Abstract
Organisms belonging to the genus Rhizobium colonize leguminous plant roots and establish a mutually beneficial symbiosis. Biofilms are structured ecosystems in which microbes are embedded in a matrix of extracellular polymeric substances, and their development is a multistep process. The biofilm formation processes of R. etli CFN42 were analyzed at an early (24-h incubation) and mature stage (72 h), comparing cells in the biofilm with cells remaining in the planktonic stage. A genome-wide microarray analysis identified 498 differentially regulated genes, implying that expression of ~8.3 % of the total R. etli gene content was altered during biofilm formation. In biofilms-attached cells, genes encoding proteins with diverse functions were overexpressed including genes involved in membrane synthesis, transport and chemotaxis, repression of flagellin synthesis, as well as surface components (particularly exopolysaccharides and lipopolysaccharides), in combination with the presence of activators or stimulators of N-acyl-homoserine lactone synthesis This suggests that R. etli is able to sense surrounding environmental conditions and accordingly regulate the transition from planktonic and biofilm growth. In contrast, planktonic cells differentially expressed genes associated with transport, motility (flagellar and twitching) and inhibition of exopolysaccharide synthesis. To our knowledge, this is the first report of nodulation and nitrogen assimilation-related genes being involved in biofilm formation in R. etli. These results contribute to the understanding of the physiological changes involved in biofilm formation by bacteria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biofilms represent a very successful mode of microbial existence (Vorachit et al. 1995) because they contribute to increased resistance to environmental stresses as well as antimicrobial tolerance, protection from protozoan predation and the opportunity for horizontal gene transfer (HGT) (Danhorn and Fuqua 2007). Bacteria interacting with plants can be pathogens, commensals or mutualists. In many of these interactions, the ability to form biofilms is likely to be advantageous, if not essential, because it maintains a critical mass of cells in a specific location for long enough periods to initiate beneficial or antagonistic interactions with hosts. Rhizobia are nitrogen-fixing symbionts of legumes that employ a variety of strategies that allow them to exist in the soil and adapt to diverse environmental conditions prior to infecting leguminous plant. In this symbiotic relationship, bacteria colonize plant roots to obtain carbon sources and provide the plant with nitrogen. The plant–bacterial interaction shows a high degree of host specificity, based on the exchange of chemical signals, and results in the development of specialized structures called nodules (Schultze and Kondorosi 1998; Karatan and Watnick 2009). The process is mediated by molecular signal exchange between the host and the bacterial cells, which often requires cell-to-cell communication.
Biofilm formation is important in both free living and symbiotic life phases of Rhizobiales. In the free living phase, biofilm formation allows these non-spore-forming soil bacteria to colonize the surrounding habitats and to survive environmental stresses such as desiccation and nutrient limitation. In symbiosis, biofilm formation is essential in the early stages of nodulation, as reported in Sinorhizobium meliloti and R. leguminosarum bv. viciae (Fujishige et al. 2006). Bacterial biofilms are widely distributed and play important roles in many environments. Environments faced by soil rhizobia range from a rhizosphere rich in nutrients and root exudates to soils deficient in nitrogen, phosphates, water and nutrients. Numerous microbial species, including rhizobia, form microcolonies or biofilms when they colonize roots (Rinaudi and Giordano 2010). S. meliloti was first reported, and the rhizobia regulatory system in this species and conditions for analyzing its ability to biofilm formation on biotic and abiotic surfaces and have been interested (Fujishige et al. 2005).
Previous reports on biofilm formation have focused on the role of specific genes, such as those involved in cellular aggregation, swarming, nitrogen metabolism in establishing biofilms on abiotic and biotic surfaces such as glass, plastic microtiter plates, sand and soil (Donlan 2002).
Another essential requirement for biofilm formation is the attachment of the cells to a surface, where adhesins, exopolysaccharides, lipopolysaccharides and surface proteins play a central role (Davey and O’Toole 2000). The production of the autoinducers like case N-acylhomoserine lactones (AHLs) has also described for Mesorhizobium tianshanense, a nitrogen-fixing symbiont (Zheng et al. 2006).
In this work, we used a microarray analysis to analyze gene expression changes in R. etli CFN42 during biofilm formation on a glass (abiotic) surface, as described previously for some pathogenic and nitrogen-fixing organisms (Christensen et al. 2007; Rinaudi and Giordano 2010). We analyzed gene expression in the planktonic and sessile phases of growth (24 and 72 h).
This is the first transcriptomic study during biofilm formation in R. etli CFN42. As opposed to the majority of previous studies in rhizobia using cells obtained in different experiments (biofilms on abiotic or biotic surfaces; and planktonic cells from shaken cultures), we used planktonic and sessile cells, collected from the same culture and performed transcriptomic analysis after carefully separating each cell type. We believe that this strategy enabled us to more reliably detect subtle metabolic and structural differences occurring during the transition from planktonic to sessile cells. Our main goal was to contribute to the understanding of even subtle metabolic, structural and regulatory changes occurring in R. etli during the process of biofilm formation, in contrast to previous reports examining only the planktonic and sessile phases. Thus, our approach focuses on differential gene expression during the change of phases, with the aim of understanding the expression changes needed for biofilm development in nature.
Methods
Biofims formation assay
Biofilm formation was assayed by the ability of R. etli cells to adhere to the wells of 96-well PVC microliter dishes (Falcon 3911 Microtest III flexible assay plate; Becton–Dickinson Labware, USA) using a modification of a previously reported protocol (O’Toole and Kolter 1998a). One-hundred microliters per well of rich medium [PY; (3 g/L yeast extract, 5-g/L bacto peptone and 7 mM CaCl2·2H2O)], or minimal medium (MM) with succinate or glucose as carbon source, was inoculated to an initial OD540 of 0.05, and the plates were without shaking incubated at 29 °C. Culture growth (OD540) was determined at intervals with a microplate reader (Bio Rad Model 3550-UV). For biofilm assays, unattached cells were removed by thoroughly rinsing with water, and then, 125 μl of a 1 % aqueous solution of crystal violet was added to each well (this dye stains cells but not PVC). The plates were incubated at room temperature for 15 min and rinsed repeatedly and thoroughly with water and scored for the formation of biofilm. Growth curves were determined at 29 °C without shaking. OD540 readings were taken over time using a Microplate Reader Bio Rad Instruments Model 3550-UV USA. The amount of CV bound to the biofilm (and therefore the amount of biofilm material) can be quantified via solubilization of CV in dimethyl sulfoxide (DMSO) and subsequently measuring the absorbance of each sample at 570 nm (O’Toole and Kolter 1998a).
Twitching and flagellar motility assays
Twitching motility was assessed by stabbing cells into a thin (2-mm) PY agar plate (1.5 % agar) according to O’Toole and Kolter (1998a). A haze of growth at the agar-plate interface and the characteristic flat spreading colony morphology indicates strains having twitching motility. Flagella-mediated motility was assessed on PY medium with 0.3 % of agar at 29 °C.
Planktonic and biofilm culture for transcriptomic and microscopy analysis
R. etli CFN42 cultures were grown in 250-ml Erlenmeyer flasks containing 15 ml liquid PY medium and 40 g of glass pearls as an abiotic adhesion surface. Cultures were inoculated at an initial OD540 of 0.2 (Beckman DU 800 spectrophotometer, USA). Cultures were incubated at 29 °C with 60 % relative humidity in a Biotronette Plant Growth Chamber (Lab-line Instruments, INC. USA), without shaking. The same culture served as a source of both planktonic and sessile cells, which were separated after 24 and 72 h of incubation by decantation. The supernatant contained the planktonic cells. The pearls containing adhered cells were gently washed with DEPC water to remove remaining planktonic cells. Sessile cells were re-suspended from the washed pearls in 15 ml DEPC water with vigorous shaking. Pools were made of 10–15 suspensions of sessile and planktonic cells to obtain a total of 150 ml of sample, in triplicate for each condition. These were centrifuged (13,000g 10 min) to concentrate the cells, and RNA later (AMBION) was added.
For confocal microscopy analyses, cells were grown in 250-ml Erlenmeyer flasks with 30 ml PY medium containing a microscope slide, which served as a surface for biofilm formation.
Microscopy analysis
Microscopic slides with biofilms attached on glass slides after 24 and 72 h of incubation the slides were removed and transferred to 50-ml conical tubes and washed once with PBS and stained for 30 min, using the BacLight LIVE/DEAD staining system according to the manufacturer’s protocol (#L7007, Molecular Probes), washed once with PBS and observed with a Zeiss LSM 510 META confocal laser scanning microscope equipped with 488-, 514-, 543- and 633-nm laser lines.
RNA extraction and cDNA synthesis
RNA was extracted from 100 ml of planktonic and sessile cell preparations. Total RNA was isolated by acid hot phenol extraction as described by de Vries et al. (1989). RNA concentrations were determined by their absorbance at 260 and 280 nm. RNA integrity was determined by running samples on a 1.3 % agarose gel. Purified RNA (10 µg) was used as a template for cDNA synthesis, which was differentially labeled with Cy3-dCTP and Cy5-dCTP using a CyScribe first-strand cDNA labeling Kit (Amersham Biosciences).
Microarray experiments
The whole-genome oligonucleotide set on the array contained 6,034 70-mers, representing all (6034) predicted R. etli open reading frames (ORFs) (Salazar et al. 2010). Microarray experiments were carried out using three biologically independent RNA samples prepared at 24 and 72 h from both biofilm formation stages (planktonic and sessile). Pairs of Cy3- and Cy5-labeled cDNA samples were mixed and hybridized on the array (Hegde et al. 2000; Locke et al. 2015). A second hybridization was performed using a dye swap, reversing the assignment of Cy3 and Cy5 to sessile or planktonic samples. The arrays were washed and scanned using a pixel size of 10 μm with a ScanArray Lite microarray scanner (Perkin-Elmer, Boston, MA). Microarray data have been deposited at the NCBI gene expression and hybridization data repository (http://www.ncbi.nlm.nih.gov/geo/), (GEO accession number GSE67656).
Statistical analysis
Spot detection, determination of mean signals and mean local background intensities, image segmentation, and signal quantification were performed on the microarray images using the Array-Pro Analyzer 4.0 software (Media Cybernetics, L.P.).
Microarray data were analyzed to assess the statistical significance of expression differences between planktonic and sessile cells after 24 and 72 h of growth in conditions favorable for biofilm maturation. The analysis consisted of three steps: background subtraction and data normalization, analysis of variance and controlling for the expected number of false positives (Quackenbush 2002). Microarray data were normalized to have comparable signal values between the different slides. After background subtraction, data were quantile-normalized using the q-spline method (Workman et al. 2002), attaining signal intensities with the same distributions between all microarray slides in the red and green channels. The signals were transformed to logarithmic scale (log2), and the resulting signals were examined using M–A plots (Supplementary Material Figure S1), where no intensity-dependent artifacts were identified.
To determine the significance of expression differences between planktonic and sessile samples, an ANOVA test was conducted for each gene at 24 and 72 h. The ANOVA model used to fit the data was:
where r gabj is the normalized intensity for gene g (g = 1,…,6038), treatment a (a = 0, 1), probe b (b = 1,…,12,076) and array j (j = 1,…,3). Treatments 0 and 1 correspond to planktonic and sessile samples, respectively. The g subindex in the model shows that the model was fitted gene by gene. T ga is the main effect of the treatment for gene g, P gb is the main effect of each probe, TPgab is the probe-specific treatment effect on each gene, A gj is the main effect of the array slide on each gene, and APgjb is the interaction effect between array and probe. We assumed that the error term γ gabj was normally distributed with zero mean and variance σγ 2. The term of interest is T ga, which expresses the differential expression between the two metabolic states, biofilm and planktonic.
The ANOVA resulted in one p value for each of the 6038 genes for each time point. These p values indicate the probability that the samples from both origins (planktonic and sessile) came from equal populations, given their differences in expression. Since thousands of hypotheses were tested, it is expected that a number of low p values were obtained only by chance (false positives), while others reflect real expression changes. The q value method (Storey and Tibshirani 2003) was used to control the fraction of false-positive results, called false discovery rate (FDR). This method allowed us to determine a p value threshold that corresponded to a FDR of 0.05. Thus, an expected 5 % of false positives were accepted.
Real-time PCR measurements
To validate the results of the microarray experiments, several genes showing expression changes were analyzed by quantitative real-time PCR (qRT-PCR), and oligonucleotides were designed using the Primer Express 3.0 program from Applied Biosystems and synthesized at the “Unidad de Síntesis del Instituto de Biotecnología, UNAM.” RNA was incubated with 1 U μg−1 RNase-free DNase I according to the manufacturer’s protocol (Fermentas Life Sciences), and the absence of DNA contamination was confirmed by PCR. Amplifications were performed with the Thermoscript RT-PCR system (Invitrogen) with a reduced number of cycles in order to avoid the reaching plateau DNA amplification reaction. The relative quantification of gene expression was done using the fixL gene. This is reference gene to use gene expression. The normalization our data is an important step to control experimental errors. In biofilm conditions, the expression fixL is constitutive and we used as an endogenous control. fixL primer sequences for forward and reverse were F<CGGGACGATCACGCACTG>, R<CGGGACGATCACGCACTG>, respectively. qRT-PCR was used to acquire an independent assessment of the expression of selected genes, under the same growth conditions as for the microarray analysis.
The following primers were used: flagellin synthesis regulator (repressor) protein-related gene (RHE_CH00683): F<GGGCAAGGACCACTCGACGG>, R<GCGTAGAGACCGCGGATGGC>; multidrug efflux system, acriflavine resistance protein-related gene (RHE_CH03348): F<GGCTTCACCAACTCCTCGAA>, R<CCAGAAAGGTCCGGTGTCTTAC>; and malate dehydrogenase (oxaloacetate decarboxylating; NADP+) protein-related gene (RHE_CH02355): F<TCTACGAGGGCCGCACCGAA>, R<CGCCAGCAGCTCCGGTTTCA>. Each reaction mixture contained 12.5 μl SYBR green PCR master mixture (Applied Biosystems), 3.5 μl H2O, forward and reverse primers in 5 μl at 5 pM concentration, and 4 μl of the template. PCRs were performed with the ABI Prism 7700 sequence detection system (Applied Biosystems) using the following program: start at 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 62 °C for 1 min. The dissociation protocol was 95 °C for 15 s and 62 °C for 20 s, followed by a ramp from 62 to 95 °C for 20 min. C t values for the target genes were normalized using fixL as reference for relative quantification. The data were analyzed using the ΔΔCT method, and the data are expressed as relative expression level. Each sample was assayed three times.
Functional classification based on COG annotations and interacting partners of gene product prediction
Differentially expressed genes in the planktonic and biofilm stages were classified using the Clusters of Orthologous Groups (COG) annotations (Tatusov et al. 2003). Prediction of interacting partners of the protein products of certain genes was done using the STRING database using default conditions.
Results and discussion
Biofilm formation
The effect of growth phase and nutritional requirements on biofilm development in R. etli was examined. Cells were grown at room temperature without shaking in PVC microtiter plates and biofilm formation quantitated by crystal violet staining as described in “Materials and methods”). The ability to form biofilms was strongly influenced by the nutritional environment. Biofilm formation occurred in PY and in minimal media containing casamino acids (CAAs), although less efficiently (Fig. 1). In contrast, minimal media without CAA (with succinate as carbon) did not support biofilm formation (Fig. 1). Rhizobia confront soil environments ranging from rhizospheres rich in nutrients and root exudates to soils deficient in nitrogen, phosphates, water and other nutrients. Our results suggest that soil containing diverse carbon sources would favor biofilm formation by R. etli. For this reason, we used rich medium (PY) to perform additional biofilm formation studies.
Nutritional effects on biofilm formation. R. etli CFN42 cells are grown at 29 °C without shaking for 48 h in PVC microtitre dishes containing: a succinate–ammonium, b glucose–casamino acids, c succinate–casamino acids and d rich medium (PY). Biofilms are stained with crystal violet as described in “Materials and methods”
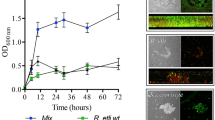
Biofilm development begins with an initial attachment of planktonic cells to a surface. This is followed by cell division and the recruitment of additional planktonic cells to form a complex three-dimensional structure consisting of exopolysaccharide-encased microcolonies punctuated by aqueous channels. This stage typically represents the mature biofilm structure. We indirectly monitored biofilm development by following the increase in CV staining over time and found R. etli initiates biofilm formation in saturated stationary-phase cultures after 24 h (Fig. 2). We chose this time point to represent the surface attachment phase for our transcriptomic analysis. In addition, we analyzed gene expression in mature 72-h biofilms (Fig. 2).
Examination of 24- and 72-h biofilms using confocal laser scanning microscopy (CLSM) showed that at 24 h, the biofilm structure is not yet stable (Fig. 3a). At this stationary growth phase, the limited availability of nutrients could be one of the triggers initiating the biofilm formation process. This is supported by previous report showing that biofilm formation was enhanced by nutrient limitation in R. leguminosarum (Fujishige et al. 2006). Moreover, it has been shown that in the symbiosis between Bradyrhizobium sp. and peanut, the attachment level varies depending on the metabolic state of the rhizobia (Rinaudi and Giordano 2010). Similarly as our case, in another study, an optimal attachment was observed when cells were harvested at the late log or the early stationary phase (Dardanelli et al. 2003). Nutrient availability thus appears to play a major role in the transition from a planktonic to a sessile mode of life, similar to our findings with in R. etli.
Confocal laser scanning microscopy and LIVE/DEAD staining system are used to identify the phases of biofilm formation in R. etli. a At 24 h after incubation, cells are visualized in initial attachment; most microscopic fields had no cells attached at all. b At 72 h after incubation, images staining showed a biofilm more structured, and biofilm maturation is accompanied by significant alterations in cell viability in the different biofilm layers. Below the original signal, a Z projection is shown
At 72 h, the CLSM obtained after LIVE/DEAD images staining showed that biofilm maturation is accompanied by significant alterations in cell viability in the different biofilm layers (Fig. 3b). A decrease in viability might be due to major restrictions in nutrient availability and accumulation of toxic metabolites as the biofilm thickens. It is expected that differential gene expression would allow the bacteria to adapt to different ecological microenvironments. Recent studies have shown that the regulated death of bacterial cells is important for biofilm development. Following cell death, a subpopulation of the dead bacteria lyse and release genomic DNA, which has a central role in intercellular adhesion and biofilm stability (Bayles 2007).
Genes expressed in the initiation of biofilm formation
As described above, the biofilm formation process starts at about 24 h. This observation was supported with the CLSM images, and where we can observe that the adhesion of microbes to the surface is beginning (Fig. 3a). Diverse genes having changes in their expression at this time point were identified (Supplementary material Table S1-S2) and could be involved in the establishment of the biofilm. One such gene that was upregulated in sessile cells is raiR, homologous to raiR of Rhizobium leguminosarum whose product is a transcriptional activator of the LuxR family. Many rhizobia use N-acyl-homoserine lactone (AHL)-based quorum sensing (QS) systems to monitor their population density and regulate their symbiotic interactions with their plant hosts (Fuqua and Winans 1994; Zarkani et al. 2013; Oh and Choi 2015). In R. etli CFN42, a knockout mutation in raiR causes lower levels of fixation activity (Zheng et al. 2015). In R. leguminosarum, the inactivation of praR has increased competitiveness for nodule infection, increased biofilms formation on pea roots and required the rap and gmsA genes (Frederix et al. 2014). Previously, the QS response has been implicated in biofilm formation in R. etli CNPAF512 (Daniels et al. 2006) and the overexpression of raiR in early steps of biofilm formation in CFN42 suggests its role in this process by enhancing QS signaling during early biofilm formation.
We observed an upregulation of RHE_PE00141, encoding a putative trifolitoxin immunity protein (TFX). TFX is homologous to a peptide antibiotic produced by R. leguminosarum bv. trifolii T24 that inhibits growth of members of the alpha proteobacteria. It has been reported that treatment of R. etli CE3 with TFX significantly increases nodule occupancy and competitiveness in field-grown plants (Robleto et al. 1998). This finding suggests that R. etli CFN42 expresses this gene, possibly in preparation for the biofilm formation.
Eight genes were overexpressed in planktonic cells, including a putative pilA pilus assembly protein (RHE_CH00202) that participates in the synthesis of pili. Although not formally assigned to this category, RHE_PE00148 encodes another putative pilus assembly protein that was overexpressed in sessile cells. It has been shown in P. aeruginosa that some type of pili is required for a form of surface-associated movement called twitching motility, which does not require flagella, but depends on the extension and retraction of the pili, allowing the cells to move across a surface (O’Toole and Kolter 1998a, b). On the other hand, previous studies with S. meliloti and other alpha proteobacteria, such as Agrobacterium, Azospirillun, Caulobacter and Rhodobacter, have demonstrated flagella-mediated motility. We found that R. etli is capable of twitching motility on PY motility medium containing 0.3 % agar (Fig. 4). This ability could be important in the first stages of biofilm formation. Pratt and Kolter (1998) suggest that pili and flagella could be directly required for attachment to abiotic surfaces, thus facilitating the initiation of biofilm formation, and also might be required for the bacteria within a developing biofilm to move along the surface.
Twitching motility assays of R. etli. Thin agar plates (1.5 %) are stab inoculated with a toothpick to the bottom of the plate and incubated for 48 h at 30 °C. The light haze of grow that the agar-plate interface is a measure of twitching motility. The smaller, denser zone represents surface colony growth
Transcriptomic analysis shows that a large number of genes are differentially expressed in sessile and planktonic cells at 72 h
Significant changes in gene expression were observed at 72 h, corresponding to a mature-stage biofilm (Fig. 3b and Supplementary material Tables S3–S4). In biofilm-attached and planktonic cells, 209 and 273 genes, respectively, were overexpressed, representing 3.3 and 4.4 % of the total genes in R. etli.
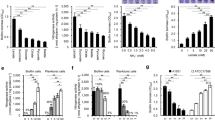
The classification of the biofilm and planktonic overexpressed genes based on their COG categories is shown in Fig. 5. The majority of genes belonged to the “General function prediction only” (18 and 26 genes), “Function unknown” (20 and 21 genes) and “Not in COGs” (62 and 74 genes). Other COGs highly represented in one or both conditions (sessile and planktonic) were transcription (K), with 7 and 14 genes, respectively; replication, recombination and repair (L), 8 and 15 genes; energy production and conversion (C), 10 and 9 genes; carbohydrate transport and metabolism (G), 16 and 23 genes; and amino acid transport and metabolism (E), 19 and 24 genes (Supplementary material Table S5). In the following sections, we describe the genes most relevant to biofilm formation that were identified in this analysis.
Differentially expresed genes classified in COG categories. All the genes in each functional group are considered, and numbers of gened with expression changes are shown as proportion. Fractions represented are overexpressed in sessile cell (black), overexpressed in planktonic cells (gray) and without changes in expression (dotted)
Information storage and processing
Translation, ribosomal structure and biogenesis (J)
In sessile cells, six genes in this category were upregulated, including RHE_CH00602, a probable translation initiation inhibitor protein. Using the STRING database, we found that RHE_CH00602 interacts with RHE_CH00601, a TetR family transcriptional regulator. This protein is involved in the transcriptional control of multidrug efflux pumps, pathways for the biosynthesis of antibiotics, response to osmotic stress and toxic chemicals, control of catabolic pathways, differentiation processes and pathogenicity (Ramos et al. 2005). STRING also predicts an interaction of this gene with the sigma factor rpoZ as well as a probable interaction with RHE_CH02879, a putative translation initiation inhibitor protein that allows the bacteria to conserve energy. However, experimental evidence of these interactions is not available so far. These results are consistent with nutrient depletion in the biofilm generating stress conditions, causing the bacteria to downregulate translation to save energy and maintain cellular homeostasis (Metselaar et al. 2015).
In planktonic cells, we identified six overexpressed translational genes (Supplementary material Table S4), including a tRNA 5-methylaminomethyl-2-thio uridylate-methyltransferase protein-related gene (mnmA), which is conserved in the anticodon wobble position of the tRNAs (Numata et al. 2006). STRING predicts an interaction of this gene with lepA, which is required for accurate and efficient protein synthesis under certain stress and with the translation initiation factor IF-1 (encoded by infA) which was also detected overexpressed. In this context, an orthologous factor seems to stimulate the activities of IF-2 and IF-3 in the bacterium Escherichia coli (Belotserkovsky et al. 2011). In E. coli, the translation initiation mechanism requires, in addition to mRNA, fMet-tRNA and ribosomal subunits, the presence of three additional proteins (initiation factors IF-1, IF-2 and IF-3) and at least one GTP molecule (Celano et al. 1988). The infA gene encodes initiation factor I in E. coli, which is a small protein involved in the initiation step of protein synthesis. The protein level of IF-1 increases when E. coli cells are subjected to cold shock (Ko et al. 2006). Because these proteins share similarity at sequence level (based on PFAM assignation), it is possible that the overexpression of infA also plays a significant role in protein synthesis in planktonic conditions. These results propose a continued protein synthesis in planktonic cells. In contrast, in P. fluorescens a continued protein synthesis is not required after the initial events of biofilm formation on an abiotic surface (O’Toole and Kolter 1998b), suggesting that the earliest events of biofilm development can be divided into two stages. The initial interaction with the abiotic surface requires protein synthesis. However, the subsequent stage (short-term maintenance of the attached cells) does not require synthesis of new proteins (O’Toole and Kolter 1998b).
Transcription (K)
In the sessile phase, seven genes related to transcription were overexpressed, including rpoA, which encodes the DNA-directed RNA polymerase alpha subunit (Jafri et al. 1995; Ribeiro et al. 2009). In Agrobacterium tumefaciens, rpoA interacts with TraR, a quorum sensing (QS) activator to activate gene expression (Qin et al. 2004). This is the first report describing a possible role for rpoA in biofilm formation in R. etli CFN42. Genes such as RHE_CH02976, encoding products belonging to the LysR family of transcriptional regulators, were also overexpressed, and further analyses are required to characterize their function.
rpoE4 is overexpressed in sessile cells and is a master regulator in bacteria, and in R. etli, it is upregulated under oxidative, saline and osmotic stress, and microaerobic and stationary-phase growth (Martinez-Salazar et al. 2009a, b). In aggregated cells, the low availability of oxygen and nutrients could provoke stress, causing rpoE4 upregulation. Probable transcriptional factors from the AsnC, LysR and TetR families were also overexpressed in biofilms (Supplementary material Table S3), and this is the first report associating these proteins with biofilm formation in R. etli CFN42.
In planktonic cells, fourteen genes were overexpressed and some of them were identified as probable transcriptional regulator, three LysR families, one LuxR, LacI and AsnC family. Among these, the rpoH1 RNA polymerase factor sigma-32 stands out. This sigma factor is associated with stress responses in diverse organisms, such as in the S. meliloti 1021 pH stress response (de Lucena et al. 2010). A R. etli rpoH1 rpoH2 double mutant had increased sensitivity to heat shock and oxidative stress when compared with the rpoH1 single mutant. This suggests that in R. etli, RpoH1 is the main heat-shock sigma factor, but a more complete protective response could be achieved with the participation of RpoH2 (Martinez-Salazar et al. 2009a, b). The overexpression of rpoH1 could be associated with stresses experienced in both metabolic states. The RNA polymerase sigma factor 24 (rpoE2) was also overexpressed in planktonic cells, and its homologs in S. meliloti 1021 are associated with osmotic stress-related genes (Flechard et al. 2010). Other probable transcription factors identified in the planktonic phase included some belonging to the LysR, AsnC and LuxR families. LysR proteins regulate diverse stress conditions, and we suggest that they may function in regulating genes for resisting stresses unique to the planktonic state.
In summary, we show that different stress-related proteins were induced in the planktonic and sessile states, both of which appear to be stressful conditions. Stresses occurring in planktonic cells are expected to include those caused by long culture age and nutrient limitation, which could predispose them to attach to surfaces. These could represent early changes in gene expression occur before the initiation of biofilm formation. On the other hand, the sessile cells also showed expression changes indicative of a stress response, such as the overexpression of rpoE4, suggesting that biofilm formation involves adjustments to specific stresses. As mentioned, limitations in nutrient availability in high cell density biofilms could play a role in triggering this response.
Cellular processes and signaling
Signal transduction mechanisms (T)
The five genes that were overexpressed in sessile cells include ntrC, encoding a global activator of nitrogen assimilation that allows cells to take up ammonia (Leigh and Dodsworth 2007; Yeom et al. 2010). Analysis with the STRING database indicated that ntrC interacts with two other genes for nitrogen regulation: the two-component sensor histidine kinase ntrB and RNA polymerase factor sigma-54 (rpoN). Vibrio vulnificus has been shown to require ntrC for mature biofilm development via controlling lipopolysaccharide and exopolysaccharide biosynthesis (Kim et al. 2009). In addition, in Rhodospirillum rubrum the phosphorylated form of NtrC acts as a transcriptional activator of nifA and glnA, and other genes involved in nitrogen fixation and assimilation (Zhang et al. 2005) and may play a similar role in R. etli.
The nodW gene was also overexpressed in sessile condition. NodW in B. japonicum positively regulates nod gene expression in response to plant-produced isoflavones (Loh et al. 1997) and is thus critical for Nod-factor production and nodulation, as has been shown in B. japonicum and other rhizobia (Jitacksorn and Sadowsky 2008). Nod factors are indispensable for the development of nitrogen-fixing nodules (Lerouge et al. 1990) and are critical for the establishment of a mature rhizobial biofilm (Fujishige et al. 2008). These dual functions of Nod factors involve different control mechanisms: one dependent on flavonoid induction (for nodulation) and the other independent of flavonoids for biofilm production (Rinaudi and Giordano 2010).
Six signal transduction genes were overexpressed under planktonic conditions, including, phoR, a two-component sensor histidine kinase required for phosphate acquisition in E. coli grown under phosphate limitation (Gardner et al. 2014). The induction of phoR in planktonic conditions probably reflects phosphorus deficiencies caused by prolonged growth in liquid medium, a condition that in other bacteria is needed prior to biofilm establishment (Israel 1987; Jayasinghearachchi and Seneviratne 2006).
Genes associated with motility (N)
Several genes associated with motility were overexpressed under sessile conditions (Supplementary material Table S3). These included flbT, which encodes the flagellin synthesis repressor protein present in most Gram-negative bacteria including Rhizobeaceae, where it promotes the degradation of flagellin mRNA (Anderson and Gober 2000). During biofilm formation, motility is required in the initial stages, while in late stages it may destabilize the biofilm structure (O’Toole and Kolter 1998a; Rasamiravaka et al. 2015). Once cells are established in biofilm, the metabolic cost of flagella synthesis is probably prohibitive, leading to its repression (Smith and Chapman 2010; Dressaire et al. 2015). We suggest that flbT could be associated with cellular aggregation because its role in post-transcriptional control of flagellin expression (Belas et al. 2009). In Rhizobia, flagellar motility allows access to attachment sites on the plant, and flagellar motility is also involved in biofilm maturation. In S. meliloti, flagellar mutants are reduced in biofilm formation and delayed in nodule formation (Fujishige et al. 2006).
Genes encoding chemotaxis methyltransferases (cheRCh2, mcpZch1 and mcpV2) were also overexpressed in sessile cells, and methyl-accepting chemotaxis proteins (MCPs) are localized in the cytoplasmic membrane and typically consist of a periplasmic ligand-binding domain, two transmembrane helices, and a highly conserved cytoplasmic signaling domain. The proteins encoded by those genes could modulate chemotaxis. According to STRING, the products of cheRCh2 and cheBch2 are interacting chemotaxis-specific methylesterase protein. In this regard, motility and chemotaxis impact different important aspects of the biofilm formation, including dispersion and re-attachment, increasing the efficiency and frequency of surface sampling by the bacteria (Schmidt et al. 2011).
Under planktonic conditions, four genes related to flagellum synthesis were overexpressed, including flaCch4 (flagellin C protein) and flgLcl (flagellar hook-associated protein). In contrast to sessile conditions, the planktonic state expression of these genes allows motility, enabling the bacteria to obtain nutrients, evade host defense systems and improve the ability to contend with hostile environments (Ottemann and Miller 1997). As mentioned, pili and flagella are involved in the attachment and microcolony formation steps of biofilm development in R. etli. Our results suggest that flagella may not be required for the maintenance of a mature biofilms but that they are involved in early steps in biofilm development.
Metabolism
Carbohydrate transport and metabolism (G)
Sixteen genes in this functional category were overexpressed in sessile cells. These include, like ndvB, encoding beta (1–>2) glucan biosynthesis protein that could contribute to the formation of the cellular matrix in biofilms. In P. aeruginosa, ndvB gene is also involved in producing glycerol-phosphorylated beta-(1–>3)-glucans that bind aminoglycosides and provide high-level antibiotic resistance (Sadovskaya et al. 2010). The gene encoding the permease protein of a trehalose maltose ABC transporter (thuG) also had increased expression in planktonic cells. thuG in S. meliloti is part of a six-gene cluster involved in trehalose transport and utilization as an energy source (Jensen et al. 2002). All of these genes are important for root colonization, but no clear participation in nodulation or in nitrogen fixation has been reported (Jensen et al. 2002). In R. etli, this operon is conserved, but only thuG overexpression was observed (Table S3). ThuG could be associated with the adhesion system of R. etli in biofilm formation.
Finally, genes related to transport systems (probable sugar ABC transporters) were overexpressed in sessile cells and their products could be used in sugar import for building the extracellular matrix, as well as for metabolism.
In planktonic cells, twenty-three genes in this category were overexpressed, including some related to sugar transport, such as gguB, encoding the permease of a xylose ABC transporter, and glgA, encoding glycogen synthase. During the transition from the planktonic to the biofilm state, the products of these genes could participate in the synthesis of matrix polysaccharides necessary for biofilm formation, import of carbohydrate for use as an energy source, or glycogen synthesis.
Downregulated metabolic genes detected during biofilm formation, included those related to carbohydrate transport and metabolism (19 out of 20 differentially expressed genes), energy conversion and production (17 out of 21 differentially expressed genes), nucleotide transport and metabolism (one out of six differentially expressed genes), and cell motility (eight out of eight differentially expressed genes). This is consistent with reports of a decreased metabolism in sessile cells (Javaherdashti et al. 2013).
Nucleotide transport and metabolism (F)
In this COG, cells in biofilms differentially expressed 6 genes, 1 upregulated and 5 downregulated), as compared to planktonic cells. Sessile cells only overexpressed gmk2, encoding guanylate (GMP) kinase (GMK). GMP kinase catalyzes the ATP-dependent phosphorylation of GMP to produce GDP and is essential for GMP recycling and the indirect regulation of cGMP levels. In prokaryotes like E. coli, lower eukaryotes, such as yeast, and in vertebrates, GMK is a highly conserved monomeric protein required for the synthesis of nucleotide precursors and thus indirectly modulating the synthesis of DNA and RNA. In P. aeruginosa, bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) controls the expression of genes required for biofilm formation and dispersal (Chua et al. 2015). The expression of this gene and its participation on the specific metabolites synthesis could be part of the metabolic predisposition on planktonic cell previous to their transformation into sessile cells.
In planktonic cells, five genes were overexpressed including, ndk (nucleoside-diphosphate kinase), which is associated with quorum sensing in P. aeruginosa (Kamath et al. 1998), and adeC2 (adenine deaminase), whose product degrades adenosine. Exogenous adenosine strongly inhibits biofilm formation in P. aeruginosa and Acidovorax sp., suggesting that adenosine could act as a biofilm formation inhibitor. Acyl homoserine lactones (AHLs) induce the expression of adenosine deaminase in these organisms (Kusada et al. 2014). We confirmed ndk expression in planktonic cells using RT-PCR and suggest that its overexpression could be the part of the initial metabolic changes in planktonic cells that occurs prior to surface attachment.
Inorganic ion transport and metabolism (P)
It is likely that bacteria in the biofilms suffer from iron limitation. In biofilm cells, afuB1, encoding the permease protein of an iron (III) ABC transporter, was overexpressed. We previously reported the central role of iron in the formation of mixed biofilms by Candida albicans and P. aeruginosa, where iron concentration alters the expression of several proteins during mixed biofilm formation (Andrade-Dominguez et al. 2014). Iron limitation also stimulates the production of AHLs, a major class of autoinducer signals used by Gram-negative bacteria to regulate biofilm formation (Modarresi et al. 2015). Thus, iron limitation could be a signal to produce more the AHLs, which would induce biofilm formation.
Poorly characterized
General function prediction only (R)
Under sessile conditions, we identified eighteen overexpressed genes in this COG, including RHE_CH03381, encoding a probable exopolysaccharide (EPS) polymerization/export protein; EPS is required for biofilm formation, for example, in S. meliloti and M. tianshanense (Fraysse et al. 2003; Wang et al. 2008). Inactivation of genes for EPS polymerization and translocation in R. leguminosarum bv. viciae (Young et al. 2006) and bv. trifolii (Laus et al. 2004) show that these genes are essential for biofilm formation. In the STRING database, the RHE_CH03381 protein associates with the PssP (exopolysaccharide polymerization) ExoY, and PssA3 (exopolysaccharide production) proteins, which could be essential for biofilm formation. pssA is required for EPS synthesis, and its inactivation in R. leguminosarum causes a drastic decrease in biofilm formation in static cultures (Russo et al. 2006). In planktonic conditions, 26 genes were overexpressed including exoR2, encoding a negative regulator of EPS production. This might act to prevent the unnecessary production of EPS by cells once they have formed biofilms.
Function unknown (S)
In sessile and planktonic cells, 20 and 21 genes, respectively, in this category were overexpressed. The analysis and study of those proteins will allow the discovery of new functions in the bacteria. We carried out a computational search for domains (http://www.uniprot.org) and found that in sessile cells at least three genes (RHE_CH01640, RHE_CH01940 and RHE_CH01503) encode proteins with transmembrane regions (Supplementary material Table S6). In planktonic cells, we identified gene products with (1) PIN domains involved in virulence, (2) peptidases, (3) vitamin synthesis, (4) toxins degradation and (5) antibiotic biosynthesis (Supplementary material Table S6). The presence of these domains in the products of genes overexpressed in sessile or planktonic cells indicates important differences in defense mechanisms or metabolic processes between the two cell types. Additional experiments are needed to determine their roles.
NOT in COG
Under sessile conditions, 61 of the overexpresed genes were assigned this COG, the majority of which were hypothetical proteins. However, the gene product of RHE_PE00148 (putative pilus assembly protein) could be important in biofilm formation, since, as mentioned, flagella and pili are necessary for the initial surface attachment (O’Toole and Kolter 1998a). Flagella or flagella-mediated motility appears to be important for the formation of a bacterial monolayer of the abiotic surface, while pili appear to play a role in downstream events such as microcolony formation and have been shown to be important for the adherence and colonization to eukaryotic cell surfaces and are thought to play a role in pathogenesis (O’Toole and Kolter 1998a, b).
The product of another notable overexpressed gene, rapB2, belongs to the RAP (Rhizobium-adhering proteins) family (Ausmees et al. 2001). RAP proteins associated with calcium promote rhizobial autoaggregation, root attachment and rhizosphere colonization (Dardanelli et al. 2003; Mongiardini et al. 2008). We found that the casA gene encoding a calcium binding protein was overexpressed, and this, along with rapB2, could have an important role during the cellular attachment to surfaces.
Validation of gene expression by qPCR
We quantified the expression of three genes showing expression changes in the microarrays using qRT-PCR. These genes were selected randomly to exclude any bias in the analysis. In all cases, we found the same differential expression as in the microarrays (Supplementary Material Figure S2).
In sessile conditions, flbT, a flagellin synthesis regulator (repressor) protein, was analyzed as a reference for sessile conditions and showed a twofold increase relative to planktonic cells. mexF1, encoding a multidrug efflux protein, was 2.5-fold overexpressed relative to planktonic condition, and malate dehydrogenase (RHE_CH00672) was twofold more highly expressed under planktonic conditions. This analysis shows that our gene expression data are robust enough to detect genes whose modified expression rates are significant in the context of biofilm formation.
Concluding remarks
Microarrays are a robust method to globally evaluate gene expression in diverse organisms. Here, we used this technology in R. etli CFN42 during biofilm formation by analyzing cells in the sessile, attached, and planktonic states.
This is the first report closely analyze gene expression during biofilm formation in R. etli, using planktonic and sessile cells collected from the same experiment culture, in contrast to previous studies where the metabolic, physiological and structural comparisons were analyzed using cells obtained under different experimental conditions. We believe that this approach allowed us to detect subtle differences in gene expression taking place throughout the transition from the planktonic to sessile phase during biofilm initiation. We identified various sets of differentially expressed genes specific for each condition. The fold changes observed in our study were small as compared to many other microarray studies. This is due to our focus in the early stages of the process, with planktonic cells that might already be preparing for attachment. Nevertheless, the changes observed were statistically significant, and independent validations were successful. Thus, we have confidence of the relevance of these expression changes, which are as subtle as the phenomenon being studied.
In the biofilm phase, overexpressed genes included those encoding proteins with diverse functions, such as membrane synthesis, transport and chemotaxis, EPS and lipopolysaccharide, AHL synthesis, autoaggregation and cellular attachments components as well as a repressor of flagellin synthesis, all of which are potentially crucial for biofilm formation. In contrast, planktonic cells overexpressed genes associated with transport, motility (flagellar and twitching) and the inhibition of EPS synthesis. The results of this work give insight into how the bacteria modify their metabolism by the expression of genes with opposing functions prior to and during the transition between planktonic and sessile stages. One of the principal contributions of this work was detecting changes in the expression of symbiosis-related genes such as nodW and ntrC. This work provides a basis for future studies that will describe gene expression changes during the early stages of the symbiotic relationship with P. vulgaris.
In summary, this study extends our understanding of the physiological, metabolic and structural requirements during the transition from the planktonic to sessile states. Further studies of transcriptional profiles with selected gene deletion mutants will elucidate the functions of these gene products during biofilm formation.
References
Anderson PE, Gober JW (2000) FlbT, the post-transcriptional regulator of flagellin synthesis in Caulobacter crescentus, interacts with the 5′ untranslated region of flagellin mRNA. Mol Microbiol 38:41–52
Andrade-Dominguez A, Salazar E, Vargas-Lagunas Mdel C et al (2014) Eco-evolutionary feedbacks drive species interactions. ISME J 8:1041–1054. doi:10.1038/ismej.2013.208
Ausmees N, Jacobsson K, Lindberg M (2001) A unipolarly located, cell-surface-associated agglutinin, RapA, belongs to a family of Rhizobium-adhering proteins (Rap) in Rhizobium leguminosarum bv. trifolii. Microbiology 147:549–559
Bayles KW (2007) The biological role of death and lysis in biofilm development. Nat Rev Microbiol 5:721–726. doi:10.1038/nrmicro1743
Belas R, Horikawa E, Aizawa SI, Suvanasuthi R (2009) Genetic determinants of Silicibacter sp. TM1040 motility. J Bacteriol 191:4502–4512. doi:10.1128/JB.00429-09
Belotserkovsky JM, Dabbs ER, Isaksson LA (2011) Mutations in 16S rRNA that suppress cold-sensitive initiation factor 1 affect ribosomal subunit association. FEBS J 278:3508–3517. doi:10.1111/j.1742-4658.2011.08272.x
Celano B, Pawlik RT, Gualerzi CO (1988) Interaction of Escherichia coli translation-initiation factor IF-1 with ribosomes. Eur J Biochem 178:351–355
Christensen LD, Moser C, Jensen PO et al (2007) Impact of Pseudomonas aeruginosa quorum sensing on biofilm persistence in an in vivo intraperitoneal foreign-body infection model. Microbiology 153:2312–2320. doi:10.1099/mic.0.2007/006122-0
Chua SL, Hultqvist LD, Yuan M et al (2015) In vitro and in vivo generation and characterization of Pseudomonas aeruginosa biofilm-dispersed cells via c-di-GMP manipulation. Nat Protoc 10:1165–1180. doi:10.1038/nprot.2015.067
Danhorn T, Fuqua C (2007) Biofilm formation by plant-associated bacteria. Annu Rev Microbiol 61:401–422. doi:10.1146/annurev.micro.61.080706.093316
Daniels R, Reynaert S, Hoekstra H et al (2006) Quorum signal molecules as biosurfactants affecting swarming in Rhizobium etli. Proc Natl Acad Sci USA 103:14965–14970. doi:10.1073/pnas.0511037103
Dardanelli M, Angelini J, Fabra A (2003) A calcium-dependent bacterial surface protein is involved in the attachment of rhizobia to peanut roots. Can J Microbiol 49:399–405. doi:10.1139/w03-054
Davey ME, O’Toole GA (2000) Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64:847–867
de Lucena DK, Puhler A, Weidner S (2010) The role of sigma factor RpoH1 in the pH stress response of Sinorhizobium meliloti. BMC Microbiol 10:265. doi:10.1186/1471-2180-10-265
de Vries S, Hoge H, Bisseling T (1989) Isolation of total and polysomal RNA from plant tissues. Springer, Netherlands
Donlan RM (2002) Biofilms: microbial life on surfaces. Emerg Infect Dis 8:881–890
Dressaire C, Moreira RN, Barahona S et al (2015) BolA is a transcriptional switch that turns off motility and turns on biofilm development. MBio 6:e02352-14. doi:10.1128/mBio.02352-14
Flechard M, Fontenelle C, Blanco C et al (2010) RpoE2 of Sinorhizobium meliloti is necessary for trehalose synthesis and growth in hyperosmotic media. Microbiology 156:1708–1718. doi:10.1099/mic.0.034850-0
Fraysse N, Couderc F, Poinsot V (2003) Surface polysaccharide involvement in establishing the rhizobium-legume symbiosis. Eur J Biochem 270:1365–1380
Frederix M, Edwards A, Swiderska A et al (2014) Mutation of praR in Rhizobium leguminosarum enhances root biofilms, improving nodulation competitiveness by increased expression of attachment proteins. Mol Microbiol 93:464–478. doi:10.1111/mmi.12670
Fujishige NA, Rinaudi L, Giordano W, Hirsch A (2005) Superficial liaisons: colonization of roots and abiotic surfaces by rhizobia. In: Sánchez F, Quinto C, López-Lara IM, Geiger O (eds) Biology of Plant-Microbe Interactions, Proceedings of the 12th International Congress on Molecular Plant–Microbe Interactions, St. Paul, MN, pp 292–299
Fujishige NA, Kapadia NN, De Hoff PL, Hirsch AM (2006) Investigations of Rhizobium biofilm formation. FEMS Microbiol Ecol 56:195–206. doi:10.1111/j.1574-6941.2005.00044.x
Fujishige NA, Lum MR, De Hoff PL et al (2008) Rhizobium common nod genes are required for biofilm formation. Mol Microbiol 67:504–515. doi:10.1111/j.1365-2958.2007.06064.x
Fuqua WC, Winans SC (1994) A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol 176:2796–2806
Gardner SG, Johns KD, Tanner R, McCleary WR (2014) The PhoU protein from Escherichia coli interacts with PhoR, PstB, and metals to form a phosphate-signaling complex at the membrane. J Bacteriol 196:1741–1752. doi:10.1128/JB.00029-14
Hegde P, Qi R, Abernathy K et al (2000) A concise guide to cDNA microarray analysis. Biotechniques 29:548–50, 552–4, 556 passim
Israel DW (1987) Investigation of the role of phosphorus in symbiotic dinitrogen fixation. Plant Physiol 84:835–840
Jafri S, Urbanowski ML, Stauffer GV (1995) A mutation in the rpoA gene encoding the alpha subunit of RNA polymerase that affects metE-metR transcription in Escherichia coli. J Bacteriol 177:524–529
Javaherdashti R, Nwaoha C, Tan H (2013) Corrosion and materials in the oil and gas industries. CRC Press, Boca Raton
Jayasinghearachchi HS, Seneviratne G (2006) Fungal solubilization of rock phosphate is enhanced by forming fungal–rhizobial biofilms. Soil Biol Biochem 38:405–408. doi:10.1016/j.soilbio.2005.06.004
Jensen JB, Peters NK, Bhuvaneswari TV (2002) Redundancy in periplasmic binding protein-dependent transport systems for trehalose, sucrose, and maltose in Sinorhizobium meliloti. J Bacteriol 184:2978–2986
Jitacksorn S, Sadowsky MJ (2008) Nodulation gene regulation and quorum sensing control density-dependent suppression and restriction of nodulation in the Bradyrhizobium japonicum-soybean symbiosis. Appl Environ Microbiol 74:3749–3756. doi:10.1128/AEM.02939-07
Kamath S, Kapatral V, Chakrabarty AM (1998) Cellular function of elastase in Pseudomonas aeruginosa: role in the cleavage of nucleoside diphosphate kinase and in alginate synthesis. Mol Microbiol 30:933–941
Karatan E, Watnick P (2009) Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev 73:310–347. doi:10.1128/MMBR.00041-08
Kim HS, Park SJ, Lee KH (2009) Role of NtrC-regulated exopolysaccharides in the biofilm formation and pathogenic interaction of Vibrio vulnificus. Mol Microbiol 74:436–453. doi:10.1111/j.1365-2958.2009.06875.x
Ko JH, Lee SJ, Cho B, Lee Y (2006) Differential promoter usage of infA in response to cold shock in Escherichia coli. FEBS Lett 580:539–544. doi:10.1016/j.febslet.2005.12.066
Kusada H, Hanada S, Kamagata Y, Kimura N (2014) The effects of N-acylhomoserine lactones, beta-lactam antibiotics and adenosine on biofilm formation in the multi-beta-lactam antibiotic-resistant bacterium Acidovorax sp. strain MR-S7. J Biosci Bioeng 118:14–19. doi:10.1016/j.jbiosc.2013.12.012
Laus MC, Logman TJ, Van Brussel AA et al (2004) Involvement of exo5 in production of surface polysaccharides in Rhizobium leguminosarum and its role in nodulation of Vicia sativa subsp. nigra. J Bacteriol 186:6617–6625. doi:10.1128/JB.186.19.6617-6625.2004
Leigh JA, Dodsworth JA (2007) Nitrogen regulation in bacteria and archaea. Annu Rev Microbiol 61:349–377. doi:10.1146/annurev.micro.61.080706.093409
Lerouge P, Roche P, Faucher C et al (1990) Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344:781–784. doi:10.1038/344781a0
Locke AE, Kahali B, Berndt SI et al (2015) Genetic studies of body mass index yield new insights for obesity biology. Nature 518:197–206. doi:10.1038/nature14177
Loh J, Garcia M, Stacey G (1997) NodV and NodW, a second flavonoid recognition system regulating nod gene expression in Bradyrhizobium japonicum. J Bacteriol 179:3013–3020
Martinez-Salazar JM, Salazar E, Encarnacion S et al (2009a) Role of the extracytoplasmic function sigma factor RpoE4 in oxidative and osmotic stress responses in Rhizobium etli. J Bacteriol 191:4122–4132. doi:10.1128/JB.01626-08
Martinez-Salazar JM, Sandoval-Calderon M, Guo X et al (2009b) The Rhizobium etli RpoH1 and RpoH2 sigma factors are involved in different stress responses. Microbiology 155:386–397. doi:10.1099/mic.0.021428-0
Metselaar KI, Saa Ibusquiza P, Ortiz Camargo AR et al (2015) Performance of stress resistant variants of Listeria monocytogenes in mixed species biofilms with Lactobacillus plantarum. Int J Food Microbiol. doi:10.1016/j.ijfoodmicro.2015.04.021
Modarresi F, Azizi O, Shakibaie MR et al (2015) Iron limitation enhances acyl homoserine lactone (AHL) production and biofilm formation in clinical isolates of Acinetobacter baumannii. Virulence 6:152–161. doi:10.1080/21505594.2014.1003001
Mongiardini EJ, Ausmees N, Pérez-Giménez J et al (2008) The rhizobial adhesion protein RapA1 is involved in adsorption of rhizobia to plant roots but not in nodulation. FEMS Microbiol Ecol 65:279–288. doi:10.1111/j.1574-6941.2008.00467.x
Numata T, Ikeuchi Y, Fukai S et al (2006) Crystallization and preliminary X-ray analysis of the tRNA thiolation enzyme MnmA from Escherichia coli complexed with tRNA(Glu). Acta Crystallogr Crystallogr F Struct Biol Cryst Commun 62:368–371. doi:10.1107/S174430910600738X
O’Toole GA, Kolter R (1998a) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304
O’Toole GA, Kolter R (1998b) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461
Oh MH, Choi CH (2015) Role of LuxIR Homologue AnoIR in Acinetobacter nosocomialis and the Effect of Virstatin on the Expression of anoR Gene. J Microbiol Biotechnol. doi:10.4014/jmb.1504.04069
Ottemann KM, Miller JF (1997) Roles for motility in bacterial-host interactions. Mol Microbiol 24:1109–1117
Pratt LA, Kolter R (1998) Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30:285–293
Qin Y, Luo ZQ, Farrand SK (2004) Domains formed within the N-terminal region of the quorum-sensing activator TraR are required for transcriptional activation and direct interaction with RpoA from agrobacterium. J Biol Chem 279:40844–40851. doi:10.1074/jbc.M405299200M405299200
Quackenbush J (2002) Microarray data normalization and transformation. Nat Genet 32(Suppl):496–501. doi:10.1038/ng1032ng1032
Ramos JL, Martinez-Bueno M, Molina-Henares AJ et al (2005) The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69:326–356. doi:10.1128/MMBR.69.2.326-356.2005
Rasamiravaka T, Labtani Q, Duez P, El Jaziri M (2015) The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res Int 2015:759348. doi:10.1155/2015/759348
Ribeiro RA, Barcellos FG, Thompson FL, Hungria M (2009) Multilocus sequence analysis of Brazilian Rhizobium microsymbionts of common bean (Phaseolus vulgaris L.) reveals unexpected taxonomic diversity. Res Microbiol 160:297–306. doi:10.1016/j.resmic.2009.03.009
Rinaudi LV, Giordano W (2010) An integrated view of biofilm formation in rhizobia. FEMS Microbiol Lett 304:1–11. doi:10.1111/j.1574-6968.2009.01840.x
Robleto EA, Kmiecik K, Oplinger ES et al (1998) Trifolitoxin production increases nodulation competitiveness of Rhizobium etli CE3 under agricultural conditions. Appl Environ Microbiol 64:2630–2633
Russo DM, Williams A, Edwards A et al (2006) Proteins exported via the PrsD-PrsE type I secretion system and the acidic exopolysaccharide are involved in biofilm formation by Rhizobium leguminosarum. J Bacteriol 188:4474–4486. doi:10.1128/JB.00246-06
Sadovskaya I, Vinogradov E, Li J et al (2010) High-level antibiotic resistance in Pseudomonas aeruginosa biofilm: the ndvB gene is involved in the production of highly glycerol-phosphorylated beta-(1- > 3)-glucans, which bind aminoglycosides. Glycobiology 20:895–904. doi:10.1093/glycob/cwq047
Salazar E, Diaz-Mejia JJ, Moreno-Hagelsieb G et al (2010) Characterization of the NifA-RpoN regulon in Rhizobium etli in free life and in symbiosis with Phaseolus vulgaris. Appl Environ Microbiol 76:4510–4520. doi:10.1128/AEM.02007-09
Schmidt J, Musken M, Becker T et al (2011) The Pseudomonas aeruginosa chemotaxis methyltransferase CheR1 impacts on bacterial surface sampling. PLoS One 6:e18184. doi:10.1371/journal.pone.0018184
Schultze M, Kondorosi A (1998) Regulation of symbiotic root nodule development. Annu Rev Genet 32:33–57. doi:10.1146/annurev.genet.32.1.33
Smith DR, Chapman MR (2010) Economical evolution: microbes reduce the synthetic cost of extracellular proteins. MBio. doi:10.1128/mBio.00131-10
Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100:9440–9445. doi:10.1073/pnas.15305091001530509100
Tatusov RL, Fedorova ND, Jackson JD et al (2003) The COG database: an updated version includes eukaryotes. BMC Bioinform 4:41. doi:10.1186/1471-2105-4-411471-2105-4-41
Vorachit M, Lam K, Jayanetra P, Costerton JW (1995) Electron microscopy study of the mode of growth of Pseudomonas pseudomallei in vitro and in vivo. J Trop Med Hyg 98:379–391
Wang P, Zhong Z, Zhou J et al (2008) Exopolysaccharide biosynthesis is important for Mesorhizobium tianshanense: plant host interaction. Arch Microbiol 189:525–530. doi:10.1007/s00203-007-0345-3
Workman C, Jensen LJ, Jarmer H et al (2002) A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol 3. doi:10.1186/gb-2002-3-9-research0048.1
Yeom S, Yeom J, Park W (2010) NtrC-sensed nitrogen availability is important for oxidative stress defense in Pseudomonas putida KT2440. J Microbiol 48:153–159. doi:10.1007/s12275-010-0075-0
Young JPW, Crossman LC, Johnston AWB et al (2006) The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol 7:R34. doi:10.1186/gb-2006-7-4-r34
Zarkani AA, Stein E, Rohrich CR et al (2013) Homoserine lactones influence the reaction of plants to rhizobia. Int J Mol Sci 14:17122–17146. doi:10.3390/ijms140817122
Zhang Y, Pohlmann EL, Roberts GP (2005) GlnD is essential for NifA activation, NtrB/NtrC-regulated gene expression, and posttranslational regulation of nitrogenase activity in the photosynthetic, nitrogen-fixing bacterium Rhodospirillum rubrum. J Bacteriol 187:1254–1265. doi:10.1128/JB.187.4.1254-1265.2005
Zheng H, Zhong Z, Lai X et al (2006) A LuxR/LuxI-type quorum-sensing system in a plant bacterium, Mesorhizobium tianshanense, controls symbiotic nodulation. J Bacteriol 188:1943–1949. doi:10.1128/JB.188.5.1943-1949.2006
Zheng H, Mao Y, Zhu Q et al (2015) The quorum sensing regulator CinR hierarchically regulates two other quorum sensing pathways in ligand-dependent and-independent fashions in Rhizobium etli. J Bacteriol 197:1573–1581. doi:10.1128/JB.00003-15
Acknowledgments
Part of this work was supported by CONACyT Grant 220790 and DGAPA-PAPIIT Grant IN213216. We are grateful to the Posgrado en Ciencias Biológicas de la Facultad de Ciencias, Universidad Nacional Autónoma de México, México; ARP was a recipient of a PhD Studentship from CONACyT. We thanks to Mario Ramírez Yáñez and Victor Bustos Zagal for technical assistance, to Rubén Paul Gaytán Colín and Eugenio López Bustos from Unidad de Síntesis, Instituto de Biotecnología-UNAM, and to Andrés Saraleguí Amaro and Xochitl Alvarado Affantranger from Unidad Laboratorio Nacional de Microscopía Avanzada-UNAM. Thanks also to Michael Dunn for comments on the manuscript and Enrique Reynaud and Veronica Narvaez for technical assistance in qPCR experiments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Erko Stackebrandt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Reyes-Pérez, A., Vargas, M., Hernández, M. et al. Transcriptomic analysis of the process of biofilm formation in Rhizobium etli CFN42. Arch Microbiol 198, 847–860 (2016). https://doi.org/10.1007/s00203-016-1241-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-016-1241-5