Abstract
A new taxon is created for the thermophilic purple nonsulfur bacterium previously designated as Rhodopseudomonas strain GI. Strain GI was isolated from a New Mexico (USA) hot spring microbial mat and grows optimally above 40 °C and to a maximum of 47 °C. Strain GI is a bacteriochlorophyll b-containing species of purple nonsulfur bacteria and displays a budding morphology, typical of species of the genus Blastochloris. Although resembling the species Blc. viridis in many respects, the absorption spectrum, carotenoid content, and lipid fatty acid profile of strain GI is distinct from that of Blc. viridis strain DSM133T and other recognized Blastochloris species. Strain GI forms its own subclade within the Blastochloris clade of purple nonsulfur bacteria based on comparative 16S rRNA gene sequences, and its genome is significantly larger than that of strain DSM133T; average nucleotide identity between the genomes of Blc. viridis and strain GI was below 85%. Moreover, concatenated sequence analyses of PufLM and DnaK clearly showed strain GI to be distinct from both Blc. viridis and Blc. sulfoviridis. Because of its unique assortment of properties, it is proposed to classify strain GI as a new species of the genus Blastochloris, as Blc. tepida, sp.n., with strain GIT designated as the type strain (= ATCC TSD-138 = DSM 106918).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Several species of phototrophic purple bacteria are thermophilic, and the first of these to be isolated was the purple sulfur bacterium Thermochromatium (formerly Chromatium) tepidum. This phototroph grows to a maximum temperature of 57 °C and has a growth temperature optimum near 50 °C (Madigan 1984, 1986; Castenholz and Pierson 1995). No purple nonsulfur bacteria are known that grow at 50 °C but several species grow optimally at 40 °C, and these include species of Rhodopseudomonas, Rhodoplanes (Rpl.), and Rubrivivax (Namsaraev et al. 2003; Okamura et al. 2007; Stadtwald-Demchick et al. 1990; Favinger et al. 1989; Hiraishi 2017a, b; Hisada et al. 2007). Rhodoplanes (formerly Rhodopseudomonas) cryptolactis (now Rpl. tepidamans) and Rhodocista centenaria (formerly Rhodospirillum centenum) are the most thermotolerant of these phototrophs, growing up to 45 °C. All of these purple bacteria contain bacteriochlorophyll (BChl) a as their chlorophyllous pigment.
Nearly 30 years ago, two of us isolated a budding purple bacterium containing BChl b from a hot spring microbial mat and reported on its basic physiology, including in particular the fact that the organism grew optimally at 42 °C and had a maximum growth temperature of 47 °C (Resnick and Madigan 1989). Since phylogenetic data were lacking at that time, this phototroph was not formally described as a new species and was simply referred to as “Rhodopseudomonas strain GI”, although it was assumed that the isolate was related to the BChl b-containing budding species Blastochloris (then Rhodopseudomonas) viridis (Resnick and Madigan 1989; Drews and Giesbrecht 1966; Hiraishi 1997). In addition to its moderately thermophilic phenotype, strain GI displayed several other properties typical of purple nonsulfur bacteria containing BChl b (Madigan and Jung 2009; Hoogewerf et al. 2003; Keppen and Gorlenko 1975) but not previously reported in thermotolerant species, including the ability to fix N2 at temperatures as high as 48 °C Resnick and Madigan 1989).
Here we complement our original study of strain GI with new data on the morphology, spectral properties, fatty acid and pigment compositions, and phylogeny and genomics of this organism, and assemble these data to support the establishment of a new species of the genus Blastochloris (Blc.), Blc. tepida, to accommodate strain GI and closely related isolates. The proposed species epithet reflects the thermophilic nature of strain GI, a phenotype unlike that of any of the currently recognized species of Blastochloris: Blc. viridis, Blc. sulfoviridis, and Blc. gulmargensis (Drews and Giesbrecht 1966; Keppen and Gorlenko 1975; Ramana et al. 2011).
Materials and methods
Isolation and microscopy
Strain GI was enriched and isolated at 45 °C from a sample of a cyanobacterial microbial mat that developed in the outflow of a small hot spring (47 °C) at Soda Dam, located within the Santa Fe National Forest (New Mexico, USA; GPS N35°793′, W106°685′); the microbial mat was dominated by species of the filamentous cyanobacteria Oscillatoria and Phormidium (Resnick and Madigan 1989). Cultures of strain GI were purified in repeated agar shake tubes and grown phototrophically (anoxic/light) in the mineral salts–malate medium described in Resnick and Madigan (1989) supplemented with 0.05% (final concentration) each of sodium ascorbate and yeast extract. Cultures of strain GI were stored at − 80 °C until accessioned into the DSMZ and ATCC in 2018.
Electron microscopy of cells of strain GI was performed as described in Cole et al. (2014) with the exception that an Orion helium ion microscope (Zeiss, Peabody, MA, USA) was used in place of a conventional scanning electron microscope. Fatty acid determinations were performed by MIDI (Newark, DE, USA) using their standard fatty acid methyl ester methods on lipids extracted from cells of strain GI grown at 43 °C. Absorption spectra were performed on membrane suspensions (chromatophores) prepared as described by Nagatsuma et al. (2019).
Phylogenetic and genomic analyses
Phylogenetic analysis of strain GI based on 16S rRNA gene sequence comparisons with other purple nonsulfur bacteria were performed as previously described in Kempher and Madigan (2012).
To obtain the genome sequence of strain GI, genomic DNA was extracted from a cell pellet using the Genome-Tip 500G DNA extraction kit (Qiagen, Venlo, The Netherlands). A sequencing library was prepared using the Accel-NGS XL Library Kit (Swift Biosciences, MI, USA) according to the manufacturer’s protocol, and DNA fragments of less than 20 kb were removed using the BluePippin size-selection system (Sage Science, MA, USA). The final gene library was run on a PacBio RSII sequencer with SMRT cell v3 and P6-C4 v2 chemistry, yielding a total of 64,481 reads and 973,705,006 bp.
PacBio reads of more than 10 kb were assembled using the HGAP3 assembler program with default settings (Chin et al. 2013). In addition, a standard Illumina 600-bp paired-end library was constructed and sequenced on a HiSeq 2500 sequencer (Illumina, CA, USA). Sequences of 250 bases comprising 1,585,689 × 2 raw reads were used to improve the assembled contig with the Pilon (version 1.22) software tool (Walker et al. 2014). Gene prediction and functional annotation were carried out using the DDBJ Fast Annotation and Submission Tool (DFAST) (Tanizawa et al. 2017).
Amino acid sequences of PufL, PufM, and DnaK proteins from Blc. viridis, Blc. sulfoviridis, and Rpl. elegans in the public sequence database were aligned with those of strain GI using MAFFT version 7.407 with default parameters (Katoh and Castresana 2014). Poorly aligned regions were excluded using GBlocks version 0.91 b with default parameters (Talavera and Castresana 2007). After concatenating the three-protein sequence alignments, a maximum likelihood tree was inferred using RAxML version 8.2.10 with the WAG + Γ model and 1000 bootstrap replicates (Stamatakis 2014).
Results and discussion
Morphology, pigments, and fatty acids
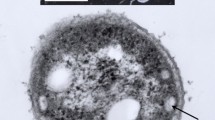
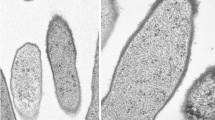
Pure cultures of strain GI were grown routinely under phototrophic conditions at 42 °C. Cells were motile and stained Gram-negative. Individual cells measured approximately 1 × 1.5–2 μm and showed a budding morphology. Although the diameter of cells was fairly constant, cell length varied depending on what stage a given cell was in during the budding process, as shown in scanning electron micrographs (Fig. 1a). Thin sections of cells revealed a lamellar-type photosynthetic membrane system running parallel to the long axis of the cell (Fig. 1b), a feature characteristic of budding purple bacteria, including Blc. viridis (Drews and Giesbrecht 1966; Imhoff et al. 1984). Negatively stained transmission electron micrographs showed cells to contain one or more polar flagella (Fig. 1c).
Phototrophic cultures of strain GI were greenish yellow in color, and absorption spectra of chromatophore preparations were similar to yet distinct from that of the type strain of Blc. viridis (strain DSM 133T), as shown in Fig. 2. The near infrared (Qy) absorption maximum of strain GI lies at 1011 nm compared with that of 1014 nm in Blc. viridis; absorption maxima beyond 1000 nm is the major characteristic of BChl b-containing phototrophs (Scheer et al. 1974; Tsukatani et al. 2019). Other major maxima in the absorption spectra of Blc. viridis and strain GI were similar, although absorbance in the carotenoid region of the spectrum of Blc. viridis was noticeably greater than that of strain GI when both spectra were normalized to absorbance at their respective Qy bands of BChl b in light-harvesting complex I around 1010 nm (Fig. 2); this suggests differences in the carotenoid composition between the two species. In this connection, a major carotenoid of strain GI was 1,2-dihdyrolycopene. Only trace amounts of this pigment are present in cells of Blc. sulfoviridis, and although detectable in Blc. viridis, 1,2-dihdyrolycopene content of this species is only one-fifth that of strain GI (Table 1). Strain GI also produced significant levels of lycopene and 1,2-dihydro-3,4-didehydrolycopene—carotenoids present in only trace amounts in Blc. viridis and Blc. sulfoviridis—and had the lowest levels of 1,2-dihydroneurosporene of all Blastochloris species (Table 1). Taken collectively, these results suggest that phytoene desaturase (CrtI), a major enzyme in the biosynthetic pathway of lycopene (which is a precursor of downstream lycopene derivatives), may be expressed at higher levels in strain GI than in other Blastochloris species.
Significant differences were also observed in the fatty acid composition of strain GI compared with other Blastochloris species. Strain GI showed over twice the C16:0 content of Blc. viridis and Blc. sulfoviridis (Kompantseva et al. 2007) and nearly twice that of Blc. gulmargensis (Table 1). Moreover, although the C18:0 fatty acid content of strain GI was notably higher than that of the other species, the unsaturated C18 content of strain GI was lower. Collectively, these differences likely reflect the thermophilic phenotype of strain GI, where saturated fatty acids would be predicted to lend more stability to membrane lipids.
Physiology
Several aspects of the physiology of strain GI were summarized in Resnick and Madigan (1989), in particular aspects of temperature requirements and tolerances, carbon and nitrogen nutrition, growth modes, and nitrogen fixation. Strain GI was isolated from an alkaline calcareous warm spring microbial mat composed primarily of cyanobacteria and Chloroflexus and in most respects displayed the physiology of a typical purple nonsulfur bacterium. That is, growth of strain GI was best under photoheterotrophic conditions at neutral pH with malate or pyruvate as carbon sources. In the presence of bicarbonate, butyrate was also a good carbon source for strain GI; this fatty acid is not used by Blc. viridis (Resnick and Madigan 1989; Trüper and Imhoff 1989). Although some aromatic compounds are used by select Blastochloris isolates (Zengler et al. 1999), benzoate was not used by strain GI or by Blc. viridis (Resnick and Madigan 1989; Trüper and Imhoff 1989). Chemotrophic dark growth on malate occurred in strain GI, but only at reduced oxygen tensions. In addition, strain GI required a reduced source of sulfur for biosynthetic purposes (thiosulfate or low levels of sulfide were preferred sources) and biotin as a growth factor (Resnick and Madigan 1989). In its natural habitat, this reduced biosynthetic sulfur source requirement is likely met by the ~ 100 µM sulfide detected in the Soda Dam spring water.
In requiring a reduced sulfur source, strain GI resembles Blc. sulfoviridis, which also has such a requirement (Keppen and Gorlenko 1975); other Blastochloris species can use sulfate for biosynthetic sulfur needs (Ramana et al. 2011; Trüper and Imhoff 1989). In requiring biotin, strain GI resembles all other Blastochloris species. All Blastochloris species other than strain GI also require para-aminobenzoic acid (PABA) and some species require pyridoxal phosphate as well (Ramana et al. 2011; Trüper and Imhoff 1989).
A defining feature of the physiology of strain GI is its growth temperature profile compared with that of other Blastochloris species and supporting data in this regard were published in Resnick and Madigan (1989). Optimal growth of strain GI occurred at 42 °C, and no growth occurred at 28 °C or 48 °C; the growth temperature range was 30–47 °C. This stands in contrast to the temperature profiles of Blc. viridis and Blc. sulfoviridis, both of whose growth temperature optima lie under 30 °C (Drews and Giesbrecht 1966; Keppen and Gorlenko 1975; Trüper and Imhoff 1989).
At 47 °C, strain GI grew on either ammonia or N2 as nitrogen sources, indicating that this Blastochloris species synthesizes a thermostable nitrogenase system (nitrogenase activity could still be detected at 48 °C, slightly above the maximum growth temperature, Resnick and Madigan 1989). Since the microbial mat from which strain GI was isolated had a temperature of 47 °C and such mats are typically N-deficient, it seems likely that strain GI fixes N2 in situ. If so, this organism may supply fixed nitrogen not only to itself but also to the filamentous nonheterocystous cyanobacteria and Chloroflexus that formed the matrix of the Soda Dam microbial mat; all of these phototrophs are nondiazotrophic (Resnick and Madigan 1989; Heda and Madigan 1986). Indeed, because of their N2-fixing abilities and their unique absorption properties, BChl b-containing purple bacteria may be widely distributed in hot spring microbial mats. The observation that strain GI-like purple bacteria exist in Japanese (Hisada et al. 2007) and Russian (Namsaraev et al. 2003) hot spring mats containing filamentous cyanobacteria and Chloroflexus, supports this hypothesis.
Phylogeny and correlation with temperature tolerances
The phylogeny of strain GI was determined by 16S rRNA gene sequencing, and the results are shown in Fig. 3. The genus Blastochloris forms a clade within the Alphaproteobacteria and its closest relatives are species of the genus Rhodoplanes (Rpl.), in particular, the thermotolerant BChl a-containing species Rpl. tepidicaeni, Rpl. tepidamans, and Rpl. azumiensis (Hiraishi 2017a, b). More distant relatives include the well-known purple nonsulfur bacteria Rhodopseudomonas palustris, and Rhodoblastus acidophilus (Fig. 3). All of these phototrophs are united by their characteristic budding growth mode, lamellar photosynthetic membrane systems, and production of BChl a (Hiraishi 2017a, b; Trüper and Imhoff 1989).
Phylogenetic tree of species of Blastochloris and Rhodoplanes and nearby relatives of purple nonsulfur bacteria based on 16S rRNA gene sequence comparisons. Approximately 1400 nucleotides were used in the analysis and bootstrap values (1000 replications) are indicated at the nodes. Strain designations are as follows: Rpi. globiformis DSM 161T; Rps. palustris ATTC 17001T; Rbl. acidophilus DSM 137T; Rpl. azumiensis NBRC 112816T; Rpl. tepidamans DSM 9987T; Rpl. tepidicaeni NBRC 112815T; Blc. viridis DSM 133T; Blc. gulmargensis DSM 19786T; Blc. sulfoviridis DSM 729T; Blc. sulfoviridis ToP1; Blc. tepida DSM 106918T; Blc. sp. TUT3225
Within the Blastochloris clade, three subclades exist, one containing Blc. viridis and Blc. gulmargensis, one containing Blc. sulfoviridis strains, and a third containing strain GI (Blc. tepida) and strain TUT3225, a Blastochloris sp. isolated from a Japanese hot spring (Hisada et al. 2007) (Fig. 3). Strain TUT3225 grows up to 45 °C with an optimum at 42 °C (Hisada et al. 2007), thus it is slightly less thermotolerant than strain GI. The three Rhodoplanes species shown in Fig. 3 all grow optimally at 40 °C and grow up to a maximum of 45 °C. However, none are capable of growth at 47 °C, as is strain GI (Resnick and Madigan 1989). In contrast to these moderately thermophilic species, Blc. viridis, Blc. sulfoviridis, and Blc. gulmargensis are all mesophilic phototrophs, with temperature optima of 25–30 °C (Drews and Giesbrecht 1966; Keppen and Gorlenko 1975; Ramana et al. 2011; Zengler et al. 1999). Thus, strain GI is the most thermophilic of all known hot spring purple nonsulfur bacteria.
Genome sequence and functional gene comparisons
The genome of strain GI was sequenced and assembled into a single contig, and a summary of the genome characteristics is presented in Table 2. Compared with the genome of Blc. viridis DSM 133T (Tsukatani et al. 2015; Liu et al. 2016), the strain GI genome is significantly larger, containing 308 additional genes (Table 2). The GC content of the two genomes also differs slightly between the two organisms, but both genomes reside on single chromosomes with no plasmids detected (Table 2).
Virtually all of the genes that encode proteins of the reaction center and light-harvesting complexes and enzymes of BChl and carotenoid biosynthesis in purple nonsulfur bacteria are grouped together in the genome in a “photosynthetic gene cluster” (PGC) (Alberti et al. 1995; Nagashima and Nagashima 2013). PGCs also exist in Blc. viridis and strain GI; however, gene synteny in the PGCs of strain GI and Blc. viridis were not identical. In particular, the bchIDO-crtIB region of the two species PGCs were inverted (Fig. 4). Moreover, an unusual characteristic of the PGC of Blc. viridis compared with that of other phototrophic purple bacteria is that some genes encoding Calvin cycle proteins (cbb and related genes) are embedded within its PGC (Tsukatani et al. 2015; Liu et al. 2016); this was also true of the strain GI PGC (Fig. 4).
Comparison of the photosynthetic gene clusters in the genomes of Blc. viridis strain DSM133T and Blc. tepida strain GI. Genes are represented by rectangles pointing in the direction of transcription. Genes for BChl (bch) and carotenoid (crt) biosynthesis are shown in green and orange, respectively. The puf and puh genes encoding the reaction center and light-harvesting 1 complexes and related products are shown in black. Calvin cycle genes are shown in blue. Open-reading frames without an assigned gene name were annotated as hypothetical proteins
Computational analysis of the potential comprehensive functions (functionome) of the Blc. viridis and strain GI genomes with the MAPLE system (Takami et al. 2016) showed no significant difference in metabolic pathway content of the two species. Many genome rearrangements and frequent repetitive sequences were observed, and overall, the syntenic structure of the two species’ genomes was only weakly conserved. This was supported by an average nucleotide identity between Blc. viridis and strain GI protein-encoding genes of 84.1%; values below 85% correlate strongly with genomic DNA:DNA hybridization values of 70% or lower, a standard used for years to delineate distinct species (Goris et al. 2007). We thus conclude that Blc. viridis and strain GI are distinct species.
The genome of Blc. sulfoviridis has not been sequenced and so whole genome comparisons of the type done between strain GI and Blc. viridis were not possible. However, because Blc. sulfoviridis, like Blc. viridis, is a close relative of strain GI (Fig. 3), additional genomic evidence was sought to differentiate strain GI from Blc. sulfoviridis. Three functional genes of phylogenetic importance have been sequenced in the type strain of Blc. sulfoviridis and are available in public databases; these include dnaK, pufL, and pufM. The two puf genes encode structural proteins in the photosynthetic reaction center of purple bacteria while dnaK encodes a key molecular chaperone in the bacterial heat shock response. Sequence comparisons of pufLM have been shown to be useful for resolving the phylogenies of purple bacteria (Swingley et al. 2009; Tank et al. 2009). The sequence of dnaK is highly conserved but also contains a variable region whose sequence has proven to be an alternative marker to 16S rRNA gene sequences for resolving the phylogenies of a broad range of Alphaproteobacteria, including phototrophic purple bacteria such as Blastochloris (Stepkowski et al. 2003).
The amino acid sequences of PufLM and DnaK deduced from the genome sequence of strain GI were, therefore, aligned with those of Blc. viridis, Blc. sulfoviridis, and Rpl. elegans in a concatenated analysis, and the results are shown in Fig. 5. The maximum likelihood tree constructed from this analysis clearly distinguishes strain GI from all other species of Blastochloris. In fact, the concatenated three-protein tree shows strain GI to be a closer relative to Blc. viridis than to Blc. sulfoviridis (Fig. 5). And, since Blc. viridis and strain GI are clearly distinct species on the basis of whole genome analyses, we conclude that strain GI and Blc. sulfoviridis are similarly distinct and separate species.
Final remarks and taxonomic conclusions
The earliest breakthroughs in studies of the structure of photosynthetic reaction center complexes occurred with the purple bacterium Blc. viridis (Michel 1982; Deisenhofer et al. 1985). This phototroph was ideal for several reasons, but in particular because it produced only a core and not a peripheral antenna complex. Now with the discovery of strain GI, a thermophilic species of Blastochloris is available that will likely produce more thermotolerant biomolecules than mesophilic species of Blastochloris. These, along with a complete genome sequence to support structural studies, suggests that BChl b-containing purple bacteria may continue to provide ideal model systems for unraveling the functional details of early events in photosynthesis.
Based on the assemblage of phenotypic properties of strain GI along with its unique phylogenetic position and genomic characteristics, this organism is proposed as a new species of the genus Blastochloris, as Blastochloris tepida sp.n. Strain GIT has been accessioned into the American Type Culture Collection as ATCC TSD-138 and into the Deutsche Sammlung von Mikroorganismen und Zellkulturen as DSM 106918.
Description of Blastochloris tepida sp. nov
Blastochloris tepida (te’pi.da. L. fem. adj. tepida lukewarm).
Cells are Gram-negative phototrophic budding rods measuring 1 × 1.5–2 µm and motile by polar flagella. Phototrophic cultures are greenish yellow in color and intracytoplasmic photosynthetic membranes are present as lamellae running parallel to the long axis of the cell. Contains BChl b and 1,2-dihydroneurosporene, 1,2-dihydrolycopene, and 1,2-dihydro-3,4-didehydrolycopene as major carotenoids; lycopene and neurosporene are minor carotenoids. Membranes from phototrophic cells show absorption maxima at 1011, 602, 521, 483, 452, and 397 nm. Photoheterotrophic growth is best in mineral media containing 0.05% yeast extract and either malate or fumarate as primary carbon sources; glucose and fructose support lesser growth. Acetate, butyrate, and pyruvate support good growth but only in the presence of bicarbonate. Succinate is a poor growth substrate, and benzoate, pentoses, short-chain alcohols, and the amino acids aspartate and glutamate are not used. A reduced source of sulfur and the B-vitamin biotin are required for growth (0.01% thiosulfate and biotin fully replace yeast extract for optimal growth). Chemotrophic dark (respiratory) growth occurs only at reduced oxygen tensions. Ammonia, glutamine, and N2 are the best nitrogen sources for growth. Growth is optimal at 42 °C and the growth temperature range is 30–47 °C. Best growth occurs at pH 6.8 and in media lacking NaCl. The type strain of Blastochloris tepida is strain GIT. Cultures of Blc. tepida strain GIT have been accessioned into the DSMZ (DSM 106918) and ATCC (TSD-138). The 16S gene sequence of Blc. tepida strain GIT has been deposited into DDBJ/Genbank as MG725814 and its complete genome sequence as AP018907.
References
Alberti M, Burke DH, Hearst JE (1995) Structure and sequence of the photosynthesis gene cluster. In: Blankenship RE, Madigan MT, Bauer CE (eds) Anoxygenic photosynthetic bacteria. Kluwer, Dordrecht, pp 1083–1106
Castenholz RW, Pierson BK (1995) Ecology of thermophilic anoxygenic phototrophs. In: Blankenship RE, Madigan MT, Bauer CE (eds) Anoxygenic photosynthetic bacteria. Kluwer, Dordrecht, pp 87–103
Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J (2013) Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569
Cole JK, Hutchison JR, Renslow RS, Kim Y-M, Chrisler WB, Engelmann HE, Dohnalkova AC, Hu D, Metz TO, Frederickson JK, Lindemann SR (2014) Phototrophic biofilm assembly in microbial-mat-derived unicyanobacterial consortia: model systems for the study of autotroph–heterotroph interactions. Front Microbiol 5:109. https://doi.org/10.3389/fmicb.2014.00109
Deisenhofer J, Epp O, Miki K, Huber R, Michel H (1985) Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3Å resolution. Nature 318:618–624
Drews G, Giesbrecht P (1966) Rhodopseudomonas viridis, nov. spec., ein neu isoliertes, obligat phototrophes bakterium. Arch Mikrobiol 53:255–262
Favinger J, Stadtwald R, Gest H (1989) Rhodospirillum centenum, sp. nov., a thermotolerant cyst-forming anoxygenic photosynthetic bacterium. Ant van Leeuwenhoek 55:291–296
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91
Heda GD, Madigan MT (1986) Utilization of amino acids and lack of diazotrophy in the thermophilic anoxygenic phototroph Chloroflexus aurantiacus. J Gen Microbiol 132:2469–2473
Hiraishi A (1997) Transfer of the bacteriochlorophyll b-containing phototrophic bacteria Rhodopseudomonas viridis and Rhodopseudomonas sulfoviridis to the genus Blastochloris gen. nov. Intl J Syst Bacteriol 47:217–219
Hiraishi A (2017a) Characterization of thermotolerant phototrophic bacteria, Rhodoplanes tepidicaeni sp. nov., and Rhodoplanes azumiensis sp. nov., isolated from a geothermal spring. Intl J Syst Evol Microbiol 67:5038–5045
Hiraishi A (2017b) Proposal of Rhodoplanes tepidamans sp. nov. to accommodate the thermotolerant phototrophic bacterium previously referred to as ‘Rhodoplanes (Rhodopseudomonas) cryptolactis’. Intl J Syst Evol Microbiol 67:1540–1545
Hisada T, Okamura K, Hiraishi A (2007) Isolation and characterization of phototrophic purple nonsulfur bacteria from Chloroflexus and cyanobacterial mats in hot springs. Microbes Environ 22:405–411
Hoogewerf GE, Jung DO, Madigan MT (2003) Evidence for limited species diversity of bacteriochlorophyll b-containing purple nonsulfur anoxygenic phototrophs in freshwater habitats. FEMS Microbiol Letts 218:359–364
Imhoff JF, Trüper HG, Pfennig N (1984) Rearrangement of the species and genera of phototrophic “purple nonsulfur bacteria”. Intl J Syst Bacteriol 34:340–343
Katoh K, Castresana J (2014) MAFFT: iterative refinement and additional methods. Meth Mol Biol 1079:131–146
Kempher ML, Madigan MT (2012) Phylogeny and photoheterotrophy in the acidophilic phototrophic purple bacterium Rhodoblastus acidophilus. Arch Microbiol 194:567–574
Keppen OI, Gorlenko VM (1975) A new species of purple budding bacteria containing bacteriochlorophyll b. Mikrobiologiya 44:258–264
Kompantseva EI, Imhoff JF, Thiemann B, Panteleeva EE, Akimov VN (2007) Comparative study of the fatty acid composition of some groups of purple nonsulfur bacteria. Microbiology (English translation of Mikrobiologiya) 76:541–551
Liu L-N, Faulkner M, Liu X, Huang F, Darby AC, Hall N (2016) Revised genome sequence of the purple photosynthetic bacterium Blastochloris viridis. Genome Announcements 4:e10152. https://doi.org/10.1128/genomeA.01520-15
Madigan MT (1984) A novel photosynthetic purple bacterium isolated from a Yellowstone hot spring. Science 225:313–315
Madigan MT (1986) Chromatium tepidum, sp.n., a thermophilic photosynthetic bacterium of the family Chromatiaceae. Intl J Syst Bacteriol 36:222–227
Madigan MT, Jung DO (2009) An overview of purple bacteria: systematics, physiology, and habitats. In: Hunter CN, Daldal F, Thurnauer MC, Beatty JT (eds) The purple phototrophic bacteria. Springer, Dordrecht, pp 1–15
Michel H (1982) Three-dimensional crystals of a membrane protein complex: the photosynthetic reaction centre from Rhodopseudomonas viridis. J Mol Biol 158:567–572
Nagashima S, Nagashima KVP (2013) Comparison of photosynthesis gene clusters retrieved from total genome sequences of purple bacteria. Adv Bot Res 66:151–178
Nagatsuma S, Gotou K, Yamashita T, Yu L-J, Shen J-R, Madigan MT, Kimura Y, Wang-Otomo Z-Y (2019) Phospholipid distributions in purple phototrophic bacteria and LH1-RC core complexes. Biochim Biophys Acta 1860:461–468
Namsaraev ZB, Gorlenko VM, Namsaraev BB, Buryukhaev SP, Yurkov VV (2003) The structure and biogeochemical activity of the phototrophic communities from the Bol’sherechenskii alkaline hot spring. Microbiology (English translation of Mikrobiologiya) 72:228–238
Okamura K, Hisada T, Hiraishi A (2007) Characterization of thermotolerant purple nonsulfur bacteria isolated from hot-spring Chloroflexus mats and the reclassification of “Rhodopseudomonas cryptolactis” Stadtwald-Demchick et al. 1990 as Rhodoplanes cryptolactis nom. rev., comb. nov. J Gen Appl Microbiol 53:357–361
Ramana VV, Kapoor S, Shobha E, Ramprasad EVV, Sasikala Ch, Ramana ChV (2011) Blastochloris gulmargensis sp. nov., isolated from an epilithic phototrophic biofilm. Intl J Syst Evol Microbiol 61:1811–1816
Resnick SM, Madigan MT (1989) Isolation and characterization of a mildly thermophilic nonsulfur purple bacterium containing bacteriochlorophyll b. FEMS Microbiol Lett 65:165–170
Scheer H, Svec WA, Cope BT, Studier MH, Scott RG, Katz JJ (1974) Structure of bacteriochlorophyll b. J Am Chem Soc 96:3714–3716
Stadtwald-Demchick R, Turner FR, Gest H (1990) Rhodopseudomonas cryptolactis, sp. nov., a new thermotolerant species of budding phototrophic bacteria. FEMS Microbiol Lett 71:117–122
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Stepkowski T, Czaplinska M, Miedzinska K, Moulin L (2003) The variable part of the dnaK gene as an alternative marker for phylogenetic studies of rhizobia and related Alpha Proteobacteria. Syst Appl Microbiol 26:483–494
Swingley WD, Blankenship RE, Raymond J (2009) Evolutionary relationships among purple photosynthetic bacteria and the origin of proteobacterial photosynthetic systems. In: Hunter CN, Daldal F, Thurnauer MC, Beatty JT (eds) The purple phototrophic bacteria. Springer, Dordrecht, pp 17–29
Takami H, Taniguchi T, Arai W, Takemoto K, Moriya Y, Goto S (2016) An automated system for evaluation of the potential functionome: MAPLE version 2.1.0. DNA Res 23:467–475
Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564–577
Tanizawa Y, Fujisawa T, Nakamura Y (2017) DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34:1037–1039
Tank M, Thiel V, Imhoff JF (2009) Phylogenetic relationship of phototrophic purple sulfur bacteria according to pufL and pufM genes. Intl Microbiol 12:175–185
Trüper HG, Imhoff JF (1989) Genus Rhodopseudomonas. In: Staley JT, Bryant MP, Pfennig N, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 3. Williams and Wilkins, Baltimore, pp 1672–1677
Tsukatani Y, Hirose Y, Harada J, Misawa N, Mori K, Inoue K, Tamiaki H (2015) Complete genome sequence of the bacteriochlorophyll b-producing photosynthetic bacterium Blastochloris viridis. Genome Announcements 3:e1006–e1015. https://doi.org/10.1128/genomeA.01006-15
Tsukatani Y, Hirose Y, Harada J, Yonekawa C, Tamiaki H (2019) Unusual features in the photosynthetic machinery of Halorhodospira halochloris DSM 1059 revealed by complete genome sequencing. Photosynth Res 140:311–319
Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963
Zengler K, Heider J, Rosselló-Mora R, Widdel F (1999) Phototrophic utilization of toluene under anoxic conditions by a new strain of Blastochloris sulfoviridis. Arch Microbiol 172:204–212
Acknowledgements
We thank Professor Aharon Oren, Hebrew University of Jerusalem, for nomenclatural advice and Dr. Ch. V. Ramana, University of Hyderabad, for cells of Blc. gulmargensis for carotenoid analyses. MTM thanks Dr. John Bauld for the mat sample from which strain GI was isolated. This work was supported in part by a grant to MTM from the NASA Exobiology Program and from JSPS KAKENHI grant number 17H05231 to YT. Electron microscopy was performed at the Environmental Molecular Sciences Laboratory (EMSL), a national scientific user facility sponsored by the United States Department of Energy Office of Biological and Environmental Research, located at the Pacific Northwest National Laboratory (Richland, WA).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no financial or other conflicts of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Genomic accession numbers Blastochloris tepida strain GIT 16S rRNA gene sequence, MG725814; Blastochloris tepida strain GIT complete genome sequence, AP018907. Culture accession numbers: Blastochloris tepida strain GIT DSM 106918; ATCC TSD-138.
Rights and permissions
About this article
Cite this article
Madigan, M.T., Resnick, S.M., Kempher, M.L. et al. Blastochloris tepida, sp. nov., a thermophilic species of the bacteriochlorophyll b-containing genus Blastochloris. Arch Microbiol 201, 1351–1359 (2019). https://doi.org/10.1007/s00203-019-01701-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-019-01701-4