60 bp); Lane 2, heterozygote *1A/*5B (c1/c2) (412, 352, 60 bp), Lane 10, undigested PCR product (412 bp). Sample in each lane correspond to the same individuals in both gels (A1 and A2). b Detection of *6 genotype by digestion with DraI. The amplified region in intron 6 bears two recognition sites for DraI, one comprises the SNP, one does not. Upon digestion, independent of the genotype, the 997 bp PCR product yields a 121 bp band. Lane 1, DNA ladder; Lanes 4–8, homozygous wild type *1A/*1A (T/T) (571, 305, 121 bp); Lanes 2 and 3, heterozygote *1A/*6 (T/A) (876, 571, 305, 121 bp); Lane 9, homozygous mutated *6/*6 (A/A) (876, 121 bp); Lane 10, undigested PCR product (997 bp). c Detection of *7B genotype by digestion with DdeI. Lane 1, DNA ladder; Lanes 2, 4, 6–9, homozygous wild type *1A/*1A (G/G) (302, 58 bp); Lane 5, heterozygote *1A/*7B (G/T) (360, 302, 58 bp); Lane 3, homozygous mutated *7B/*7B (T/T) (360 bp); Lane 10, undigested PCR product (360 bp)
In case of CYP2E1*6 (T7632A) polymorphism, digestion of corresponding PCR product with DraI yielded the homozygous wild type genotype *1A/*1A (T/T), the heterozygous genotype *1A/*6 (T/A) and the homozygous mutated variant genotype *6/*6 (A/A) as shown in Fig. 1b. The distribution of CYP2E1*6 genotypes and allele frequencies are presented in Table 1. The genotype frequencies in 206 subjects were as follows; 83.98% homozygous wild type *1A/*1A (T/T), 15.53% heterozygote *1A/*6 (T/A) and 0.49% homozygous mutated variant *6/*6 (A/A). The frequency of the mutated allele *6 (A) was found to as 8.25% and of the wild type allele *1A (T) as 91.75%, as shown in Table 1.
For the CYP2E1*7B (G-71T) polymorphism, DdeI digestion of amplified PCR product resulted in the wild type genotype *1A/*1A (G/G), the heterozygous genotype *1A/*7B (G/T), and the homozygous mutated genotype *7B/*7B (T/T) (Fig. 1c). As presented in Table 1, the genotype frequencies were 86.89% for homozygous wild type *1A/*1A (G/G), 12.62% for heterozygote *1A/*7B (G/T), and 0.49% for homozygous mutated variant *7B/*7B (G/G) in 206 subjects. The corresponding frequency for the mutated variant allele *7B (T) and the wild type allele *1A (G) was 6.80 and 93.20%, respectively (Table 1).
The expected genotype distributions calculated by the Hardy–Weinberg equation for CYP2E1*5B, *6 and *7B are shown in Table 1. Accordingly, all genotype frequencies were found to fit Hardy–Weinberg equilibrium.
The study comprised of 81 male and 125 female subjects, as mentioned in “Materials and methods” section. Genotype and allele frequencies for CYP2E1*5B, *6 and *7B polymorphisms in male and female subjects were compared with Chi-Square test, and no significant difference were found between the two genders in terms of genotype and allele frequencies for all polymorphisms studied (data not shown).
Discussion
Owing to its particular substrate spectrum and inducibility by toxic and carcinogenic compounds, the cytochrome P450 isozyme CYP2E1 is of great interest to industrial and environmental medicine and toxicology. CYP2E1 gene possess several polymorphisms, some of which have been associated with increased genetic susceptibility to several types of chemical-induced diseases, including several types of cancer (El Zein et al. 1997; Wu et al. 1998; Farker et al. 1998; Liu et al. 2001).
CYP2E1 polymorphism frequencies show variability in different races and ethnicities, as most other xenobiotic-metabolizing enzymes do. The genotype and allele frequencies of CYP2E1*5B polymorphism in the Turkish population is given in Table 1. In a total of 206 subjects studied, 96.12% were found to be homozygous wild type (*1A/*1A), and 3.88% heterozygote (*1A/*5B). On the other hand, no subject with homozygous mutated genotype (*5B/*5B) was detected. Accordingly, in this study, the allele frequency of the mutated allele, CYP2E1*5B, was calculated to be 1.94% (Table 1). Similar to what is observed in this study, the frequency of CYP2E1*5B was recently reported as 1.96% in 153 Turkish subjects by Omer et al. (2001). The frequency of the mutant CYP2E1*5B allele is determined to be 25–36% in Eastern Asians and 2–8% in Caucasians (Kato et al. 1992; Oyama et al. 1997; Wang et al. 1999; Garte et al. 2001). As can be seen from Table 2, the allele and genotype frequencies of CYP2E1*5B in Turkish population were similar to Caucasian populations while they were significantly different from Chileans, Brazilians, Mexican-Americans, African-Americans and Eastern Asian populations—such as Chinese, Taiwanese and Japanese.
On the other hand, the mutated allele frequency of CYP2E1*6 was determined as 8.25% for Turkish population (Table 1). As presented in Table 3, the genotype and allele frequencies of CYP2E1*6 in Turkish population were significantly different from Chilean, Indian, Mexican-American, and Asian populations, but they were similar to other Caucasian populations. In agreement with the results obtained in this study, the mutated allele frequency of CYP2E1*6 polymorphism for the control Turkish population was reported to be 8.17% by Omer et al. (2001).
So far, most of the studies on CYP2E1 polymorphisms focused on two sites—CYP2E1*5B and *6 polymorphisms. On the other hand, it has been suggested that CYP2E1*7B (G-71T) polymorphism has functional activity during gene transcription as it is located 9 bp upstream from the consensus TATA box, and, therefore may enhance binding of transcription factor TFIID (Fairbrother et al. 1998). There have been few studies and limited data on CYP2E1*7B (G-71T) polymorphism frequencies and, so far, no information has been available for Turkish population. This study presented the genotype frequencies of CYP2E1*7B (G-71T) in the Turkish population for the first time. The genotype frequencies of CYP2E1*7B (G-71T) from various populations is given in Table 4. As can be seen from Table 4, the heterozygosity for the G-71T polymorphism was found to be 7.4–14.3% in the studied Caucasian populations and 12.6% in Turkish population which are also Caucasians. As far as we know, the available data concerning the allele frequency of CYP2E1*7B (G-71T) polymorphism is limited in the world and comprise less than 600 people (Table 4). Thus, this study with a total number of 206 Turkish subjects, would add a valuable data to the pool which could make it possible to more precisely define the population frequency of G-71T polymorphism in a healthy Caucasian population.
In view of significant inter-ethnic differences of CYP2E1 polymorphisms, it was an important issue to establish its genotype distribution and allele frequencies in Turkish population. Such studies on control populations would contribute to the attempts to more precisely define population-frequencies of polymorphisms in control populations. Population studies like this could be useful in assessing the susceptibility of different populations to diseases related to CYP2E1 polymorphisms and in determining whether it is necessary to design different therapeutic and toxicological protocols to reduce the risk of population.
References
Ada AO, Süzen SH, Iscan M (2004) Polymorphisms of cytochrome P450 1A1, gluthatione S-transferases M1 and T1 in a Turkish population. Toxicol Lett 151:311–315
Aktas D, Ozen H, Atsu N, Tekin A, Sozen S, Tuncbilek E (2001) Glutathione S-transferase M1 gene polymorphism in bladder cancer patients: a marker on invasive bladder cancer? Cancer Genet Cytogen 125:1–4
Arinç E, Adali O (1983) Solubilization and partial purification of 2 forms of cytochrome P-450 from trout liver microsomes. Comp Biochem Phys B 76:653–662
Arinç E, Adali O, Gencler-Ozkan AM (2000a) Induction of N-nitrosodimethylamine metabolism in liver and lung by in vivo pyridine treatments of rabbits. Arch Toxicol 74:329–334
Arinç E, Adali O, Gençler-Özkan AM (2000b) Stimulation of aniline, p-nitrophenol and N-nitrosodimethylamine metabolism in kidney by pyridine treatments of rabbits. Arch Toxicol 74:527–532
Arinç E, Arslan S, Adali O (2005) Differential effects of diabetes on CYP2E1 and CYP2B4 proteins and associated drug metabolizing enzyme activities in rabbit liver. Arch Toxicol 79(8):427–433
Arinç E, Arslan S, Bozcaarmutlu A, Adali O (2007) Effects of diabetes on rabbit kidney and lung CYP2E1 and CYP2B4 expression and drug metabolism and potentiation of carcinogenic activity of N-nitrosodimethylamine in kidney and lung. Food Chem Toxicol 45(1):107–118
Arinç E, Philpot RM (1976) Preparation and properties of partially purified pulmonary cytochrome P-450 from rabbits. J Biol Chem 25:3213–3220
Aynacioglu AS, Brockmoller J, Bauer S, Sachse C, Guzelbey P, Ongen Z, Nacak M, Roots I (1999) Frequency of cytochrome P4502C9 variants in a Turkish population and functional relevance for phenytoin. Br J Clin Pharmacol 48:409–415
Balta G, Yuksek N, Ozyurek E, Ertem U, Hicsonmez G, Altay C, Gurgey A (2003) Characterization of MTHFR, GSTM1, GSTT1, GSTP1, and CYP1A1 Genotypes in Childhood Acute Lymphoblastic Leukemia. Am J Hematol 71:154–160
Bolt HM, Roos PH, Thier R (2003) The cytochrome P-450 isoenzyme CYP2E1 in the biological processing of industrial chemicals: consequences for occupational and environmental medicine. Int Arch Occup Environ Health 76:174–185
Bouchardy C, Hirvonen A, Coutelle C, Ward PJ, Dayer P, Benhamou S (2000) Role of alcohol dehydrogenase 3 and cytochrome P-4502E1 genotypes in susceptibility to cancers of the upper aerodigestive tract. Int J Cancer 87:734–740
Brockmöller J, Cascorbi I, Kerb R, Roots I (1996) Combined analysis of inherited polymorphisms in arylamine N-acetyltransferase 2, glutathione S-transferases M1 and T1, microsomal epoxide hydrolase, and cytochrome P450 enzymes as modulators of bladder cancer risk. Cancer Res 56(17):3915–3925
El-Zein R, Zwischenbergher JB, Wood TG, Abdel Rahman SZ, Brekelbaum C, Au WW (1997) Combined genetic polymorphism and risk for development of lung cancer. Mutat Res 381:189–200
Ernstgard L, Warholm M, Johanson G. (2004) Robustness of chlorzoxazone as an in vivo measure of cytochrome P450 2E1 activity. Br J Clin Pharmacol 58(2):190–200
Fairbrother A, Grove J, de Waziers I, Steimel DT, Day CD, Crespi CL, Daly AK (1998) Detection and characterization of novel polymorphisms in the CYP2E1 gene. Pharmacogenetics 8:543–552
Farker K, Lehmann MH, Oelschlagel B, Haerting J, Hoffmann A, Janitzky V, Schubert J (1998) Impact of CYP2E1 genotype in renal cell and urothelial cancer patients. Exp Toxicol Pathol 50:425–431
Garte S, Gaspari L, Alexandrie A-K, Ambrosone C, Autrup H, Autrup JI, Baranova H, Bathum L, Benhamou S, Boffetta P, Bouchardy C, Breskvar K, Brockmöller J, Cascorbi I, Clapper ML, Coutelle C, Daly A, Dell’Omo M, Dolzana V, Dresler CM, Fryer A, Haugen A, Hein DW, Hildesheim A, Hirvonen A, Hsich L-L, Ingelman-Sundberg M, Kalina I, Kang D, Kihara M, Kiyohara C, Kremers P, Lazarus P, Le Marchand L, Lechner MC, Lieshout EMM, London S, Manni JJ, Maugard CM, Morita S, Nazar-Stewart V, Noda K, Oda Y, Parl FF, Pastorelli R, Persson I, Peters WHM, Rannug A, Rebbeck T, Risch A, Roelandt L, Romkes M, Ryberg D, Salagovic J, Schoket B, Seidegard J, Shileds PG, Sim E, Sinhet D, Strange RC, Stücker I, Sugimura H, To-Figueras J, Vineis P, Yu MC, Taioli E (2001) Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol. Biomarkers Prev 10:1239–1248
Guengerich FP, Kim DH, Iwasaki M (1991) Role of human cytochrome P450 IIE1 in the oxidation of many low molecular weight cancer suspects. Chem Res Toxicol 4:168–179
Haufroid V, Buchet JP, Gardinal S, Lison D (2002) Cytochrome P4502E1 phenotyping and the measurement of the chlorzoxazone metabolic ratio: assessment of its usefulness in workers exposed to styrene. Int Arch Occup Environ Health 75:453–458
Hayashi S, Watanabe J, Kawajiri K (1991) Genetic polymorphisms in the 5′-flanking region change transcriptional regulation of the human cytochrome P450IIEI gene. J Biochem 110:559–565
Hong J-Y, Pan J, Gonzales FJ, Gelboin HV, Yang CS (1987) The induction of a specific form of cytochrome P-450 (P-450j) by fasting. Biochem Biophys Res Commun 142:1077–1083
Ingelman-Sundberg M, Johansson I, Yin H, Terelius Y, Eliason E, Clot P, Albano E (1993) Ethanol-inducible cytochrome P4502E1: Genetic polymorphism, regulation, and possible role in the etiology of alcohol-induced liver disease. Alcohol 10:447–452
Johansson I, Ingelman-Sundberg M (1988) Benzene metabolism by ethanol-, acetone-, and benzene-inducible cytochrome P-450 (IIE1) in rat and rabbit liver microsomes. Cancer Res 48(19):5387–5390
Kato S, Shields PG, Caporaso NE, Hoover RN, Trump BF, Sugimura H, Weston A, Harris CC (1992) Cytochrome P4502E1 genetic polymorphisms, racial variation, and lung cancer risk. Cancer Res 52:6712–6715
Kharasch ED, Thummel KE (1993) Identification of cytochrome P450 2E1 as the predominant enzyme catalyzing human liver microsomal defluorination of sevoflurane, isoflurane, and methoxyflurane. Anesthesiol 79:795–807
Konishi T, Calvillo M, Leng A-S, Feng J, Lee T, Lee H, Smith JL, Sial SH, Berman N, French S, Eysselein V, Lin K-M, Wan Y-JY (2003) The ADH3*2 and CYP2E1 c2 alleles increase the risk of alcoholism in Mexican American men. Exp Mol Pathol 74:183–189
Koop DR, Casazza JP (1985) Identification of ethanol-inducible P-450 isozyme 3a as the acetone and acetol monooxygenase of rabbit microsomes. J Biol Chem 260:13607–13612
Lieber CS (1997) Cytochrome P-4502E1: its physiological and pathological role. Physiol Rev 77(2):517–543
Liu S, Park JY, Schantz SP, Stern JC, Lazarus P (2001) Elucidation of CYP2E1 5′ regulatory RsaI/PstI allelic variants and their role in risk for oral cancer. Oral Oncol 37:437–445
Lu AYH, Levin W (1974) The resolution and reconstitution of the liver microsomal hydroxylation system. Biochem Biophys Acta 344:205–240
Neuhaus T, Ko Y-D, Lorenzen K, Fronhoffs S, Harth V, Bröde P, Vetter H, Bolt HM, Pesch B, Brüning T (2004) Association of cytochrome P450 2E1 Polymorphisms and head and neck squamous cell cancer. Toxicol Lett 151:273–282
Nishimoto IN, Hanaoka T, Sugimura H, Nagura K, Ihara M, Li X-J, Arai T, Hamada GS, Kowalski LB, Tsugane S (2000) Cytochrome P450 2E1 polymorphism in gastric cancer in Brazil: cas-control studies of Japanese Brazilians and non-Japanese Brazilians. Cancer Epidem Biomark Prev 9:675–680
Omer B, Ozbek U, Akkose A, Kilic G (2001) Genetic polymorphism of cytochrome P450 2E1 in the Turkish population. Cell Biochem Funct 19:273–275
Oyama T, Kawamato T, Mizoue T, Sugio K, Kodama Y, Mitsudomi T, Yasumoto K (1997) Cytochrome P4502E1 polymorphism as a risk factor for lung cancer: in relation to p53 gene mutation. Anticancer Res 17:583–588
Persson I, Johansson I, Lou YC, Yue QY, Duan LS, Bertilsson L, Ingelman-Sundberg M (1999) Genetic polymorphism of xenobiotic metabolizing enzymes among Chinese kung cancer patients. Int J Cancer 81:325–329
Peter H, Deutschmann S, Reichel C, Hallier E (1989) Metabolism of methyl chloride by human erythrocytes. Arch Toxicol 72:454–455
Peter R, Böker R, Beaune PH, Iwasaki M, Guengerich FP, Yang CS (1990) Hydroxylation of chlorzoxazone as a specific probe for human liver cytochrome P-450IIEI. Chem Res Toxicol 3:566–573
Pinarbasi H, Silig Y, Seyfikli Z, Celik VK, Pinarbasi E (2001) Genetic polymorphism of GSTM1 in Turkish lung cancer patients. Toxicol Lett 123(Suppl 1):115
Quinones L, Lucas D, Godoy J, Caceres D, Berthou F, Varela N, Lee K, Acevedo C, Martinez L, Aguilera AM, Gil L (2001) CYP1A1, CYP2E1 and GSTM1 genetic polymorphisms. The effect of single and combined genotypes on lung cancer susceptibility in Chilean people. Cancer Lett 174:35–44
Sikdar N, Mahmud ASk, Paul RR, Roy B (2003) Polymorphism in CYP1A1 and CYP2E1 genes and susceptibility to leukoplakia in Indian tobacco users. Cancer Lett 195(1):33–42
Song BJ, Matsunaga T, Hardwick JP, Park SS, Veech RL, Yang CS, Gelboin HV, Gonzales FJ (1987) Stabilization of cytochrome P450j messenger ribonucleic acid in the rat. Mol Endocrinol 1:542–547
Thier R, Lewalter J, Selinski S, Bolt HM (2002) Possible impact of human CYP2E1 polymorphisms on the metabolism of acrylonitrile. Toxicol Lett 128:249–255
Uematsu F, Ikawa S, Kikuchi H, Sagami I, Kanamaru R, Abe T, Satoh K, Motomiya M, Watanabe M (1994) Restriction fragment length polymorphism of the human CYP2E1 (cytochrome P450IIE1) gene and susceptibility to lung cancer: possible relevance to low smoking exposure. Pharmacogenet 4(2):58–63
Uematsu F, Kikuchi H, Motomiya M, Abe T, Sagami I, Ohmachi T, Wakui A (1991) Association between restriction fragment length polymorphism of the human cytochrome P450 2E1 gene and susceptibility to lung cancer. Jpn J Cancer Res 82:254–256
Umeno M, McBridge OW, Yang CS, Gelboin HV, Gonzales FJ (1988) Human ethanol-inducible P450IIEI: complete gene sequence, promoter characterization, chromosome mapping and cDNA-directed expression. Biochemistry 27:9006–9013
Vodicka P, Soucek P, Tates AD, Dusinska M, Sarmanova J, Zamecnikova M, Vodickova L, Koskinen M, Zwart FA, Natajaran AT, Hemminki K (2001) Association between genetic polymorphisms and biomarkers in styrene-exposed workers. Mutat Res 482:89–103
Wang S-L, Lee H, Chen K-W, Tsai K-J, Chen C-Y, Lin P (1999) Cytochrome P4502E1 genetic polymorphisms and lung cancer in a Taiwanese population. Lung Cancer 26:27–34
Watanabe J, Hayashi S, Nakachi K, Imai K, Suda Y, Sekine T, Kawajiri K (1990) PstI and RsaI RFLPs in complete linkage disequilibrium at the CYP2E gene. Nucleic Acids Res 18(23):7194
Wu X, Amos CI, Kemp BL, Shi H, Jiang H, Wan Y, Spitz MR (1998) Cytochrome P450 2E1 DraI polymorphism in lung cancer in minority populations. Cancer Epidemiol Biomark Prev 7:13–18
Wu X, Shi H, Jiang H, Kemp B, Hong WK, Delclos GL, Spitz MR (1997) Associations between cytochrome P4502E1 genotype, mutagen sensitivity, cigarette smoking and susceptibility to lung cancer. Carcinogenesis 18(5):967–73
Yang B-M, O’Reilly DA, Demaine AG, Kingsnorth AN (2001) Study of polymorphisms in the CYP2E1 gene in patients with alcoholic pancreatitis. Alcohol 23:91–97
Acknowledgments
The authors thank to volunteers donating blood, Dr. Nusret Taheri, Dr. Sibel Yıldız and Middle East Technical University Health Center for their help in collecting blood samples. This study was supported in part by Middle East Technical University Research Project Fund (Project number: BAP-2003-07-02-00-40).
Author information
Authors and Affiliations
Corresponding author
Additional information
An account of this work has been presented at the 31st Federation of European Biochemical Societies (FEBS) Congress, in Istanbul, Turkey, on June 24–29, 2006.
Rights and permissions
About this article
Cite this article
Ulusoy, G., Arinç, E. & Adali, O. Genotype and allele frequencies of polymorphic CYP2E1 in the Turkish population. Arch Toxicol 81, 711–718 (2007). https://doi.org/10.1007/s00204-007-0200-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-007-0200-y