Abstract
Appearance of drug-resistant microorganisms prompted researchers to unravel new environments for development of novel antimicrobial agents. Culture-supported analysis of heterotrophic bacteria associated with seaweeds yielded 152 strains, in that larger share of the isolates was embodied by Bacillus atrophaeus SHB2097 (54%), B. velezensis SHB2098 (24%), B. subtilis SHB2099 (12%), and B. amyloliquefaciens SHB20910 (10%). One of the most active strains characterized as B. atrophaeus SHB2097 (MW821482) with an inhibition zone more than 30 mm on spot-over-lawn experiment, was isolated from a seaweed Sargassum wightii, was selected for bioprospecting studies. Significant antibacterial potential was displayed by bacterial organic extract against vancomycin-resistant Enterococcus faecalis, Pseudomonas aeruginosa, methicillin-resistant Staphylococcus aureus, and Klebsiella pneumonia with minimum inhibitory concentration 6.25 µg/mL and comparable to the antibiotics ampicillin and chloramphenicol. The genes of type 1 pks (MZ222383, 700 bp) and hybrid nrps/pks (MZ222389, 1000–1400 bp) of B. atrophaeus MW821482 could be amplified. The bacterium displayed susceptibility to the commercially available antibiotic agents, and was negative for the pore-forming non-hemolytic hemolysin BL (hbl) and enterotoxin (nhe) genes, and therefore, was not pathogenic. The bacterium was found to possess genes (1000–1400 bp) involved in the biosynthesis of siderophore-class of compounds (MZ222387 and MZ222388) that showed 99% of similarity in BLAST search, and showed production of siderophore. Noteworthy antibacterial activities against clinically important pathogenic bacteria in conjunction with occurrence of genes coding for antimicrobial metabolites inferred that the marine heterotrophic bacterium B. atrophaeus SHB2097 could be used for the development of antibacterial agents against the emerging antibiotic resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacterial communities associated with seaweeds are valuable biotechnological resources as they yield a wide-ranging diversity of pharmaceutically active compounds (Zubia et al. 2009; Le Lann et al. 2016). The increase in antibiotic-resistant microorganisms and the necessity for new antimicrobials prompted researchers to look into novel habitations for bioactive substance production. The capacity of marine heterotrophic bacteria to provide biochemical protection to host eukaryotes in the face of predators and pathogenic organisms has been recognized (Kanagasabhapathy et al. 2006; Penesyan et al. 2009; Ali et al. 2012). While chemical defences have a propensity for displaying a significant part to establish the relationship between the marine eukaryotic host and their associated microorganisms, antimicrobial properties of marine organisms have received little attention. Biotechnological and pharmacological sectors highlighted the ability of marine heterotrophic isolates as promising antimicrobial agents (Soria-Mercado et al. 2012; Kizhakkekalam and Chakraborty 2020).
Marine Bacillus were described to produce various bioactive metabolites (Fickers 2012), such as polyketides, non-ribosomal peptides, macrolactones, among others (Cherian et al. 2019). A number of previous works envisaged to demonstrate antibiotic and antifouling activities of seaweed-related bacterial species (Penesyan et al. 2009; Wiese et al. 2009; Chakraborty et al. 2017). Bacillus subtilis, with 252 products so far, is among the leading producers of bioactive metabolites and products. The bioactives are mostly antibacterial in nature, such as bacitracin and rhizocticin, accounting for 61% of the amino acids and peptides (Chassagne et al. 2019). Bacillus velezensis was previously recognized as one of the biocontrol agents against phytopathogen (Kim et al. 2017; Chen et al. 2018; Francis and Chakraborty 2021; Chakraborty et al. 2021), even though it has recently developed into an antagonist against pathogens (Chen et al. 2018; Yi et al. 2018; Li et al. 2020). Nonribosomal peptide synthetases (NRPSs) and polyketide synthetases (PKSs), which are ubiquitous in Proteobacteria, Firmicutes and Actinobacteria, were found to catalyze the biosyntheses of bioactive natural products encompassing distinct structural architecture, and have been widely used for the development of biotechnological tools to evaluate bioactive bacteria (Kubanek et al. 2003; Wang et al. 2014). Previous reports of literature demonstrated the presence of biosynthetic genes, namely non-ribosomal peptide synthetase (nrps) and polyketide synthetase-I (pks-I) in heterotrophic bacteria (Chakraborty et al. 2014; Thilakan et al. 2016).
Herein, we have isolated the culturable heterotrophic bacteria related with seaweeds available at the Gulf-of-Mannar of peninsular India isolated by culture-reliant method, and assessed those for antimicrobial properties. Further, the selected bioactive isolate Bacillus atrophaeus SHB2097 (GenBank accession number MW821482) were characterized by morphological, biochemical and molecular techniques. Characteristic nrps and pks-I genes were analyzed by PCR amplification with degenerate primers, exploiting their conserved nature other than profiling its antimicrobial and hemolytic activities. Pathogenicity was assessed by in vitro hemolytic assay and presence of pore-forming non-hemolytic enterotoxin and hemolysin genes. B. atrophaeus SHB2097 was found to possess potential antimicrobial activities, and thus, its organic extract was assessed for antibacterial properties against selected pathogens coupled with its ability of siderophore production.
Materials and methods
Isolation of seaweed-associated heterotrophs
Seaweeds (class Phaeophyceae, Rhodophyceae, and Chlorophyceae) were accumulated by hand picking from the Gulf-of-Mannar region in Mandapam (9°17′0″ North, 79°7′0″ East), at low tide, and stored in seawater under sterile condition, as described earlier (Lemos et al. 1985). The brown seaweeds collected were Sargassum wightii, Turbinaria conoides, Turbinaria ornata, and Turbinaria decurrens, red seaweeds were Gracilaria salicornia and Gracilaria edulis, whereas Ulva lactuca was the green seaweed. Heterotrophic bacteria associated with the seaweeds were isolated by a formerly described method (Thilakan et al. 2016). Concisely, thallus parts of the seaweed samples (~ 1 g) were cleansed in sterile distilled water to eliminate the dirt and surface-associated microorganisms before being aseptically homogenized under suspended condition in sterile seawater (~ 100 mL) in a laminar airflow hood. Serial dilutions were made with the suspension, wherein sterile distilled water (9 mL) was added to the microbial suspension (1 mL), and various dilutions were prepared and plated on nutrient agar (NA) and Zobell Marine Agar (ZMA) supplemented with sodium chloride (NaCl, 1% w/v). The plates were incubated in the dark at 37 ± 1 °C for 1 week, and pure colonies were acquired by succeeding purification on NA added with common salt (1% w/v) (Chakraborty et al. 2014). While over 113 different colony phenotypes were observed, we have succeeded to isolate 75 strains as pure cultures. Colony morphology and pigments of these isolates in the incubated plates were recorded, and a total of 31 well-grown and morphologically distinct bacterial cultures were selected and sub-cultured into the sterile NA plates. The bacterial cultures were further quadrant-streaked to obtain the pure cultures, and single colonies were selected and streaked on NA slants. The isolates were incubated at 34 °C, and further stored at 20 °C in the BOD incubator (Labline, India). The pure cultures were inoculated into NA (with 1% NaCl) by stab method, and glycerol was added onto it. These glycerol stab stocks were stored in − 20 °C in the BOD incubator for long-term storage of the cultures (Ismail et al. 2018; Singh et al. 2011).
Preliminary antibacterial screening
The test pathogenic organisms used for preliminary screening of the heterotrophic bacteria were Aeromonas salmonicida (ATCC 27013), Aeromonas caviae (ATCC 15468), vancomycin-resistant Enterococcus faecalis (ATCC 51299), Aeromonas salmonicida (ATCC 27013), Aeromonas hydrophila (ATCC 7966), methicillin-resistant Staphylococcus aureus (ATCC 33592), and Vibrio parahaemolyticus (ATCC 17802), which were procured from the American Type Culture Collection (Manassas, VA). Vibrio parahaemolyticus (MTCC 451), Escherichia coli (MTCC 443), Yersinia enterocolitica (MTCC 859), Streptococcus pyogenes (MTCC 1924), and Edwardsiella tarda (MTCC 2400) were obtained from the Microbial Type Culture Collection and Gene Bank (Institute of Microbial Technology, Chandigarh of India), whereas Vibrio harveyi (LMG 4044) type strain was procured from Sigma-Aldrich (St. Louis, MO). Upon receipt of the cultures, pathogenicity was judged by using PCR amplification with their explicit virulent features, whereas their characteristics were established by microbiological/biochemical and genotypic experiments, such as 16S rRNA sequence likeness (Armstrong et al. 2001). The pathogenic bacterial turf was grown on the plates of Mueller Hinton agar (MHA) (HiMedia, PA) and NA, over which the isolates were spotted using sterile swab. The plates were incubated at 37 °C, and the antibacterial activities were recorded by assessing the inhibition zone diameter using an antibiotic zone scale on the plates after 24, 48, and 72 h of incubation, by the spot-over-lawn antibacterial assay (Chakraborty et al. 2014; Kizhakkekalam and Chakraborty 2019).
Biochemical and molecular characterization of antibacterial isolates
The bacterial isolates with noteworthy antibacterial potential were characterized by microbiological and biochemical experiments (Krieg and Holt 1984). Morphological evaluation of the bacterial clusters was assessed on the agar plates, and standard checks, such as Gram staining, motility, reduction of nitrate/indole, Voges–Proskauer, citrate utilization, hydrolysis of gelatin/starch, etc. were performed. HiMedia Bacillus Identification kit including tests for malonate, ortho-nitrophenyl-β-galactoside, Voges–Proskauer, nitrate reduction, citrate, catalase, arginine, glucose, sucrose, trehalose, arabinose, and mannitol were used (supplementary material S1). The characteristics of the strains were substantiated by 16S rRNA gene system-centered phylogenetic examination supported by BLAST-likeness examination, and the biotyping was substantiated by matrix-assisted laser desorption/ionization-time of flight mass spectral analysis (MALDI/ToF by using a MALDI-TOF MS Biotyper, Microflex, Bruker Daltonics, Germany; version 3.1) and biolog characterization (GEN III Microplate and turbidimeter, Biolog) (Kanagasabhapathy et al. 2008) (supplementary material S1).
Genomic DNA of the bacteria was isolated using GenElute™bacterial genomic DNA isolation kit (Sigma-Aldrich, MO), whereas the quality of DNA was judged using a nanodrop biospectrophotometer (Eppendorf, Hamburg, Germany) by quantifying the absorbance at 260/280 nm, and the concentration of the sample (ng/µL) were observed. The 16S rRNA gene sequencing supported by BLAST resemblance search was used for molecular characterization. PCR reaction primer sequences were listed in Table 1. PCR was performed in the reaction mixture of PCR/(2X) master mix (12.5 µL), forward/reverse primer (1 µL both), DNA (1 ng), and the volume was adjusted to 0.025 mL using sterile water. PCR reactions were carried out in the thermo cycler (Biorad, CA) with the programming condition as follows: initial denaturation (94 °C for 5 min) subsequently 35 cycles (95 °C and 58 °C for 1 min each and 72 °C for 2 min), and an extension at 72 °C for 5 min. The 1 kb ladder on a 1.5% (w/v) agarose gel (in 1X TBE buffer) was used to assess molecular sizes of the amplified fragments (Thilakan et al. 2016), whereas the amplicons were sequenced for molecular identification before submitting in the GenBank. The sequenced data were analyzed against standard ones by BLAST program before being phylogenetically analyzed by MEGA X (Bioedit) (Kumar et al. 2018). Evolutionary history was resolved by neighbor-joining process centered on Kimura model and bootstrap study. Partial 16S rRNA gene sequences of the bacterial isolates were deposited in the GenBank (NCBI accession of MW821482, MW821799).
Molecular identification of functional genes
Identification of functional genes for pks-I, nrps, and siderophore in the most active strain was executed by PCR amplification of the specific genes using degenerate primers (Table 1). PCR was carried out in the reaction mixture comprehended in the earlier section as follows: denaturation at 94 °C for 5 min, subsequently 35 cycles at 95 °C for 1 min, annealing at 45 °C for 1 min for pks, and 55 °C for 1 min for nrps, subsequently extension at 72 °C for 5 min. PCR conditions for siderophore gene were fixed as primary denaturation 94 °C for 5 min, subsequently 30 cycles of 30 s/94 °C, 30 s/56 °C, 60 s/72 °C, and an extension at 72 °C for 5 min. The amplified PCR products were subjected to agarose gel electrophoretic (1.5%) analysis. The products (expected size of 700 bp for pks-I and 1000 bp for nrps/siderophore) were purified by an extraction kit (Gel Elute™, Sigma). The pks-I (MZ222383, 700 bp)/hybrid nrps/pks (MZ222389, 1000–1400 bp)/siderophore sequenced products (MZ222387 and MZ222388 1000–1400 bp) were deposited under the accession number MT394492 (in NCBI GenBank). The partial pks-I, nrps and siderophore sequences for B. atrophaeus SHB2097 were submitted to the NCBI GenBank. Amino acid sequence alignments with GenBank reference sequences were performed for phylogenetic study (Zhu et al. 2009) and evolutionary history was deduced (Whelan and Goldman 2001; Tamura et al. 2011; Jones et al. 1992).
Antibiotic susceptibility, siderophore production, and toxicity assessment
Antibiotic sensitivity of B. atrophaeus SHB2097 was found out using commercially available antibiotic-permeated octadiscs (HiMedia Laboratories LLC, PA) (Kizhakkekalam et al. 2020, CLSI, 2009). Chrome azurol sulfonate (CAS) analysis was used to deduce the presence of siderophore (Louden et al. 2011) (supplementary material S2). Siderophore biosynthetic genes were characterized in the bacterial genome of B. atrophaeus SHB2097, and the sequences were deposited in the NCBI GenBank (MZ222387 and MZ222388). Toxicity of B. atrophaeus SHB2097 was assessed in vitro hemolytic assay (Gao et al. 2000) by streaking pure culture of the isolated bacterium on sheep blood agar plates (Kizhakkekalam and Chakraborty 2019), and by inspecting the presence of enterotoxin genes in the candidate bacterium (supplementary material S2) with reference to a Streptococcus pyogenes MTCC1924 as a control. Presence of pore-forming non-hemolytic enterotoxin (nhe A–C), and hemolysin BL (hbl A–D) genes was ascertained using PCR. The primers used to amplify the enterotoxin genes are shown in Table 2.
Sporulation efficiency
The culture of B. atrophaeus SHB2097 was prompted to sporulate by the nutrient depletion technique (Nicholson and Setlow 1990), wherein bacterial suspension (300 µL) was filled into a multi-well plate, and the absorbance was evaluated at 600 nm for every half an hour for a period of 48 h using a microplate reader (Multiskan-GO model 1510, Thermo Scientific, MA). The time was noted at which the bacterial culture reached their death phase in the exhaustion medium, as the commencement of sporulation. Following 48 h of incubation, the cultures were acquired by centrifugation (9500×g, 15 min), and the lysozyme treatment followed by salt and detergent washes were used for the purification of spores. Further, sterile deionized water was used to suspend the bacterial spores before being proceeded with the final purification step, wherein the spore suspension was subjected to heating at 80 °C for 10 min, to confirm the removal of vegetative cells. The spore suspension was diluted by tenfold with maximum recovery diluent (Merck, Darmstadt, Germany), and plated on the brain heart infusion agar by spread plating method before incubating at 37 °C for two days so as to determine the total viable counts. Sporulation efficacy of the studied heterotrophic Bacillus was evaluated as percentage survival after heating.
Development of bacterial crude extracts and bioactivity assessment
B. atrophaeus SHB2097 possessing potential antimicrobial properties against pathogens, was extracted with ethyl acetate to produce extracellular bacterial extract. Briefly, B. atrophaeus was cultured over NA with 1–2% NaCl (at pH 8, 30 °C for 3 days), and the incubation period was ascertained from the bacterial growth kinetics to produce secondary metabolites. The symbiotic bacteria discharge the bioactive compounds in the agar growth medium, the agar containing extracellular metabolites was extracted with solvent ethyl acetate (~ 2500 mL) under reflux at a temperature below 80 °C on a water bath. The extract (~ 5.4 L) was subjected to dehydration by using anhydrous sodium sulfate (0.5 kg) before being concentrated by a rotary vacuum evaporator (Heidolph, Schwabach, Germany) at 40 °C (Kizhakkekalam and Chakraborty 2019). The bacterial organic extract (~ 5 g) was analyzed for in vitro bioactive potential. Antibacterial properties of the organic extract against clinically acknowledged pathogens were assessed by disc diffusion on Mueller Hinton agar plates and minimum inhibitory concentration (MIC) by microdilution method (Kizhakkekalam and Chakraborty 2019; Bauer et al 1966; Chakraborty et al. 2021). Iron chelation properties of the solvent extract of B. atrophaeus SHB2097 were assessed using ferrous ion (Fe2+) chelating method (Gülçin 2007). The concentration of bacterial extract required to realize 50% Fe2+ chelating capacity was deliberated as IC50 (effective concentration) (supplementary material S3).
Data analysis and accession numbers
Statistical Program for Social Sciences (SPSS Inc, CA; ver. 10.0) was used for analyzing the data. Experiments were carried out in triplicate, and means of the parameters were computed for significance (p ≤ 0.05) by analysis of variance. Partial 16S rRNA gene sequences were deposited in the GenBank (accession numbers of MW821482–821799). Partial pks sequences for B. atrophaeus MW821482 were deposited under the accession numbers MZ222383, MZ222384, MZ222385, MZ222386, whereas nrps gene sequence was deposited in the GenBank as MZ222389. Coding sequences for siderophore biosynthetic genes were deposited under the accession numbers MZ222387 and MZ222388.
Results
Isolation of seaweed-associated bacteria and antimicrobial assessment
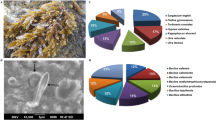
Seven different intertidal macroalgae (otherwise termed as seaweeds) representing Sargassum wightii, Turbinaria conoides, Gracilaria salicornia, Ulva lactuca, Turbinaria ornata, Turbinaria decurrans, and Gracilaria edulis belonging to the families of Phaeophyceae, Chlorophyceae, and Rhodophyceae were harvested from the intertidal zones (Fig. 1A). One hundred and fifty-two heterotrophic bacterial isolates were purified to homogeneity by culture-dependent studies, whereas a total of 21% of those was isolated from S. wightii and T. conoides (Fig. 1B). The heterotrophic bacteria belonged to B. atrophaeus SHB2097 (54%), B. velezensis SHB2098 (24%), B. subtilis SHB2099 (12%), and B. amyloliquefaciens SHB20910 (10%) that were assessed for antibacterial properties against a wide range of drug-resistant pathogens. From the isolates, three strains of B. atrophaeus and one each of B. velezensis isolated from S. wightii and T. conoides were purified to homogeneity, showed potential antibacterial properties {inhibition zone diameter of 32–40 mm, against MRSA (Fig. 2A) and V. parahaemolyticus (Fig. 2B)}. Among various macroalgal species considered in the present study, two bioactive isolates B. atrophaeus (MW821482) and B. velezensis (MW821799), which were capable of enduring sub-culturing for laboratory culture conditions, showed general antagonistic activities against the studied pathogens (Fig. 2B). Pure bacterial colonies, which were susceptible to antibiotics (Fig. S1), were preserved at −80 °C under sterile glycerol stab.
A Antibacterial activity displayed by B. atrophaeus SHB2097 (GenBank accession number MW821482) and B B. velezensis SHB2098 (GenBank number: MW821799) against V. parahaemolyticus and MRSA (diameter of zone of growth inhibition, 40 mm and 32 mm, respectively) as visualized on MHA plates by spot-on-lawn assay. C Phylogenetic tree from partial 16S rRNA sequences between the studied isolates and related species. The isolates were categorized as B. atrophaeus (SHB2097) and B. velezensis (SHB2098)
Characterization of potential bioactive strains and 16S rRNA-based phylogeny
Morphological, microbiological and biochemical attributes of the studied bacteria are listed in Table S1–S2, which enabled the classification under the genus of Bacillus, whereas 16S rRNA gene sequencing led to the characterization at species level as B. atrophaeus SHB2097 and B. velezensis SHB2098 (Fig. 2C). The bacteria were classified as Gram positive by potassium hydroxide/Gram staining experiments. White lobed margins with casein hydrolyzing and nitrate reduction properties characterized B. atrophaeus SHB2097 (Table S1–S2). B. velezensis SHB2098 was found to develop a dark brown pigmentation at the later stage of exponential growth phase (Turick et al. 2008). B. velezensis SHB2098 grown at its optimum with 1–4% NaCl at a temperature and pH of about of 30 °C and 8.0, respectively, whereas B. atrophaeus SHB2097 was found to grow at its optimum at the pH range of 7–9, when incubated at 37 °C. Features of biochemical reactions for biolog characteristics of B. atrophaeus SHB2097 and B. velezensis SHB2098 were also detailed (Table S3-S4). MALDI–TOF biotyping certainty score of 2.152 and 1.953 for high abundance ribosomal proteins (Seng et al. 2009) of B. velezensis SHB2098 and B. atrophaeus SHB2097, respectively, further corroborated the characterization (Fig. S2-S3). The above strains were subjected to 16S rRNA genotyping (GenBank accession numbers of MW821482 for B. atrophaeus SHB2097 and MW821799 for B. velezensis SHB2098) (Fig. S4), and the sequences were harmonized with the contiguous lineages recorded in the GenBank to build a phylogenetic tree. Evolutionary lineage was construed by maximum likelihood mode (Tamura et al. 2011), and the tree with maximum logarithm of probability was presented.
Analyses of pks, nrps and phylogenetic structure
Type-1 pks and nrps genes were amplified in B. atrophaeus SHB2097 and B. velezensis SHB2098 (Fig. S5), and the sequence showing substantial BLAST-likeness was deposited in the GenBank (accession number MZ222383) (Fig. 3). The PKS-specific primers could successfully obtain the PCR amplicons (~ 700 bp) displaying substantial analogy to the sequences submitted in the GenBank. The positive amplicons exhibited 99% likeness in sequence with those in hybrid nrps/pks gene of B. velezensis SHB2098 and type 1 pks gene of B. atrophaeus SHB2097, as observed in the blast analysis. Phylogeny of the KS domains in B. atrophaeus SHB2097 categorized the inferred sequences of amino acid as bacterial PKSs type I and for B. velezensis SHB2098 as NRPS/PKS hybrid (Fig. 3).
A Polyketide synthase and B non-ribosomal peptide synthetase gene (nrps) products (~ 700 bp) of the antimicrobial bacterium B. atrophaeus SHB2097 associated with S. wightii. (S1) type I polyketide synthase gene (pks-I) product (~ 700 bp) of B. velezensis SHB2098 associated with T. conoides (T1). The amplified gene sequences for type I pks and nrps were submitted in the GeneBank with accession numbers of MZ222383, MZ222384, MZ222385 and MZ222386 for pks and MZ222389 for nrps, respectively. C, D Molecular phylogeny of ketosynthase domains as pks types I–III, nrps and hybrid of genes
Siderophore production and ferrous ion chelation by B. atrophaeus SHB2097
A yellow hallow of about 32 mm diameter on CAS agar experiment recognized the production of siderophore by B. atrophaeus SHB2097 (Fig. 4B). Presence of siderophore biosynthetic genes was predicted, wherein one of the genes (1626 bp, GenBank accession number of MZ222387) could decode to (2,3-dihydroxybenzoyl) adenylate synthetase, while other gene (786 bp, MZ222388) was turned into dihydro-dihydroxybenzoate-NAD+ oxidoreductase. Evolutionary pedigree was deduced, and the tree with maximum logarithm of probability was illustrated (Fig. 4D). Extracellular extract of B. atrophaeus SHB2097 showed potential ferrous ion chelation capacity (IC50 4.2 mg/mL).
A–B Siderophore genes PF, PR and SF, SR amplified for B. atrophaeus SHB2097 (S1) were shown. C Indicative photograph showing the production of siderophore by B. atrophaeus SHB2097 on CAS agar plate with zone of clearance of 32 mm. D Molecular phylogeny analysis of the siderophore genes PF, PR and SF, SR by B. atrophaeus SHB2097 were submitted in the GenBank with the respective accession numbers as MZ222387, MZ222388
Toxicity assessment and sporulation efficiency
Presence of enterotoxin genes nhe (A–C) and hbl (A–D) were ascertained using PCR. Amplification for nhe (A–C) and hbl (A–D) were not observed proving that the bacterium B. atrophaeus SHB2097 did not have the hemolysis capacity. The pathogenic strains were amplified due to the presence of nhe and hbl genes, and therefore, displayed hemolysis activity. List of genes used for the amplification is summarized in Table 2. It was recognized that B. atrophaeus SHB2097 possessed spore-forming ability (Fig. 5E).
Antimicrobial properties of the bacterial extracts of B. atrophaeus SHB2097 against A methicillin-resistant S. aureus, B S. pyogenes, and C V. parahaemolyticus. D Indicative photograph representing the antibiotic susceptibility test (with octadisc) result of bacteria B. atrophaeus. The octadisc comprised of eight antibiotics, ofloxacin, penicillin-G, clindamycin, cephalothin, trimoxazole, vancomycin, gentamicin, and erythromycin positioned on the center of the MHA plate with the culture of candidate bacteria (after the guidelines of Clinical and Laboratory Standard Institute guidelines, CLSI, 2009). E, F Graphs representing bacterial growth kinetics and sporulation efficiency of the selected isolate B. atrophaeus SHB2097 at 660 nm
Antibacterial potential of B. atrophaeus SHB2097 organic extract
Growth kinetics of B. atrophaeus SHB2097 for expression of bioactivity showed that antibacterial activity peaked (38 mm zone of inhibition against MRSA) following 56 h of incubation (Table 3, Fig. 5). Consequently, extracellular metabolites were recovered by incubating the cells for 60 h before being extracted with ethyl acetate to acquire the organic extract of B. atrophaeus (~ 5 g). Antimicrobial potential of the organic extract displayed 38 and 28 mm of inhibition zone against MRSA ATCC 33952 and S. pyogens MTCC 1924, respectively, and 32 mm against V. parahaemolyticus (Fig. 5A–C) in comparison to chloramphenicol (15 mm inhibition zone against V. parahaemolyticus). It was apparent that the MIC of B. atrophaeus SHB2097 organic extract was 6.25 μg/mL against majorities of these pathogens (Table 4). MBC was noted with 6.25 µg/mL for B. atrophaeus SHB2097 against MRSA.
Discussion
Occurrence of microbial resistance to the prevalent antimicrobial agents and antibiotics led to the search for novel antimicrobial agents. Constant pursuit for newer pharmacophore leads could pave the system to discover high-value compounds and drugs from microbes (Pandey 2019). Seaweeds or marine macroalgae were found to have chemical defense mechanisms against pathogens, and the substantial share of seaweed-associated bioactive heterotrophs shape a treasured source of pharmaceutical leads (Zheng et al. 2005; Vijayalakshmi et al. 2008). Lately, marine macroalgae-associated heterotrophs have attracted the marine microbiologists and biochemists in view of their potentials to biosynthesize bioactive compounds of various classes with pharmacological implications (Winter et al. 2013).
Bacteria associated with seaweeds were obtained from the Gulf-of-Mannar, as the intertidal heterotrophs were described to biosynthesize distinctive specialized metabolites in reaction to the stress conditions. Comprehensive findings of surface-associated bacteria isolated from the seaweeds were firstly described before the nineteenth century (Penesyan et al. 2009). It was found that the percentage of bioactive bacteria with potential antibacterial properties, associated with seaweeds and other marine organisms was more (11%) compared to those living in sediments and seawater (Zheng et al. 2005; Gram et al. 2010). Between the total of 152 bacterial isolated strains in the present study, Gammaproteobacteria and Firmicutes presented the largest communities. The phylum Proteobacteria was found to be the present in large quantities as hetrotrophic association with seaweeds, followed by Actinobacteria, Bacteroidetes, Firmicutes and Chlorofexi (Armstrong et al. 2001; Longford et al. 2007). Among the different seaweed species considered in this study, Sargassum wightii and Turbinaria conoides contributed bacterial isolates with potential bioactivities against the test pathogens. The share of bioactive isolates from seaweed hosts has been depicted in Fig. 2. Remarkably, the genera belonging to Firmicutes were plentiful, and majority of which were B. atrophaeus SHB2097 followed by B. velezensis SHB2098 (54 and 24% of the active isolates, respectively). Bacillus subtilis SHB2099 and B. amyloliquefaciens SHB20910 contributed the shares of 12 and 10% of the collective number of bioactive isolates associated with the seaweeds (Fig. 1).
Previous literature reports described the potential antimicrobial properties of seaweed-associated heterotrophic bacteria against drug-resistant pathogens, such as methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecalis (VREfs) (Thilakan et al. 2016; Kizhakkekalam et al. 2020). With the aim of assessing the bioactive capability of the seaweed-associated isolates, they were evaluated for antibacterial properties by spot-over-lawn assay, wherein 57% of isolates were selected initially, and those isolates showed positive result towards the preliminary screening. The bacteria isolated from brown seaweeds, S. wightii and T. conoides showed greater activities among all the isolates. The bacterial isolates displaying broad-continuum of antimicrobial activities against nosocomial pathogens were selected for advanced studies (Fig. 2; Table 1). B. atrophaeus SHB2097 and B. velezensis SHB2098 (family Firmicutes), isolated from the brown seaweeds S. wightii and T. conoides, respectively, showed significant antagonistic potential against MRSA, VREfs, A. salmonicida and V. parahaemolyticus with zones of inhibition of 26–40 mm. Conspicuously, Bacillus sp. were ubiquitous on the surface of marine macroalgae, and were known to produce bioactive metabolites of biotechnological and pharmaceutical importance (Goecke et al. 2010).
Marine-derived Bacillus sp. were known to biosynthesize potential antimicrobial substances, for example cyclic peptides, polyketides, bacteriocins, and polyketide–peptide hybrids (Goecke et al. 2010) by non-ribosomal peptide synthetase (nrps), polyketide synthase (pks), and nrps-pks hybrid (Brakhage 2012; Wang et al. 2014). The β-lactam antibiotics, lipopeptide antibiotic daptomycin, and cyclosporine were produced by nrps (Felnagle et al. 2008; Doekel et al. 2008). Herein, the pks gene amplified for B. atrophaeus SHB2097 (GenBank ID MZ222383, MZ222384, MZ222385, MZ222386) revealed likeness to type I pks (Fig. 3). These cluster of genes were accountable to produce bacterial polyketide and lipopeptide metabolites (Aleti et al. 2015). Recently elucidated whole genome mining study showed that more than 30% of the Bacillus species were assessed to accommodate in the pks/nrps gene metabolite groups, while nrps and hybrid nrps/pks or pks shared a proportion of 7:3 of these encoded genes (Wang et al. 2014). Previous reports in bacteria associated with sponges also deduced the presence of pks gene (Zhu et al. 2009; Zhang et al. 2009). Presently, KS domain sequence-built phylogeny bundled four sequences with B. atrophaeus SHB2097. Hence, the sequences of KS domain facilitated to distinguish B. atrophaeus SHB2097 from the B. velezensis SHB2098 strains that could not be realized by 16S rRNA-built phylogenetic method.
For analyzing the presence of antimicrobial compounds, spectroscopic features of the solvent extract of B. atrophaeus SHB2097 were evaluated by proton nuclear resonance spectroscopic (1H NMR) study. 1H NMR spectrum were apportioned to different characteristic sections, wherein a greater proton integral (> 150) at δ 0.5–2.0 in the organic extract was recorded. The number of protons (~ 50) at δ 2.1–2.5 were apportioned to acetyl or allylic groups, whereas weaker proton integrals (~ 23) at δ 2.6–3.5 displayed the appearance of alkoxyl functionality. Marked proton integrals at olefinic region (δ 4.6–6.0, 58.49) also depicted the presence of unsaturation in the compounds present in the organic extract (Fig. S6). Detailed purification and structure characterization of the organic extract will form a part of separate study.
Hemolysis activity of the isolate B. atrophaeus SHB2097 was analyzed with different set of hemolysis primers, wherein negative result recognized the isolate as non-pathogenic. Sporulation assessment indicated that the isolate B. atrophaeus SHB2097 has spore-forming capacity (1.29 × 108 cfu/mL).
Siderophores are bacterial iron-chelating metabolites synthesized by nrps or pks domains, to scavenge iron from surroundings (Kramer et al. 2020), thereby augmenting their antagonistic properties against competing pathogens. The current study revealed that the heterotrophic B. atrophaeus SHB2097 could produce siderophore on the CAS agar plates (Fig. 4) with a hallow of about 32 mm. Presence of siderophore biosynthetic gene in the genome assembly of B. atrophaeus SHB2097 further corroborated the CAS assay results (Fig. 4). Reactive oxygen species (ROS) are intermediates in metabolic reactions embodying the mitochondrial electron transport, in which reduced form of Fe2+ ion could promulgate the radical chain reaction by losing/acquiring the electrons. As a result, the attenuation to develop ROS could be carried out by Fe2+ chelation property of the organic extract of B. atrophaeus SHB2097 (IC50 4.2 µg/mL), thus contributing to its antioxidant property. Therefore, assembly of siderophores and their biosynthetic genes (accession numbers of MZ222387 and MZ222388) in tandem with Fe2+ chelation ability acknowledged its potential to scavenge Fe2+ ion from ambiences, and as a result could effectively antagonize the pathogens in consort with attenuating the buildup of ROS. Moreover, siderophore antibiotics could be transported into the Gram-negative bacterial cells at ease by using Fe2+ accretion pathways (Gram et al. 1999).
The bacterial growth curve of B. atrophaeus SHB2097 was found to be proportional to the inhibition curve, which recognized that the presence of greater number of cells could result in the production of more secondary metabolites. The crude organic (ethyl acetate) extract of B. atrophaeus SHB2097 displayed antagonism against a wide spectra of nosocomial pathogens, for example MRSA (skin infection), E. coli (gastrointestinal), S. pyogenes (skin), E. tarda (myonecrosis and gastrointestinal), V. parahemolyticus (gastrointestinal), and VREfs (sepsis and meningitis). Organic extract of B. atrophaeus SHB2097 revealed noteworthy antibacterial potential against the pathogens (zone of inhibition 28–38 mm), compared to that exhibited by chloramphenicol (8–15 mm) (Fig. 4B–D). Notably, MIC and MBC of the organic extracts were comparable or better than those displayed by chloramphenicol. The MIC of the organic extract was 6.25 µg/mL against V. parahemolyticus, MRSA, A. salmonicida (ATCC 27,013), Y. enterocolitica (MTCC 859), and A. hydrophila (ATCC 7966) (Table 4), wherein chloramphenicol displayed a comparable MIC (6.25 µg/mL). The crude extract fared better than the standard antibiotics (MIC 12.5 µg/mL) against vancomycin-resistant S. aureus, E. coli (MTCC 443), and A. hydrophila (ATCC 7966) showing the MIC of 6.25 µg/mL.
Conclusion
Need for newer sources of antibiotics due to the increasing antibiotic resistance has resulted into the expanded use of bioactives from novel natural origins. Seaweeds are prospective host organisms for the associated heterotrophic bacteria. In this study, heterotrophic B. atrophaeus SHB2097 was isolated from the marine macroalga exhibited potential iron-chelating and antimicrobial activities, which were reinforced by the presence of pks, nrps and siderophore biosynthetic genes, and therefore, could be studied for cutting-edge pharmacological applications. It could be inferred that antimicrobial properties could not be exclusively judged by metagenomic analyses, considering that a number of strains might get away with the amplification of the anticipated genes. It follows that the antibacterial properties could solely be weighed by evaluating the attenuation potential of the anticipated indicator organism. We also vouch for metabolite gene-centered screening of heterotrophs for using biosynthetic gene clusters. Future studies regarding isolation and characterization of antimicrobial compounds from B. atrophaeus SHB2097 would be attempted for development of potential antibiotic agents.
GenBank accession numbers
Partial 16S rRNA gene sequences of B. atrophaeus and B. velezensis were submitted in NCBI GenBank (accession of MW821482, MW821799). Partial pks sequences corresponding to B. atrophaeus MW821482 were deposited under the accession of MZ222383, MZ222384, MZ222385, MZ222386, whereas nrps gene sequence was deposited in the Genbank as MZ222389. Coding sequences for siderophore biosynthetic genes were deposited under the accession numbers MZ222387 and MZ222388.
References
Aleti G, Sessitsch A, Brader G (2015) Genome mining: prediction of lipopeptides and polyketides from Bacillus and related Firmicutes. Comput Struct Biotechnol J 13:192–203. https://doi.org/10.1016/j.csbj.2015.03.003
Ali AIB, Bour ME, Ktari L, Bolhuis H, Ahmed M, Boudabbous A, Stal LJ (2012) Jania rubens-associated bacteria molecular identification and antimicrobial activity. J Appl Phycol 24(3):525–534
Armstrong E, Yan L, Boyd KG, Wright CP, Burgess JG (2001) The symbiotic role of marine microbes on living surfaces. Hydrobiologia 461:37–40
Bauer AW, Kirby WM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45(4):493–496
Brakhage AA (2012) Regulation of fungal secondary metabolism. Nat Rev Microbiol 11(1):21–32
Chakraborty K, Thilakan B, Raola VK (2014) Polyketide family of novel antibacterial 7-O-methyl-5’-hydroxy-3’-heptenoate-macrolactin from seaweed-associated Bacillus subtilis MTCC 10403. J Agric Food Chem 62(50):12194–12208
Chakraborty K, Thilakan B, Kizhakkekalm VK (2017) Antibacterial aryl-crowned polyketide from Bacillus subtilis associated with seaweed Anthophycus longifolius. J Appl Microbiol 124(1):108–125
Chakraborty K, Francis A, Chakraborty RD, Asharaf S, Kizhakkekalam VK, Paulose SK (2021) Marine macroalga-associated heterotrophic Bacillus velezensis: a novel antimicrobial agent with siderophore mode of action against drug-resistant nosocomial pathogens. Arch Microbiol 203(9):5561–5575
Chassagne F, Cabanac G, Hubert G, David B, Marti G (2019) The landscape of natural product diversity and their pharmacological relevance from a focus on the Dictionary of Natural Products®. Phytochem Rev 18(3):601–622
Chen L, Heng J, Qin S, Bian K (2018) A comprehensive understanding of the biocontrol potential of Bacillus velezensis LM2303 against Fusarium head blight. PLoS ONE 13(6):e0198560
Cherian T, Yalla S, Mohanraju R (2019) Antimicrobial potential of methanolic extract of Bacillus aquimaris isolated from the marine waters of Burmanallah coast, South Andaman. Int J Biopharm Res 8(12):2806–2813
Datta B, Chakrabartty PK (2014) Siderophore biosynthesis genes of Rhizobium sp. isolated from Cicer arietinum L. 3 Biotech 4(4):391–401
Doekel S, Coëffet-Le Gal MF, Gu JQ, Chu M, Baltz RH, Brian P (2008) Non-ribosomal peptide synthetase module fusions to produce derivatives of daptomycin in Streptomyces roseosporus. Microbiology 154:2872–2880. https://doi.org/10.1099/mic.0.2008/020685-0
Felnagle EA, Jackson EE, Chan YA, Podevels AM, Berti AD, McMahon MD, Thomas MG (2008) Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol Pharm 5:191–211. https://doi.org/10.1021/mp700137g
Fickers P (2012) Antibiotic compounds from Bacillus: why are they so amazing? Am J Biochem Biotechnol 8(1):40–46
Francis A, Chakraborty K (2021) Marine macroalgae-associated heterotroph Bacillus velezensis as prospective therapeutic agent. Arch Microbiol 203(4):1671–1682
Gao Q, Tomlinson G, Das S, Cummings S, Sveen L, Fackenthal J, Schumm P, Olopade OI (2000) Prevalence of BRCA1 and BRCA2 mutations among clinic-based African American families with breast cancer. Hum Genet 107(2):186–191
Goecke F, Labes A, Wiese J, Imhoff JF (2010) Chemical interactions between marine macroalgae and bacteria. Mar Ecol Prog Ser 409:267–299
Gram L, Melchiorsen J, Spanggaard B, Huber I, Nielsen TF (1999) Inhibition of Vibrio anguillarum by Pseudomonas fluorescens AH2, a possible probiotic treatment of fish. Appl Environ Microbiol 65(3):969–973
Gram L, Melchiorsen J, Bruhn JB (2010) Antibacterial activity of marine culturable bacteria collected from a global sampling of ocean surface waters and surface swabs of marine organisms. Mar Biotechnol 12(4):439–451
Gülçin I (2007) Comparison of in vitro antioxidant and antiradical activities of L-tyrosine and L-dopa. Amino Acids 32(3):431–438
Hwang JY, Park JH (2015) Characteristics of enterotoxin distribution, hemolysis, lecithinase, and starch hydrolysis of Bacillus cereus isolated from infant formulas and ready-to-eat foods. J Dairy Sci 98:1652–1660. https://doi.org/10.3168/jds.2014-9042
Ismail AF, Ktari L, Ahmed M, Bolhuis H, Bouhaouala-Zahar B, Stal LJ, Boudabbous A, Bour ME (2018) Heterotrophic bacteria associated with the green alga Ulva rigida: identification and antimicrobial potential. J Appl Phycol 30(5):2883–2899
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8(3):275–282
Kanagasabhapathy M, Sasaki H, Haldar S, Yamasaki S, Nagata S (2006) Antibacterial activities of marine epibiotic bacteria isolated from brown algae of Japan. Ann Microbiol 56(2):167–173
Kanagasabhapathy M, Sasaki H, Nagata S (2008) Phylogenetic identification of epibiotic bacteria possessing antimicrobial activities isolated from red algal species of Japan. World J Microbiol Biotechnol 24(10):2315–2321
Kim SY, Lee SY, Weon HY, Sang MK, Song J (2017) Complete genome sequence of Bacillus velezensis M75, a biocontrol agent against fungal plant pathogens, isolated from cotton waste. J Biotechnol 241:112–115
Kizhakkekalam VK, Chakraborty K (2019) Pharmacological properties of marine macroalgae-associated heterotrophic bacteria. Arch Microbiol 201(4):505–518
Kizhakkekalam VK, Chakraborty K (2020) Marine macroalgae-associated heterotrophic Firmicutes and Gammaproteobacteria: prospective anti-infective agents against multidrug resistant pathogens. Arch Microbiol 202(4):905–920
Kizhakkekalam VK, Chakraborty K, Joy M (2020) Oxygenated elansolid-type of polyketide spanned macrolides from a marine heterotrophic Bacillus as prospective antimicrobial agents against multidrug-resistant pathogens. Int J Antimicrob Agents 55(3):105892
Kramer J, Özkaya Ö, Kümmerli R (2020) Bacterial siderophores in community and host interactions. Nat Rev Microbiol 18(3):152–163
Krieg NR, Holt JG (1984) Bergey’s manual of systematic bacteriology, 1st edn. Williams and Wilkins Co, Baltimore, pp 161–172
Kubanek J, Jensen PR, Keifer PA, Sullards MC, Collins DO, Fenical W (2003) Seaweed resistance to microbial attack: a targeted chemical defense against marine fungi. Proc Natl Acad Sci USA 100(12):6916–6921
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549
Le Lann K, Surget G, Couteau C, Coifard L, Cerantola S, Gaillard F, Larnicol M, Zubia M et al (2016) Sunscreen, antioxidant, and bactericide capacities of phlorotannins from the brown macroalga Halidrys siliquosa. J Appl Phycol 28(6):3547–3559
Lemos ML, Toranzo AE, Barja JL (1985) Antibiotic activity of epiphytic bacteria isolated from intertidal seaweeds. Microb Ecol 11(2):149–163
Li X, Gao X, Zhang S, Jiang Z, Yang H, Liu X, Jiang Q, Zhang X (2020) Characterization of a Bacillus velezensis with antibacterial activity and inhibitory effect on common aquatic pathogens. Aquaculture 523:735165
Longford S, Tujula N, Crocetti GR, Holmes AJ, Holmstrom C, Kjelleberg S, Steinberg P, Taylor MW (2007) Comparisons of diversity of bacterial communities associated with three marine eukaryotes. Aquat Microb Ecol 48(3):217–229
Louden BC, Haarmann D, Lynne AM (2011) Use of blue agar CAS assay for siderophore detection. J Microbiol Biol Educ 12:51–53. https://doi.org/10.1128/jmbe.v12i1.249
Nicholson WL, Setlow P (1990) Dramatic increase in negative superhelicity of plasmid DNA in the forespore compartment of sporulating cells of Bacillus subtilis. J Bacteriol 172(1):7–14
Pandey A (2019) Pharmacological potential of marine microbes. In: Arora D, Sharma C, Jaglan S, Lichtfouse E (eds) Pharmaceuticals from microbes. Environmental chemistry for a sustainable world, 28th edn. Springer, Cham, pp 1–25
Penesyan A, Marshall-Jones Z, Holmstrom C, Kjelleberg S, Egan S (2009) Antimicrobial activity observed among cultured marine epiphytic bacteria reflects their potential as a source of new drugs. FEMS Microbiol Ecol 69(1):113–124
Sastalla I, Fattah R, Coppage N, Nandy P, Crown D, Pomerantsev AP, Leppla SH (2013) The Bacillus cereus hbl and nhe tripartite enterotoxin components assemble sequentially on the surface of target cells and are not interchangeable. PLoS One 8:e76955. https://doi.org/10.1371/journal.pone.0076955
Schirmer A, Gadkari R, Reeves CD, Ibrahim F, DeLong EF, Hutchinson CR (2005) Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl Environ Microbiol 71(8):4840–4849
Seng P, Drancourt M, Gouriet F, La-Scola B, Fournier PE, Rolain JM, Raoult D (2009) Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-fight mass spectrometry. Clin Infect Dis 49(4):543–551
Singh RP, Mantri VA, Reddy CRK, Jha B (2011) Isolation of seaweed-associated bacteria and their morphogenesis-inducing capability in axenic cultures of the green alga Ulva fasciata. Aquat Biol 12(1):13–21
Soria-Mercado IE, Villarreal-Gómez LJ, Rivas GG, Sánchez NEA (2012) Bioactive compounds from bacteria associated to marine algae. Biotechnology molecular studies and novel application for improved quality of human life. InTech, Croatia, pp 25–44
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Thilakan B, Chakraborty K, Chakraborty RD (2016) Antimicrobial properties of cultivable bacteria associated with seaweeds in the Gulf of Mannar on the southeast coast of India. Can J Microbiol 62(8):668–681
Turick CE, Caccavo F Jr, Tisa LS (2008) Pyomelanin is produced by Shewanella algae BrY and affected by exogenous iron. Can J Microbiol 54(4):334–339
Vijayalakshmi S, Ramasamy MS, Murugesh S, Murugan A (2008) Isolation and screening of marine associated bacteria from Tamil Nadu, Southeast coast of India for potential antibacterial activity. Ann Microbiol 58(4):605–609
Wang H, Fewer DP, Holm L, Rouhiainen L, Sivonen K (2014) Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci USA 111(25):9259–9264
Whelan S, Goldman N (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18(5):691–699
Wiese J, Thiel V, Nagel K, Staufenberger T, Imhof JF (2009) Diversity of antibiotic-active bacteria associated with the brown alga Laminaria saccharina from the Baltic Sea. Mar Biotechnol 11(2):287–300
Winter JM, Chiou G, Bothwell IR, Xu W, Garg NK, Luo M, Tang Y (2013) Expanding the structural diversity of polyketides by exploring the cofactor tolerance of an inline methyltransferase domain. Org Lett 15(14):3774–3777
Xia J, Sinelnikov IV, Han B, Wishart DS (2015) MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res 43(W1):W251–W257
Yi Y, Zhang Z, Zhao F, Liu H, Yu L, Zha J, Wang G (2018) Probiotic potential of Bacillus velezensis JW: antimicrobial activity against fish pathogenic bacteria and immune enhancement effects on Carassius auratus. Fish Shellfish Immunol 78:322–330
Zhang W, Zhang F, Li Z, Miao X, Meng Q, Zhang X (2009) Investigation of bacteria with polyketide synthase genes and antimicrobial activity isolated from South China Sea sponges. J Appl Microbiol 107(2):567–575
Zhao K, Penttinen P, Guan T, Xiao J, Chen Q, Xu J, Lindström K, Zhang L, Zhang X, Strobel GA (2011) The diversity and anti-microbial activity of endophytic actinomycetes isolated from medicinal plants in Panxi plateau, China. Curr Microbiol 62(1):182–190. https://doi.org/10.1007/s00284-010-9685-3
Zheng L, Han X, Chen H, Lin W, Yan X (2005) Marine bacteria associated with marine macro organisms: the potential antimicrobial resources. Anal Microbial 55(2):119–124
Zhu P, Zheng Y, You Y, Yan X, Shao J (2009) Molecular phylogeny and modular structure of hybrid NRPS/PKS gene fragment of Pseudoalteromonas sp. NJ6–3–2 isolated from marine sponge Hymeniacidon perleve. J Microbiol Biotechnol 19(3):229–237
Zubia M, Fabre MS, Kerjean V, Lann KL, Pouvreau VS, Fauchon M, Deslandes E (2009) Antioxidant and antitumoural activities of some Phaeophyta from Brittany coasts. Food Chem 116(3):693–701
Acknowledgements
This work was funded by the Indian Council of Agricultural Research (ICAR) under the project titled as “Development of Bioactive Pharmacophores from Marine Organisms” (grant number MBT/HLT/SUB23). The authors thank the Director, Central Marine Fisheries Research Institute and Head, Marine Biotechnology Division, Central Marine Fisheries Research Institute for facilitating the research activities. The authors thank the Dean, School of Biotechnology, Amrita Vishwa Vidyapeetham for supporting CV.
Funding
This study was funded by the Indian Council of Agricultural Research (ICAR).
Author information
Authors and Affiliations
Contributions
KC and CV conceived and designed research, acquired funds, and conducted experiments. KC, RDC, and SA analyzed data. All authors drafted and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No potential conflict of interest was reported by the authors. This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Jorge Membrillo-Hernández.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chakraborty, K., Varghese, C., Asharaf, S. et al. Antibiotic-active heterotrophic Firmicutes sheltered in seaweeds: can they add new dimensions to future antimicrobial agents?. Arch Microbiol 204, 183 (2022). https://doi.org/10.1007/s00203-022-02784-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-02784-2