Abstract
The development of drug-resistant bacteria and the necessity for unique antimicrobial agents, directed to the search of new habitats to screen the production of anti-infective substances. Culture-dependent studies of heterotrophic bacteria from the intertidal macroalgae thriving along the Southern coast of India resulted in the isolation of 148 strains, which were assayed for antibacterial activities against wide spectrum of pathogens including drug-resistant pathogens, methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecalis (VRE). Two of the most active strains with a zone of inhibition ≥ 30 mm on spot over lawn assay, belonging to the phyla Firmicutes and Gamma-proteobacteria, isolated from a Rhodophycean marine macroalga, Hypnea valentiae, were selected for bioprospecting studies. They were further characterized as Shewanella algae MTCC 12715 and Bacillus amyloliquefaciens MTCC 12716, based on integrated phenotypic and genotypic analysis. The bacterial extracts exhibited significant antibacterial activities against MRSA and VRE with minimum inhibitory concentrations of 6.25–12.5 µg/mL. Time kill kinetic profiles of these bacteria revealed rapid bactericidal activity against both MRSA and E. coli, showing a ≥ 3log10 decline in viable cell count compared to the initial. In BacLight™ live/dead staining technique, the propidium iodide uptake results appropriately attributed that the components in the B. amyloliquefaciens extract might compromise the integrity of the cytoplasmic membrane of the pathogenic bacteria. Type-1 pks gene (MH157093) of S. algae and hybrid nrps/pks gene (MH157092) of B. amyloliquefaciens could be amplified. Antibacterial activity study combined with the results of amplified genes coding for polyketide synthase and nonribosomal peptide synthetase showed that these marine symbiotic bacteria had a promising broad-spectrum activity, and therefore, could be used against the emerging dilemma of antibiotic-resistant bacterial infections.
Graphic abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Marine macroalgae and their symbiotic bacterial communities are potential sources in the field of biotechnology as a result of their production of a wide diversity of metabolites with wide range of bioactivities (Zubia et al. 2009; Le Lann et al. 2016). Microbes with inhibitory traits were found to reside symbiotically on the surface of marine macroalgae, providing it with a microorganism-mediated defence community (Kumar et al. 2011; Penesyan et al. 2009; Albakosh et al. 2016; Xiong et al. 2018). The interest in bacterial populaces living in relationship with marine macroalgae has expanded amid the recent decades to control food-borne pathogens as a result of the rise of antibiotic-resistant bacteria, and the need for new antimicrobial compounds. The retrieval of marine macroalgae associated bacterial strains with antimicrobial activity suggests that marine macroalgae represent an ecological niche, which harbours a specific microbial diversity with potential biotechnological and pharmaceutical interests (Kanagasabhapathy et al. 2008). The search for new antibiotics and pharmacophores possessing potential anti-infective properties is necessary because of the evolving trend of resistance in infectious microbes to the existing compounds (Strobel and Daisy 2003). Marine microorganisms have developed exceptional metabolites and physiological competencies, which not only make sure survival in extreme habitats, but also offer the potential for the production of metabolites not apparent in their terrestrial counterparts (Garson 2001). Moreover, numerous researches have proved that the actual producers of majority of the marine bioactive compounds are not the eukaryotes themselves, but the symbiotic microorganisms associated with them (Kubanek et al. 2003; Li 2009). From a biotechnological point of view, the production of signalling molecules, toxins and other secondary metabolites as effective antagonism and defence propositions by symbiotic microorganisms compose an incomparable repository for the discovery of novel bioactive compounds, pharmaceutical agents and drug molecules, with applications in medical, environmental and industrial areas. Even though, the great biotechnological prospective of marine symbiotic microorganisms remains generally untapped (Egan et al. 2008).
Marine macroalgae constitute one of the vast and diverse ecosystems contributing a vital role in marine environment in global primary production and provides protected and nutrient rich environment for range of organisms. Hence, compared to other eukaryotic hosts, macroalgae are rich in diversity of associated microbes (Armstrong et al. 2001). In particular, the surface-associated bacterial species have been stated as essential for the growth and development of macroalgae, and the bacteria with antagonistic potentials are supposed to shield the macroalgae from pathogens as well as other surface colonizing competitors (Avendaño-Herrera 2005). Some bacterial strains display bactericidal property and host specificity against particular pathogens involving complex biochemical connections between macroalgae and microbes (Strobel and Daisy 2003).
Various researches were focused on the antifouling and antibiotic activities of macroalgae-associated bacterial species (Zheng et al. 2005; Kanagasabhapathy et al. 2008; Penesyan et al. 2009; Wiese et al. 2009; Chakraborty et al. 2018). In 1997, Imamura et al. reported the production of a phenazine compound with antimicrobial activity, by a strain of Pelagiobacter variabilis isolated from the brown marine macroalgae Pocockiella variegata. Antibacterial activity of Bacillus licheniformis associated with intertidal macroalgae Fucus serratus against MRSA and VRE were described earlier by Jamal et al. (2006). Culture-based studies indicated that symbiotic bacteria display a great potential to produce bioactive compounds, which were found to inhibit the growth of environmental pathogens, laboratory strains and also human pathogens (Kizhakkekalam and Chakraborty 2019). Highly competitive environment as well as space and nutrient limitation, forces surface inhabiting microorganisms to evolve specific adaptive methods as well as antagonistic tactics to prevent colonization or growth of possible competitors (Egan et al. 2008).
Polyketides and nonribosomal peptides were found to account for a significant share of identified natural products, and have been the major source of potential pharmacophore candidates (Walsh 2004). Intensive research on nrps (nonribosomal peptide synthetase) and pks-I (type I polyketide synthase) provide a great avenue to the identification and application of novel secondary metabolites in biotechnology and biomedicine (Amoutzias et al. 2008). An increasing number of gene clusters encoding nrps, type I pks and their hybrids were recognized for biosynthesis of complicated natural products (Wang et al. 2014). Also, Wang et al. (2014) reported the widespread dispersal of nrps and modular pks biosynthetic genes in higher numbers among Proteobacteria, Actinobacteria, and Firmicutes.
The major difficulties, which have hindered the development of drugs from marine natural products are the supply issues (Martins et al. 2014). Structure complexity and rich in centres of asymmetry are limiting the large-scale preparation of antibiotics by chemical synthesis, of majority of the promising marine compounds (Davidson 1995). Moreover, the yield from harvesting and isolating from microorganisms might be quite low because the microorganisms are rare or the active compound is a minor constituent (Kasanah and Hamann 2004). In the meantime it is hard to collect infinite amounts of marine animal or plant material, whereas the isolation and culturing of associated microorganisms are important for an adequate and constant supply of bioactive natural products. Host associated marine microbes, which could be cultivated would aid in future development of marine natural product leads if the compound could be produced by a symbiotic microorganism (Kobayashi and Ishibashi 1993).
With the expanding requirement for novel medication against antibiotic-resistant pathogens, macroalgae-associated heterotrophic bacteria with significant antimicrobial activities intends the macroalgae as a perfect ecosystem harboring definite microbial diversity demonstrating a generally unexplored origin of novel antibacterial secondary metabolites. A larger percentage of marine host-associated isolates having antagonistic activities likewise emphasised the biotechnological prospective for targeted isolation of symbiotic bacteria of marine origin (Ismail-Ben et al. 2012). Herein, we have implemented a culture-dependent technique to evaluate the variety of cultivable bioactive bacterial strains in association with intertidal macroalgae along the Southern Coast of Indian peninsular, to discover them as a source for possibly useful antibacterial agents with broad spectrum activity against multidrug-resistant pathogens. The prospective of macroalgae-associated Firmicutes and Gamma-proteobacteria to bring about bioactive metabolites was analyzed by PCR with degenerate primers of the nrps and pks-I genes, utilizing their conserved features.
Materials and methods
Isolation of intertidal marine macroalgae-associated heterotrophic bacteria
Samples of marine macroalgae were collected from the intertidal region at low tide, from Mandapam situated at 9° 17′ 0″ North, 79° 7′ 0″ East, of the Penninsular India and Vizhinjam located at 8.3932° North, 77.0046° East, in the Arabian Sea. Macroalgae belonging to the family Chlorophyceae, Phaeophyceae and Rhodophyceae, which include red algae Hypnea valentiae, Kappaphycus alvarezii, brown macroalgae, Sargassum myricocystum, Padina gymnospora, Turbinaria ornata and green algae Ulva reticulata and Caulerpa racemosa were collected by hand picking and scuba diving. The macroalgal samples were sorted out, and the symbiotic bacteria were isolated by following an established methodology (Kizhakkekalam and Chakraborty 2019). The colonies were differentiated according to their morphology, and were made axenic by restreaking them on nutrient agar plates.
Scanning electron microscopy analysis of microfouling
Scanning Electron Microscopy was performed after Vairappan and Suzuki (2000) with appropriate modifications. Briefly, the algal thalli of approximately 0.5 cm × 0.5 cm size were cut into pieces and fixed in 4% glutaraldehyde, in 0.1 mol L−1 sodium cacodylate buffer (pH 7.2) for 24 h. The specimens were then rinsed with cacodylate buffer (0.1 mol L−1) before post fixation in OsO4 (1%) for 2 h at 4 °C, followed by dehydration through a series of graded alcohol, before being subjected to critical-point drying. Dehydrated algae specimens were mounted on stubs and coated with a 10–30 nm layer of gold before being observed with a scanning electron microscope (JEOL JSM-6380LV, Japan).
Screening for antibacterial activity
The isolated strains were assessed for their antagonistic activity against wide spectrum of pathogenic strains including aquatic and human pathogens, such as Plesiomonas shigelloides (ATCC 14029), Photobacterium phosphoreum (MTCC 11040), Yersinia enterocolitica (MTCC 859), Streptococcus pyogenes (MTCC 1924), Escherichia coli (MTCC 443), Edwardsiella tarda (MTCC 2400), Aeromonas caviae (ATCC 15468), Shigella flexneri (MTCC 1457), Vibrio harveyi (LMG 4044), Aeromonas hydrophila (ATCC 7966), Aeromonas salmonicida (ATCC 27013), Vibrio parahaemolyticus (MTCC 451) and multidrug-resistant pathogens, such as methicillin-resistant Staphylococcus aureus (ATCC 33592) and vancomycin-resistant Enterococcus faecalis (ATCC 51299). The inhibitory potential of each strains were screened by spot over lawn assay. In short, the pathogenic bacterial strains were grown up to the exponential phase in NB (nutrient broth) with 1–2% NaCl. Thereafter, under sterile conditions, a lawn was made on Mueller–Hinton agar (MHA, HiMedia, Mumbai, India) plates by swabbing them, upon which pure colonies of the bacteria were spotted in 6 mm diameter. The plates were incubated for 24–72 h at 30 °C, and the inhibition zone observed around the isolates was recorded using antibiotic zone scale (HiMedia, Mumbai, India).
Characterization of the bioactive isolates
Seven bacterial strains exhibiting significantly greater antibacterial activity against the tested pathogens were further identified by classical biochemical methods followed by Biolog characterization (GEN III Microplate), Matrix Assisted Laser Desorption/Ionization (MALDI) biotyping (IMTECH, India) and 16S rRNA gene sequencing. The identification of isolated bacteria was first performed by new GEN III MicroPlate™ test panel of the Biolog system, which provided information on the phenotypic fingerprint of the microorganism, and could be used to identify them in species level. Fresh overnight cultures of the isolates were tested according to the manufacturer’s protocol. Bacterial suspensions were prepared by taking pure bacterial colonies from the plate surface by a sterile cotton swab and mixing it in 5 mL of 0.85% saline solution. Bacterial suspension was adjusted in IF-0a to achieve a 90–98% transmittance (T90) using a Biolog turbidimeter. Briefly, about 150 µL of the suspension was distributed into each well of a Biolog GEN III microplate, and the plates were then incubated at 26 °C in an Omnilog Reader/Incubater (Biolog). After incubation, the phenotypic fingerprints of purple wells were compared to the Biolog’s extensive species library for species level identification of the isolates.
Further the characteristic identification was performed with the bacterial surface protein analysis using MALDI-TOF biotyping, where small pure colonies of 12–24 h grown cultures were suspended in sterile distilled water (300 μL). It was then washed thrice with absolute ethanol (900 μL) and 70% formic acid (50 μL) followed by centrifugation (at 12000 rpm for 2 min). The supernatant and pellet were separated, whereas the pellet was dissolved in acetonitrile (~ 50 μL) and centrifuged at 12,000 rpm for 2 min. A volume of 1 mL of the supernatant was loaded on target and dried followed by analysis using MALDI-Biotyper (Microflex, Bruker, Germany).
The identities of the heterotrophic bacteria were furthermore affirmed by sequencing their 16S rRNA fingerprint regions. Isolates showing significant and consistent inhibitory activity against the pathogens tested were selected, and DNA was extracted from the bacteria grown in NB (nutrient broth) with 1–2% NaCl by phenol–chloroform method. Further, the quality of these bacterial DNA was assessed by agarose gel electrophoresis, and their concentrations were recorded using a Biophotometer (Applied Biosystems, USA). A total volume of 25 μL of PCR mix was prepared with 10× buffer (Sigma), 0.25 mM of dNTPs (Sigma), 0.5 mM of forward and reverse primers (Sigma), 1 ng of DNA and 0.3 U of Taq DNA polymerase enzyme (Sigma) with 3 mM of MgCl2 for amplifying the specific regions. The following cycling conditions were used: initial denaturation for 5 min at 94 °C, then 30 cycles of 95 °C for 1 min, 58 °C for 1 min, and 72 °C for 2 min, with a 5 min final extension at 72 °C (Applied Biosystems, USA). Universal primer sequences used for the amplification of 16S rRNA region was listed in Table 3 (Weisburg et al. 1991). The bands on gel with the expected size on the agarose gel were purified by following the manufacturer’s protocol of GelElute™-gel extraction kit (Sigma), and sequenced. Similar sequences were searched in the gene database with BLAST search tool and further deposited in GenBank. Each sequence was aligned with CLUSTALW Software (Bioedit program) against reference sequences, and the aligned sequence set was given as input for phylogenetic tree construction program. The neighbour-joining method based on the Kimura two-parameter model and bootstrap analysis with 1000 replications was used to infer the evolutionary history. MEGA6 was used to perform evolutionary analyses.
Antibiotic sensitivity of the selected bacteria
Antibiotic sensitivity of the organisms was determined using commercially available antibiotic impregnated octadiscs (HiMedia) consisting of eight antibiotics—cephalothin, clindamycin, trimoxazole, erythromycin, gentamicin, ofloxacin, penicillin-G and vancomycin placed on the centre of the Mueller–Hinton Agar plates with the culture of candidate bacteria, according to the guidelines set forth by the Clinical and Laboratory Standard Institute guidelines (CLSI 2009).
Extraction of bacterial metabolites
The bacteria were cultured on nutrient agar plates with 1–2% NaCl at pH 8. As the symbiotic bacteria Shewanella algae (MTCC 12715) and Bacillus amyloliquefaciens (MTCC 12716) secrete the bioactive compounds to the surrounding medium, the solid agar was separated aseptically by removing the bacteria at stationary phase and were extracted comprehensively with solvent ethyl acetate for 5 h (three times) at a temperature not beyond the boiling point of the solvent before being concentrated in vacuo with a rotary vacuum evaporator (Heidolph, Schwabach, Germany) at about 40 °C. The residues obtained were stored for further analyses at 4 °C.
Antibacterial activity of organic extract
Disc diffusion method
The disc diffusion method of Bauer et al. (1966) was initially used to study the antibacterial activity of the crude extracts against clinical as well as aquatic pathogens. Sterile discs (6 mm) containing 30 µg of each extract was prepared from a 1 mg stock solution. MHA plates were inoculated with overnight grown pathogenic organisms of 107 cfu mL−1 (cfu: colony forming unit). The discs with bacterial extracts were then placed on the surface of these MHA plates and incubated for 24–72 h at 37 °C. The results were noted down by measuring the size of zone of inhibition using antibiotic zone scale (HiMedia). Ethyl acetate was used as negative control and commercially available antibiotic discs (HiMedia) as positive control.
Immersion bioautography
The crude organic extracts (10 μL, 1.0 mg/mL) were loaded onto TLC plates in a thin band and eluted using 60% ethyl acetate/n-hexane, as the mobile solvent system. The developed plates were then dried in a fast moving air stream in order to remove solvent traces on the plates. The overnight grown bacterial cultures of pathogens were used to inoculate the MHA plates. The prepared chromatograms were kept in sterile petriplates and MHA mixed with each culture broth was poured at a temperature lesser than 40 °C. Further, the plates were incubated overnight with 100% relative humidity, at 35 °C, and in the dark. After 24 h of incubation, the plate was sprayed with a solution of 2 mg/mL tetrazolium salt 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma®, USA), before being incubated for 2 h (Suleimana et al. 2009). The white bands indicated the absence of reduction of MTT to the coloured formazan because of the presence of compounds that inhibited the growth of the test organisms.
Minimum inhibitory concentration and minimum bactericidal concentration
Minimum inhibitory concentration (MIC) calculation was carried out according to the Clinical and Laboratory Standard Institute guidelines (CLSI 2012). The MIC test was performed using MH broth, which was supplemented with 1–2% NaCl. In a 96 well plate, 11 pathogens and the crude extracts were added in different dilutions. Chloramphenicol antibiotic was used as the positive control and ethyl acetate as the negative control. Briefly, the antibacterial extracts were serially diluted (1:1), through sterile MH broth in a microwell plate, and the standardized inoculums, which were identical to the volume of the diluted crude extract, were added to each dilution well, bringing the microbial concentration to about 500,000 cells/mL. The blank was set with the nutrient broth and the microplate was incubated at 35 °C for 18 h. Further, it was observed for microbial growth, which was indicated by measuring the absorbance followed by addition of a tetrazolium salt MTT, where MTT (0.5%) was used as the indicator stain of growth. The last wells in the dilution series, which did not exhibit growth, were found to correspond with the MIC of the antibacterial agent. The minimum bactericidal concentration (MBC) was determined by swabbing 10 μL of the dilutions from the incubated plates to spot the colony formation, and the lowest concentration at which the colony formation was not observed, were recorded.
Time-kill assay
Time-kill assay was performed to define the primary mode of action of the antibacterial agents present in the extracellular metabolites of macroalgae associated bacteria under the study. The assay was carried out by macro-dilution method as per the Clinical and Laboratory Standards Institute guidelines (CLSI 2012). Approximately 105 cfu/mL of exponentially growing bacterial cells in inoculum suspensions (MR S. aureus and E. coli) were used in this test. The crude extracts were added to 10 mL of inoculum with final concentrations corresponding to ½ × MIC, 1 × MIC and 2 × MIC. A control containing the bacterial strain without the extract was incorporated in each trial. Further, the inoculum cultures were incubated on an orbital shaker at 200 rpm at 37 °C. Aliquots were removed from the inoculum after timed intervals of incubation (i.e., 0, 0.5, 1, 2, 4, 8, 12 and 24 h), and serial tenfold dilutions were prepared in saline, as needed. The numbers of viable cells were observed by the plate count technique, briefly, plating 25 µL of each dilution on a MHA plate, whereas all plates were incubated for 24 h at 37 °C. Resulting data were evaluated as killing curves by plotting the log10 cfu/mL versus time (h), and the change in bacterial concentration was determined. Bactericidal activity, i.e., the viable bacterial cell count for the time-kill end point determination, was defined as a reduction of ≥ 3log10 cfu/mL comparative to the initial inoculum, whereas, bacteriostatic activity relating to < 3log10 cfu/mL was found to decrease relative to the initial inoculum suspensions (Petersen et al. 2007).
Live/dead assay
The bacteria exhibiting higher antagonistic potential and complete bactericidal activity were further tested with live/dead assay. The viability assay was executed using the live/dead BacLight™ bacterial viability staining kit L7012 (Invitrogen, Molecular Probes, USA), which included two nucleic acid-binding stains, i.e., SYTO 9 and propidium iodide (PI). These stains when used in a suitable mixture, the live bacteria with intact membranes fluoresce green, whereas bacteria with damaged membranes fluoresce red (Singh 2006). The bacterial strains of E. coli and MRSA were grown overnight at 30 °C in MH broth before being centrifuged and washed with NaCl, (0.85%). Afterwards, the bacteria were suspended in saline solution to attain an OD600 nm of 0.150 ± 0.02, and 1 mL of each cell suspension was kept in contact with the crude extract (at equivalent MIC) for 1 h. Control was set with cell suspension in 10% DMSO, without the extract. Further, pathogenic bacterial cultures were diluted in saline (1:10) and each diluted suspension (300 μL) was filtered through a Nucleopore™ (Whatman) black polycarbonate membrane of 0.22 mm pore size and stained with 250 mL of diluted SYTO 9, a green-fluorescent nuclear counterstain and equal amount of diluted PI. Further, the dyes were kept to react in the dark at 27 ± 3 °C, for 15 min. The membrane was then mounted on BacLight™ mounting oil, as mentioned in the manufacturer’s protocol. A LEICA DMLB2 microscope with mercury lamp HBO/100W/3 incorporating a color digital camera to acquire images using IM50 software (LEICA) and a 100× oil immersion fluorescence objective was used to observe the stained bacteria. The optical filter combination for optimal viewing of stained mounts was consisted with a 480–500 nm excitation filter bearing a 485 nm emission filter (Chroma 61000-V2 DAPI/FITC/TRITC).
Identification of secondary metabolite genes
The genes encoding pks-I and nrps were amplified with different sets of degenerate primers in order to screen the secondary metabolite genes responsible for the biosynthetic potential of bacterial isolates. The primers used for the analysis were given in Table 3. Polymerised chain reaction was performed in a total volume of 25 µL having 10× buffer with MgCl2 (Sigma), 0.25 mM dNTPs (Sigma), 1.0 mM of each primer (Sigma), 1 ng DNA, and 0.3 U Taq polymerase (Sigma). PCR conditions were set as initial denaturation time of 5 min at 94 °C followed by 30 cycles of 30 s at 94 °C, 30 s at 56 °C, 1 min at 72 °C, and a final extension of 5 min at 72 °C. Agarose gel (1.5%) was used to detect the amplified products as fragments, by agarose gel electrophoresis, and bands of 700–800 bp and 1000–1400 bp were considered as products of pks-I and nrps genes, respectively. The products of the estimated size (pks-I ~ 700 bp and nrps ~ 1000 bp) were then purified with Gel Elute™ Gel Extraction Kit (Sigma). The sequences obtained were further submitted in the NCBI GenBank.
Statistical analysis
The variances (ANOVA) between various results were analyzed by Statistical Program for Social Science (SPSS, ver. 13.0, SPSS Inc., Quarry Bay, HK), and was used for assessing the significant differences between the means. Data were expressed as mean ± standard deviation of three replicates, and significant differences were represented as P < 0.05.
Results
Isolation of heterotrophic bacterial strains associated with intertidal macroalgae and antibacterial screening
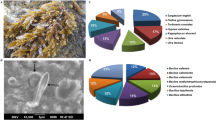
Bacteria associated with intertidal macroalgae (Figure S1) belonging to Phaeophyceae, Rhodophyceae and Chlorophyceae along the Southern coast of India, were isolated and screened for their bioactive potentials. A total of 148 bacterial strains were isolated from 33 different macroalgae, and 77 of them (53%) showed significant antibacterial activity in the preliminary screening. Single, pure colonies of the active strains were made and preserved with sterile glycerol at − 80 °C. Among the above-mentioned 77 isolates, only 23 were thrived in withstanding further laboratory sub-culturing, and could conserve the bioactivity. High-resolution scanning electron microscopy images (Fig. 1a) inferred the symbiotic interaction between macroalgae and bacteria especially on the thalli surface. Among them, seven strains with greater and consistent bioactivities were selected for the present work, and the antibacterial properties of these isolates have been enlisted in Table 1.
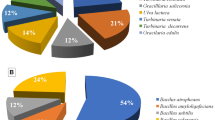
a Scanning electron micrographs of the macroalgae on which different microorganisms were attached. b Distribution of macroalgae-associated bacteria contributed by the representative algal hosts screened for the study. c Contribution (as percent share towards the total number of bacterial isolates with potentially greater antibacterial properties against the pathogens) of the individual representative bacterium as marine macroalgal association
The green algae, U. reticulata contributed a share of 37% of the bioactive strains of bacteria, followed by brown algae S. myriocystum (23%), where, P. gymnospora and H. valentiae gave an equal share of 17% towards the total of bacterial symbionts with potential antibacterial properties against the test pathogens. The aggregate share of the macroalgae K. alvarezii, T. ornata and C. racemosa towards the total number of bioactive isolates was found to be considerably smaller (17% of the total symbiotic bacteria) (Fig. 1b). Majority (83%) of the bioactive strains were the representatives of Gamma-proteobacteria and Firmicutes. The former was found to include Shewanella algae, Photobacterium phosphoreum, Pseudomonas putida, Vibrio sp. and Aeromonas sp. The largest bacterial communities belonging to Firmicutes were found to be Bacillus subtilis and Bacillus amyloliquefaciens followed by Bacillus cereus and Bacillus marinus. Individual share contributed by each bacteria to the total number of macroalgae-associated bioactive isolates were given in Fig. 1c.
Identification of potential strains and 16S rRNA-based phylogeny
The bacteria, which exhibited significant antagonistic properties, were categorized using diverse biochemical tests according to Bergey’s Manual of Determinative Bacteriology protocol (Chakraborty et al. 2014). Phenotypic and genotypic analyses grouped the potential bacteria into two major phyla of Gamma-proteobacteria and Firmicutes. The GEN III MicroPlates™ could facilitate testing of Gram-negative and Gram-positive bacteria in the similar test panel, which comprised of 71 carbon sources and 23 chemical sensitivity assays. GEN III dissected and examined the capability of the cell to metabolize all major classes of compounds, besides defining other significant physiological properties, such as pH, salt and lactic acid tolerance, reducing power and chemical sensitivity. Characteristics of biochemical reactions obtained from GEN III and basic cellular morphological properties were described in the supplementary material (Table S1–S3). The isolated Shewanella spp. and Bacillus spp. were identified as Shewanella algae, Bacillus subtilis and Bacillus amyloliquefaciens by the Biolog system. The identities of the bacteria were further affirmed with MALDI –TOF biotyping and Biolog characterization (Bruker Daltonik GmbH, Bremen, Germany). MALDI Biotyper identification of bacteria used the ribosomal protein fingerprint of the cell and depends on high abundance proteins (Seng et al. 2009). The producer strains were identified by MALDI biotyping as Bacillus amyloliquefaciens with a score of 1.90 and Shewanella algae as 2.2, which showed the high level confidence of identification (Figure S2Ab-B). Amongst the active strains, seven most potential strains were selected and further characterized by 16s rRNA genotyping and submitted in GenBank with accession numbers of KX272632–KX272638. The 16S rRNA sequences of the isolated strains were matched with the most similar species in the GenBank, and a phylogenetic tree was then made by linking the sequences of the seven bioactive strains with their closest relatives (Fig. 2). The bioactive strains were deposited in Microbial Type Culture Collection and Gene Bank (MTCC) at the Institute of Microbial Technology (IMTECH), Chandigarh (India), an International Depository Authority (IDA) under the Budapest Treaty as S. algae MTCC12715 and B. amyloliquefaciens MTCC12716. The selected strains were found to be susceptible to the antibiotics used in this study (Fig. 3d, e).
Phylogenetic tree obtained from 16S rRNA gene sequences of the selected strains, showing evolutionary relationships between bacterial isolates associated with intertidal macroalgae. The evolutionary history was inferred by using the Maximum Likelihood Method based on the Kimura 2-parameter model. The tree with highest log likelihood (− 6375.4197) was shown. The percentage of trees in which associated taxa were clustered together, was shown next to the branches. There were a total of 1577 positions in the final dataset. Evolutionary analyses were conducted in MEGA6. Candidate strains of B. amyloliquefaciens (KX272634) and S. algae (KX 272635) were highlighted
Antibacterial activities of the bacteria and their crude extracts assessed by a Spot on lawn assay, b Immersion bio autography and c disc diffusion method, against MRSA (Inset: B = B. amyloliquefaciens and S = S. algae). Ampicillin (30 µg) was used as the positive control. Antibiotic sensitivity of the candidate bacteria tested dS. algae MTCC12715 and eB. amyloliquefaciens MTCC12716, with eight commercially available antibiotics (Octadisc, HiMedia)
Antagonistic activity of the organic extract of heterotrophic bacteria
The antibacterial activity of the organic extracts of S. algae MTCC12715 and B. amyloliquefaciens MTCC12716 were assessed by various means. The zone of inhibition obtained from spot on lawn assay of the selected strains and disc diffusion assay with the organic extracts were summarized in Table S4 (Fig. 3a, c). Further, it was observed that the lowest concentration of the antibacterial agent inhibiting the growth of pathogenic bacteria as 3.125–25 µg/mL and 3.125–12.5 µg/mL for S.algae MTCC 12715 and B. amyloliquefaciens MTCC12716, respectively. MBC was noted with 10 µg/mL for S. algae and 6.25 µg/mL for B. amyloliquefaciens against MRSA. The results were summarized in Table 2.
In immersion bioautography, the white bands specified where reduction of the tetrazolium salt, MTT to the coloured formazan did not occur due to the presence of compounds that inhibited growth of tested organisms. The white spot against the purple background (Fig. 3b) displayed on the petridish specified the fractions of symbiotic bacterial extract possessing antibacterial activity against MR Staphylococcus aureus. The Rf of the spots were intended as 0.83 and 0.54 for S. algae MTCC 12715 and 0.97 and 0.63 for B. amyloliquefaciens MTCC 12716 organic extracts.
Time-kill evaluation of bacterial extracts
To further evaluate the antibacterial effect of the extracts in greater depth, the killing curve analysis, which offer information on the type of the inhibition (bacteriostatic or bactericidal) (concentration-dependent or time-dependent), was implemented. Time-kill analyses have been broadly used for in vitro studies of new antimicrobial agents as these provide qualitative information on the pharmacodynamics of antibacterial agents (Tam et al. 2005). The kinematic interaction between pathogenic bacteria and extracts was observed at concentrations of one-half of the MIC (½ × MIC), MIC and two times the MIC (2 × MIC). Rapid bactericidal activity towards both pathogenic strains, MRSA and E. coli were displayed on the kill kinetic profiles, showing a ≥ 3log10 decrease in viable cell count relative to the original. The time-kill assays, as expected from the determined MIC ratios, for B. amyloliquefaciens MTCC12716 towards MRSA and E. coli, were consistent with bactericidal characteristic at all tested concentrations within 4 h of exposure (Fig. 4). The organic extracts from S. algae MTCC12715 showed both bactericidal and bacteriostatic activity depending on time of exposure and concentration of the extract. For instance, Vidaillac et al. (2011) revealed that oritavancin displayed rapid bactericidal activity against MRSA after 9 h of exposure. In the present study, the kill kinetic profiles of both the extracts displayed higher bactericidal activity, i.e., within 4 h, toward MRSA and E. coli, compared to oritavancin.
Bactericidal efficiency
The combined usage of SYTO 9 and propidium iodide in a commercially available kit (BacLight™—Molecular Probes®) was designated as a rapid and reliable method for the assessment of bacterial viability (Borges et al. 2013). The assay evaluated membrane integrity by selective stain exclusion to live and dead cells (Simões et al. 2005). B. amyloliquefaciens extract (MTCC 12716) with greater activity was tested with live/dead assay kit against representatives of Gram-positive and Gram-negative pathogens, MRSA and E. coli. From the results it could be observed that intact live cells emit a green fluorescence, whereas dead cells emit red fluorescence upon staining with propidium iodide. The increase in red fluorescence could, in turn, be considered as increased bactericidal activity of the bacterial extracts. SYTO9 is a green-fluorescent (510–540 nm) nucleic acid stain that is able to penetrate both healthy and damaged bacterial cytoplasmic membrane. After incubation with SYTO9, almost all bacteria emit green fluorescence upon excitation with blue light facilitating counting total cells. In contrast, propidium iodide is incapable to penetrate healthy bacteria and its red emission (620–650 nm) is only associated to permeabilized bacteria (Figure S3). When both stains are there in the same cell, the emission of SYTO9 has been reduced as a result of the displacement of one stain by the other and apparent quenching by Förster resonance energy transfer (FRET) (Stocks 2004). Live cells were observed as green, caused by SYTO 9 and dead cells were fluoresced in red. As shown in the Figure S3, the pathogens treated with crude extracts of B. amyloliquefaciens MTCC 12716 were almost red with fluorescence, indicating the high permeability of propidium iodide dye towards them, and most of the cells were dead. The control stains showed strong green fluorescence (Figure S3), revealing that the bacteria were viable. In this way, the potential of bacterial organic extract to interfere with membrane integrity after 1 h exposure was analysed. The propidium iodide uptake results appropriately attributed that the components in the B. amyloliquefaciens MTCC 12716 organic extract might compromise the integrity of the cytoplasmic membrane of both pathogenic bacteria tested.
Analysis of pks and nrps gene and phylogenetic sequence analysis
The pks-I and nrps specific primers were positive in detecting the PCR amplicons in the DNA of S. algae MTCC12715 and B. amyloliquefaciens MTCC12716, displaying significant homology to the sequences deposited in the NCBI GenBank. The primers used in this work were listed in the Table 3. To verify the amplified product (band size of ~ 700 bp and ~ 1000 bp) (Fig. 5), the sequenced results acquired through cloning were then subjected to blastx in the GenBank. Afterwards, the nucleotide sequences were translated and the deduced amino acid sequences were also analysed through blastp program of NCBI. In the blast analysis report, it was observed that the positive amplicons had sequence similarity with S. algae type 1 pks gene (99%) and hybrid nrps/pks gene of B. amyloliquefaciens strains (99%), and they were submitted in GenBank as MH157093 and MH157092, respectively. The maximum likelihood method based on the JTT matrix-based model was used to infer the evolutionary history (Jones et al. 1992). The tree with the highest likelihood (− 3511.9966) was shown, and the percentage of trees in which the associated taxa clustered together was shown next to the branches. Initial tree for the heuristic search were obtained automatically by applying neighbour-join and BioNJ algorithms to a matrix of pairwise distance using JTT model, and then selecting the topology with superior log likelihood value. There were 152 positions in the final dataset of evolutionary analysis, conducted in MEGA6.
a (Ba) Polyketide synthase/nonribosomal peptide synthetase hybrid gene (nrps/pks) product (~ 1000 bp) of the bioactive bacterium B. amyloliquefaciens MTCC 12716 associated with Hypnea valentiae. (Sa) type I polyketide synthase gene (pks- I) product (~ 700 bp) of S. algae MTCC 12715 associated with Hypnea valentiae. The amplified gene sequences for type I pks and hybrid nrps/pks have been submitted at the NCBI GeneBank with accession numbers of MH157092 and MH157093, respectively. b Molecular phylogeny analysis of the diverse range of ketosynthase domains as type I, II and III, nrps and hybrid of nrps/pks genes. The evolutionary history was inferred by using the maximum likelihood method based on the JTT matrix-based model. There were 152 positions in the final dataset. Evolutionary analysis was conducted in MEGA6. The sequences in the experiment were headed by circles
The phylogeny of the sequenced KS domains of S. algae and amino acid adenylation domain-containing protein of B. amyloliquefaciens, categorized the presumed amino acid sequences as bacterial pks type I and hybrid nrps/pks, respectively. The deduced amino acid sequence analysis verified that the sequences shared a conserved motif EAHGTGAAGDVAEF in KS domain and SGSTGKPKG in hybrid nonribosomal protein synthetase/type 1 polyketide synthetase.
Discussion
In the last decades, the incidence of human pathogens resistant to several antibiotics has increased worldwide according to Centre for Disease Control and Prevention (CDC 2013). Hence, it is essential to find new alternative remedies to conflict or decrease the incidents of infectious diseases caused by drug-resistant bacteria. The interest in antibiotics derived from nature has increased due to the established safe status of certain natural products (Cowan 1999).
In the current work, heterotrophic bacteria associated with the intertidal macroalgae were collected along the Southern coast of Indian peninsular region, as the intertidal symbionts were reported to produce unique secondary metabolites in response to various stresses (Penesyan et al. 2009). Microorganisms from the intertidal zones must be capable to tolerate fast and frequent fluctuations in environmental conditions including temperature, light and salinity; and are exposed to ultraviolet radiation, wave action, as well as phases of drought (Dionisi et al. 2012). Descriptive studies of surface-associated bacteria isolated from the marine macroalgae were reported as early as 1875, and several new bacterial species and genera have been discovered from the macroalgae suggesting that these species might represent an interesting biotic environment for unearthing new bacterial communities (Goecke et al. 2010). Microorganisms of different species were observed distinctly attached to the surface of intertidal macroalgae collected through scanning electron micrographs. Previously, Duan et al. (1995) reported the use of SEM to study the algae–bacteria interaction.
Among a total of 148 bacterial strains isolated, Gamma-proteobacteria and Firmicutes were the predominant communities present. Microbial communities living on macroalgae vary in composition and number of species from those occurring in seawater (Longford et al. 2007). The most abundantly represented phylum on macroalgae being the Proteobacteria (Armstrong et al. 2001) followed by the Firmicutes, Bacteroidetes, Actinobacteria and Chloroflexi (Longford et al. 2007). Furthermore, the bacteria residing on the surfaces of marine organisms were known to comprise a percentage of antibiotic generating bacteria greater than that detected in free-living bacteria isolated from marine environs (Zheng et al. 2005).
Among the different macroalgal species collected, Padina gymnospora contributed the major share of 24% of the active bacterial isolates, followed by Sargassum myricocystum (17%). Turbinaria ornata and Caulerpa racemosa contributed equally (14% each) to the total of bacterial isolates. Hypnea valentiae contributed 13% of the isolates with significantly greater antibacterial activities against the pathogenic bacteria used. The collective share of the macroalgae Kappaphycus alvarezii and Ulva reticulata towards the total active isolates was found to be suggestively lesser (9% of the total symbiotic bacteria). The contribution of bioactive isolates from the different macroalgal host was given in Fig. 1b. Interestingly, the representatives of the Gamma-proteobacteria were abundant (47% of the active isolates) and majority of which were allied with Shewanella algae (Fig. 1c). Bacillus subtilis followed by Bacillus amyloliquefaciens (32 and 17% of the active isolates, respectively) belonging to Firmicutes were found to be the second predominant bacterial groups. Bacillus cereus and Vibrio alginolyticus gave an individual share of 2% of the cumulative number of macroalgae-associated active isolates (Fig. 1c). Simultaneously, according to some findings, common strains of B. subtilis and B. pumilus persist as the most plentiful surface-associated bacilli found in marine environment besides proteobacteria (Ivanova et al.1999).
Since the first antibiotic from a marine bacterium was described in 1966 (Burkholder et al. 1966), the antibiotic nature of marine bacteria gained attention of the marine research world. In order to screen the bioactive capacities of the isolates under study, they were initially screened for antibacterial activities by spot on lawn assay, and 53% of the total bacteria possessed antibacterial activity against at least one of the pathogenic organism tested. The most active bacteria representing Gamma-proteobacteria and Firmicutes were further used for the bioprospecting studies. The bacteria isolated from the red marine macroalga Hypnea valentiae with greater activities were identified with standard biochemical methods, Biolog characterization and MALDI biotyping followed by 16s rRNA sequencing. These bioactive strains were deposited in the Microbial Type Culture Collection and Gene Bank (MTCC) as Shewanella algae MTCC 12715 and Bacillus amyloliquefaciens MTCC 12716.
The bacterial metabolites were then extracted in ethyl acetate and antibacterial activities were primarily assessed with disc diffusion method. Significant activities were observed with an inhibition zone of 16–32 mm for B. amyloliquefaciens MTCC 12716 and 11–27 mm for S. algae MTCC 12715, against wide spectrum of pathogenic organisms including MRSA and VRE. Burgess et al. (2003) isolated numerous bacteria with antifouling activity from macroalgae Ulva lactuca, and found that most of these bacteria belonged to the genus Bacillus, such as B. licheniformis and B. subtilis. A previous report of literature demonstrated that a heterotrophic Shewanella sp. isolated from macroalga Ulva lactuca exhibited antibacterial activity against pathogens with an inhibitory zone up to 9.3 mm (Kumar et al. 2011). Also, it was interesting to note that the MIC and MBC of the bacterial extracts were comparable with the commercially available antibiotic chloramphenicol (MIC 6.25 µg/mL). The macroalga-associated heterotrophic bacterial extracts (Bacillus and Shewanella) inhibited the growth of pathogens at concentration of 3.125–25 µg/mL including multi drug-resistant strains of MRSA and VRE.
The TLC-bioautography was established to be a simple, effective and cost-effective technique for the separation of a complex mixture, and simultaneously, it localized the active constituents on the TLC plate, has found extensive application in the search for new antibiotic leads (Wagman and Weinstein 1973). From the bioautography results, well-defined inhibition bands were obtained with S. algae MTCC12715 as well as B. amyloliquefaciens MTCC12716 extracts, in correspondence with the active compounds. The method was convenient in isolating antibacterial compounds because the Rf of the active compound could be used for further bioassay directed fractionation as an alternative of requiring labour intensive determination of activity of fractions. This also aids to comprehend that the compound isolated has been identical to that was present in the extract, and was not an artefact of the isolation procedure (Suleimana et al. 2009).
The killing curve assay discloses a time-dependent or a concentration-dependent antibacterial effect of the extracts (Pfaller et al. 2004), whereas for bacteria, this test has been standardized and described in M26-A document of CLSI (2012). The kill kinetic profiles displayed quick bactericidal activity against both pathogenic strains, i.e., MRSA and E. coli, showing a ≥ 3log10 decrease in viable cell count comparative to the initial. B. amyloliquefaciens MTCC12716 extract killed MRSA cells after 2 h exposure, and it was clearly visible after 4 h, and 100% inhibition was achieved within 6–8 h. In the case of E. coli, the bactericidal activity was consistent for Bacillus extract at all tested concentrations, within 4 h exposure. Notably, the extracts from S. algae MTCC12715 exhibited bacteriostatic activity with MRSA strains at a concentration half its MIC, and at the same time the extract inhibited 100% MRSA cells within 8 h at MIC and 2 × MIC concentrations, and therefore, the bactericidal activity of S. algae MTCC12715 extract was found to depend on time of exposure and concentration of the extract (Fig. 4).
In the live/dead Baclight™ assay, survival rate has been expressed in terms of preservation of membrane integrity as the percentage of bacterial cells with intact membranes (stained with SYTO-9) to the total number of cells (Berney et al. 2007). The decease of a bacterial cell has long been defined as the incapability of a cell to grow to a visible colony on bacteriological media. However, with traditional culture methods the bacterial death could be observed only in retrospect (Postgate 1989). This assay enabled the differentiation between bacteria with intact and impaired cytoplasmic membranes; it has often been used to distinguish between active and dead cells (Sachidanandham et al. 2005). From the microscopic images observed, more than 70% of the red cells on both red filter and merged images indicated the complete growth inhibition of both the pathogenic bacteria, MRSA and E. coli by B. amyloliquefaciens MTCC 12716 extract. The permeability of propidium iodide to the bacterial cells exposed to B. amyloliquefaciens MTCC 12716 organic extract ascribed that the components in the extract might affect the integrity of the cytoplasmic membrane of the pathogenic bacteria tested, and thereby stained by this selective dye.
Non-ribosomal peptide synthetase (nrps) and polyketide synthase (pks), which catalyze the elongation of polyketides and synthesis of oligopeptides, respectively, are the key enzymes of secondary metabolite biosynthesis (Wang et al. 2014). Many medically relevant secondary metabolites were found to be produced by nrps, including the β-lactam antibiotics and cyclosporine (Felnagle et al. 2008) and the lipopeptide antibiotic daptomycin (Doekel et al. 2008). The pks produced secondary metabolites have also resulted in several marketed pharmaceuticals, including lovastatin. In addition, some secondary metabolites are produced by a combination of nrps and pks activities (Brakhage 2013). The pks gene sequence amplified from S. algae MTCC 12715 was found to resemble 99% to the type 1 pks of Gamma proteobacteria in BLAST search. Type I polyketide synthase were identified in marine S. algae, and they were found to code for the production of eicosapentaenoic acid in the bacteria (Yazawa 1996). Based on a recent whole genome mining study, 31% of the Bacillus species were estimated to harbour nrps and pks secondary metabolite gene clusters, whereas 70% of these encoded nrps, and 30% as hybrid nrps/pks or pks (Wang et al. 2014). The sequence obtained from B. amyloliquefaciens MTCC 12716 showed similarity with the hybrid nrps/pks genes in the databank. These gene clusters were found to be responsible for the production of various polyketide and lipopeptide secondary metabolites in the bacteria (Aleti et al. 2015).
Conclusions
Presently, microbial infections have developed as a vital clinical threat, with associated morbidity and mortality, which are primarily due to the emergence of microbial resistance to the prevailing antimicrobial drugs. Thus, searches for antimicrobial susceptibility testing and discovering novel antimicrobial agents have been widely used and continued to be developed. Marine heterotrophic microorganisms provide a wide range of diversity in chemical structure. In addition to the molecular novelties and potential bioactivities, marine microbial metabolites present a renewable supply of targeted compounds for scientific investigations and commercial development. Herein, the activity of extracts derived from macroalgae-associated heterotrophic bacteria B. amyloliquefaciens MTCC 12716 and S. algae MTCC12715 were comparable with the existing antibiotics, and therefore, could address the problem of resistance and re-emergence in infectious diseases. Interestingly, the extracellular secondary metabolites were bactericidal in nature. According to the results, these marine symbiotic bacteria, more specifically, B. amyloliquefaciens MTCC 12716, with secondary metabolite genes pks-1 and nrps, could be considered as a promising solution against the threat of antibiotic resistance.
References
Albakosh MA, Naidoo RK, Kirby B, Bauer R (2016) Identification of epiphytic bacterial communities associated with the brown alga Splachnidium rugosum. J Appl Phycol 28:1891–1901
Aleti G, Sessitsch A, Brader G (2015) Genome mining: prediction of lipopeptides and polyketides from Bacillus and related Firmicutes. Comput Struct Biotechnol J 13:192–203
Amoutzias GD, Van de Peer Y, Mossialos D (2008) Evolution and taxonomic distribution of nonribosomal peptide and polyketide synthases. Future Microbiol 3(3):361–370
Armstrong E, Yan L, Boyd KG, Wright PC, Burgess JG (2001) The symbiotic role of marine microbes on living surfaces. Hydrobiologia 461:37–40
Avendaño-Herrera RE (2005) Avances en el conocimiento del patógeno de peces "Tenacibaculum maritimum": implicaciones en el diagnóstico y prevención de la enfermedad. Ph.D. thesis, Universidad de Santiago de Compostela
Bauer AW, Kirby WM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45(4):493–496
Berney M, Hammes F, Bosshard F, Weilenmann HU, Egli T (2007) Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight Kit in combination with flow cytometry. Appl Environ Microbiol 73(10):3283–3290
Borges A, Ferreira C, Saavedra MJ, Simões M (2013) Antibacterial activity and mode of action of ferulic and gallic acids against pathogenic bacteria. Microb Drug Resist 19(4):256–265
Brakhage AA (2013) Regulation of fungal secondary metabolism. Nat Rev Microbiol 11:21–32
Burgess JG, Boyd KG, Armstrong E, Jiang Z, Yan L, Bergren M, May U, Pisacane T, Granmo A, Adams DR (2003) The development of a marine natural product-based antifouling paint. Biofouling 19(1):197–205
Burkholder PR, Pfister RM, Leitz FH (1966) Production of a pyrrole antibiotic by a marine bacterium. Appl Microbiol 14:649–653
Chakraborty K, Thilakan B, Raola VK (2014) Polyketide family of novel antibacterial 7-O-methyl-5’-hydroxy-3’-heptenoate-macrolactin from seaweed-associated Bacillus subtilis MTCC 10403. J Agric Food Chem 62:12194–12208
Chakraborty K, Thilakan B, Kizhakkekalm VK (2018) Antibacterial aryl-crowned polyketide from Bacillus subtilis associated with seaweed Anthophycus longifolius. J Appl Microbiol 124:108–125
Clinical and Laboratory Standards Institute (2009) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved Standard M07-A10. Clinical and Laboratory Standards Institute, Wayne
Clinical and Laboratory Standards Institute (2012) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard-9th ed. CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne
Cowan MM (1999) Plant products as antimicrobial agents. Clin Microbiol Rev 12(4):564–582
Davidson BS (1995) New dimensions in natural products research: cultured marine microorganisms. Curr Opin Biotechnol 6:284–291
Dionisi HM, Lozada M, Olivera NL (2012) Bioprospection of marine microorganisms: biotechnological applications and methods. Rev Argent Microbiol 44:49–60
Doekel S, Coeffët-Le Gal MF, Gu JQ, Chu M, Baltz RH, Brian P (2008) Non-ribosomal peptide synthetase module fusions to produce derivatives of daptomycin in Streptomyces roseosporus. Microbiology 154:2872–2880
Duan D, Xu L, Fei X, Xu H (1995) Marine organisms attached to seaweed surfaces in Jiaozhou Bay, China. World J Microbiol Biotechnol 11:351–352
Egan S, Thomas T, Kjelleberg S (2008) Unlocking the diversity and biotechnological potential of marine surface associated microbial communities. Curr Opin Microbiol 11:219–225
Felnagle EA, Jackson EE, Chan YA, Podevels AM, Berti AD, McMahon MD, Thomas MG (2008) Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol Pharm 5:191–211
Garson MJ (2001) Ecological perspectives on marine natural product biosynthesis. In: McClintock JB, Baker BJ (eds) Marine chemical ecology. CRC Press, Boca Raton, pp 71–114
Goecke F, Labes A, Wiese J, Imhoff JF (2010) Chemical interactions between marine macroalgae and bacteria. Mar Ecol Prog Ser 409:267–299
Imamura N, Nishijima M, Takadera T, Adachi K, Sakai M, Sano H (1997) New anticancer antibiotics pelagiomicins, produced by a new marine bacterium Pelagiobacter variabilis. J Antibiot 50(1):8–12
Ismail-Ben Ali A, El Bour M, Ktari H, Bolhuis H, Ahmed M, Boudabbous A, Stal LJ (2012) Jania rubens-associated bacteria: molecular identification and antimicrobial activity. J Appl Phycol 24:525–534
Ivanova EP, Vysotskii MV, Svetashev VI, Nedashkovskaya OI, Gorshkova NM, Mikhailov VV, Yumoto N, Shigeri Y, Taguchi T, Yoshikawa S (1999) Characterization of Bacillus strains of marine origin. Int Microbiol 2:267–271
Jamal MT, Morris PC, Hansen R, Jamieson DJ, Burgess JG, Austin B (2006) Recovery and characterization of a 30.7- kDa protein from Bacillus licheniformis associated with inhibitory activity against methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococci and Listeria monocytogenes. Mar Biotechnol 8:587–592
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Kanagasabhapathy M, Sasaki H, Nagata S (2008) Phylogenetic identification of epibiotic bacteria possessing antimicrobial activities isolated from red algal species of Japan. World J Microbiol Biotechnol 24:2315–2321
Kasanah N, Hamann MT (2004) Development of antibiotics and the future of marine microorganisms to stem the tide of antibiotic resistance. Curr Opin Investig Drugs 5(8):827–837
Kizhakkekalam VK, Chakraborty K (2019) Pharmacological properties of marine macroalgae-associated heterotrophic bacteria. Arch Microbiol 201:505–518
Kobayashi J, Ishibashi M (1993) Bioactive metabolites of symbiotic marine microorganisms. Chem Rev 93(5):1753–1769
Kubanek J, Jensen PR, Keifer PA, Sullards MC, Collins DO, Fenical W (2003) Seaweed resistance to microbial attack: a targeted chemical defense against marine fungi. Proc Natl Acad Sci USA 100(12):6916–6921
Kumar V, Rao D, Thomas T, Kjelleberg S, Egan S (2011) Antidiatom and antibacterial activity of epiphytic bacteria isolated from Ulva lactuca in tropical waters. World J Microbiol Biotechnol 27:1543–1549
Le Lann K, Surget G, Couteau C, Coiffard L, Cerantola S, Gaillard F, Larnicol M, Zubia M et al (2016) Sunscreen, antioxidant, and bactericide capacities of phlorotannins from the brown macroalga Halidrys siliquosa. J Appl Phycol 28:3547–3559
Li Z (2009) Advances in marine microbial symbionts in the China Sea and related pharmaceutical metabolites. Mar Drugs 7(2):113–129
Longford S, Tujula N, Crocetti GR, Holmes AJ, Holmstrom C, Kjelleberg S, Steinberg P, Taylor MW (2007) Comparisons of diversity of bacterial communities associated with three marine eukaryotes. Aquat Microb Ecol 48:217–229
Martins A, Vieira H, Gaspar H, Santos S (2014) Marketed marine natural products in the pharmaceutical and cosmeceutical industries: tips for success. Mar Drugs 12:1066–1101
Penesyan A, Marshall-Jones Z, Holmstrom C, Kjelleberg S, Egan S (2009) Antimicrobial activity observed among cultured marine epiphytic bacteria reflects their potential as a source of new drugs. FEMS Microbiol Ecol 69(1):113–124
Petersen PJ, Jone CH, Bradford PA (2007) In vitro antibacterial activities of tigecycline and comparative agents by time-kill kinetic studies in fresh mueller-hinton broth. Diagn Microbiol Infect Dis 59(3):347–349
Pfaller MA, Sheehan DJ, Rex JH (2004) Determination of fungicidal activities against yeasts and molds: lessons learned from bactericidal testing and the need for standardization. Clin Microbiol Rev 17:268–280
Postgate JR (1989) A microbial way of death. New Sci 122:43–47
Sachidanandham R, Gin KY, Poh CL (2005) Monitoring of active but non-culturable bacterial cells by flow cytometry. Biotechnol Bioeng 89:24–31
Schirmer A, Gadkari R, Reeves CD, Ibrahim F, DeLong EF, Hutchinson CR (2005) Metgenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl Environ Microbiol 71(8):4840–4849
Seng P, Drancourt M, Gouriet F, La-Scola B, Fournier PE, Rolain JM, Raoult D (2009) Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49:543–551
Simões M, Pereira MO, Vieira MJ (2005) Validation of respirometry as a short-term method to assess the efficacy of biocides. Biofouling 21(1):9–17
Singh MP (2006) Rapid test for distinguishing membrane-active antibacterial agents. J Microbiol Method 67:125–130
Stocks SM (2004) Mechanism and use of the commercially available viability stain, BacLight. Cytometry A 61:189–195
Strobel G, Daisy B (2003) Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev 67(4):491–502
Suleimana MM, McGaw LJ, Naidoo V, Eloff JN (2009) Detection of antimicrobial compounds by bioautography of different extracts of leaves of selected South African tree species. Afr J Tradit Complement Altern Med 7(1):64–78
Tam VH, Schilling AN, Nikolaou M (2005) Modelling time-kill studies to discern the pharmacodynamics of meropenem. J Antimicrob Chemother 55:699–706
Vairappan CS, Suzuki M (2000) Dynamics of total surface bacteria and bacterial species counts during desiccation in the Malaysian sea lettuce, Ulva reticulata (Ulvales, Chlorophyta). Phycol Res 48(2):55–61
Vidaillac C, Para-Ruiz J, Rybak MJ (2011) In vitro time kill of oritavancin against clinical isolates of methicillin-resistant Staphylococcus aureus with reduced susceptibility to daptomycin. Diagn Microbiol Infect Dis 71:470–473
Wagman G, Weinstein M (1973) Chromatography of antibiotics. Elsevier, Amsterdam, p 7
Walsh CT (2004) Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science 303(5665):1805–1810
Wang H, Fewer DP, Holm L, Rouhiainen L, Sivonen K (2014) Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci USA 111(25):9259–9264
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173(2):697–703
Wiese J, Thiel V, Nagel K, Staufenberger T, Imhoff JF (2009) Diversity of antibiotic-active bacteria associated with the brown alga Laminaria saccharina from the Baltic Sea. Mar Biotechnol 11:287–300
Xiong Y, Yang R, Sun X, Yang H, Chen H (2018) Effect of the epiphytic bacterium Bacillus sp. WPySW2 on the metabolism of Pyropia haitanensis. J Appl Phycol 30(2):1225–1237
Yazawa K (1996) Production of eicosapentaenoic acid from marine bacteria. Lipids 31:S297–S300
Zhao K, Penttinen P, Guan T, Xiao J, Chen Q, Xu J, Lindström K, Zhang L, Zhang X, Strobel GA (2010) The diversity and antimicrobial activity of endophytic actinomycetes from medicinal plants in Panxi Plateau, China. Curr Microbiol 62(1):182–190
Zheng L, Han X, Chen HM, Lin W, Yan XJ (2005) Marine bacteria associated with marine macroorganism: the potential antimicrobial resources. Ann Microbiol 55(2):119–124
Zhu P, Zheng Y, You Y, Yan X, Shao J (2009) Molecular phylogeny and modular structure of hybrid NRPS/PKS gene fragment of Pseudoalteromonas sp.NJ6-3-2 isolated from marine sponge Hymeniacidon perleve. J Microbiol Biotechnol 19(3):229–237
Zubia M, Fabre MS, Kerjean V, Lann KL, Pouvreau VS, Fauchon M, Deslandes E (2009) Antioxidant and antitumoural activities of some Phaeophyta from Brittany coasts. Food Chem 116:693–701
Acknowledgements
This work was supported by funding under the Kerala State Council for Science, Technology and Environment (Grant No. 040/FSHP-LSS/2014/KSCSTE). The authors are thankful to Indian Council of Agricultural Research (ICAR), New Delhi for providing facilities to carry out the work. The authors thank the Director, Central Marine Fisheries Research Institute and Dean, Faculty of Marine Sciences, Lakeside Campus, Cochin University of Science and Technology for the support. Thanks are due to the Head, Marine Biotechnology Division, Central Marine Fisheries Research Institute and Scientist-in-Charge, Central Marine Fisheries Research Institute, Mandapam for facilitating the research activity.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Shuang-Jiang Liu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kizhakkekalam, V.K., Chakraborty, K. Marine macroalgae-associated heterotrophic Firmicutes and Gamma-proteobacteria: prospective anti-infective agents against multidrug resistant pathogens. Arch Microbiol 202, 905–920 (2020). https://doi.org/10.1007/s00203-019-01800-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-019-01800-2