Abstract
TrmB of Pyrococcus furiosus was discovered as the trehalose/maltose-specific repressor for the genes encoding the trehalose/maltose high-affinity ABC transporter (the TM system). TrmB also represses the genes encoding the high affinity maltodextrin-specific ABC transporter (the MD system) with maltodextrin and sucrose as inducers. In addition, TrmB binds glucose leading to an increased repression of both, the TM and the MD system. Thus, TrmB recognizes different promoters and depending on the promoter it will be activated or inactivated for promoter binding by different sugar effectors. The TrmB-like protein TrmBL1 of P. furiosus is a global regulator and recognizes preferentially, but not exclusively, the TGM (for Thermococcales–glycolytic motif) sequence that is found upstream of the MD system as well as of genes encoding enzymes involved in the glycolytic and the gluconeogenic pathway. It responds to maltose and maltotriose as inducers and functions as repressor for the genes encoding the MD system and glycolytic enzymes, but as activator for genes encoding gluconeogenic enzymes. The TrmB-like protein TrmBL2 of P. furiosus lacks the sugar-binding domain that has been determined in TrmB. It recognizes the MD promoter, but not all TGM harboring promoters. It is evolutionary the most conserved among the Thermococcales. The regulatory range of TrmBL2 remains unclear.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pyrococcus furiosus is a strictly anaerobic hyperthermophilic archaeon that is able to metabolize peptides, polysacccharides as well as pyruvate. For sugar metabolism the organism relies on a modified Embden–Meyerhof pathway. It uses an ADP-dependent glucokinase and phosphofructokinase. It utilizes the classical enzyme couple (GAPDH and PGK) for anabolic function and in addition two catabolic enzymes GAPOR and GAPN (non-phosphorylating GAPDH) (Siebers and Schonheit 2005). Enzymes in the gluconeogenic pathway are expressed to a higher level in cells grown on pyruvate than when grown on carbohydrates, and glycolytic enzymes are elevated in cells grown on carbohydrates (Schäfer and Schönheit 1993). Thus, the allosteric regulation of classical Embden–Meyerhof enzymes of bacteria does not seem to play a major role (Verhees et al. 2003). Instead, Pyrococcus species seem to rely on transcriptional control. Gene expression experiments using microarrays of cells grown with different carbon sources support this view (Schut et al. 2003). Surprisingly, P. furiosus is unable to transport and use glucose as a carbon source even though they do harbor an ADP-dependent glucokinase (Kengen et al. 1995). However, two sugar-specific and binding protein-dependent ABC transporters were identified and characterized in P. furiosus.
One is specific for trehalose and maltose (the TM system) (Xavier et al. 1996; Horlacher et al. 1998) Fig. 1. The operon encoding this system is nearly identical in P. furiosus and Thermoccus litoralis due an obvious lateral gene transfer of a 16 Kb DNA fragment that is flanked by transposon elements (DiRuggiero et al. 2000). The operon consists of the genes malE malF malG treT trmB malK (Horlacher et al. 1998; (Greller et al. 1999; Qu et al. 2004). It is induced by growth in the presence of maltose and trehalose (Xavier et al. 1996). The specificity and the high substrate affinity of the TM system for trehalose and maltose (Km about 20 nM) are carried by the lipid anchored binding protein, the product of the malE gene. Interestingly, despite the low sequence identity, the structure of the protein is nearly superimposible on the E. coli maltose binding protein (Diez et al. 2001). The same is true for MalK, the corrresponding ABC subunit of the system (Diederichs et al. 2000). malF and malG encode the membrane spanning subunits of the transporter (Horlacher et al. 1998). treT encodes the only enzyme of the operon. It catalyzes the reversible transfer of glucose from ADPglucose onto glucose to form trehalose. Rather than being an enzyme involved in the synthesis of trehalose, we picture this enzyme as a trehalose metabolizing enzyme forming glucose and ADPglucose from trehalose. ADPglucose would then be used to form glycogen or linear maltodextrins, which, via glycogen or maltodextrin phosphorylase, would yield glucose-1-P for glycolysis (Qu et al. 2004). There are several enzymes that might be involved in the metabolism of maltose and maltodextrins. The cellular extract contains the activity of an α-glucosidase splitting maltose to glucose (Costantino et al. 1990; Xavier et al. 1999), probably identical to PF0132 (Shockley et al. 2003). Other enzymes are amylopullulanase (PF1935), 4-alpha-glucanotransferase (PF0272) forming glucose and larger maltodextrins from maltose, and maltodextrin phosphorylase (PF1535) that are induced after growth on maltose and starch (Schut et al. 2003).
The gene cluster of the TM and the MD system. a The gene cluster encoding the binding protein-dependent ABC transporter for trehalose/maltose (the TM system), trehalose synthase and TrmB is shown. Divergent to malE, frk encoding ATP-dependent fructokinase is located. b The gene cluster encoding the binding protein-dependent ABC transporter for maltodextrins (the MD system). c The TM promoter harbors a perfect palindrome (underlined), whereas the MD system contains only the first half of this palindrome. Bold letters indicate the TrmB binding sites; +1 indicates the transcription start site
The second identified sugar-specific and binding protein-dependent ABC transporter is homologous to the TM system, but recognizes with high specificity maltodextrins, but not maltose or trehalose, the MD system (Koning et al. 2002; DiRuggiero et al. 2000) (Fig. 1). The structure of the cognate-binding protein has also been determined with maltotriose bound to it (Evdokimov et al. 2001). The MD system is induced by growth on starch and maltodextrins, but surprisingly also by maltose, even though it does not transport maltose. In a recent review, the evolutionary origin of prokaryotic maltose and maltodextrin ABC transporters has been analyzed (Noll et al. 2008) The authors come to the conclusion that the TM and the MD system in P. furiosus have different origins. Whereas the MD system arose within the archaea, the TM system appears to be derived from bacterial lineage.
The first step in maltodextrin metabolism is most likely the 4-alpha-glucanotransferase (PF0272) mediated disproportionation to glucose and longer maltodextrins followed by maltodextrin phosphorylase (PF1535) to form glucose-1-P (Xavier et al. 1999). From these observations, it is clear that maltose and maltodextrin transport and metabolism in P. furiosus is under transcriptional control. In contrast to bacterial transcription, archaeal transcription is mediated via a multicomponent RNA polymerase homologous to the eucaryotic RNA polymerase II (Thomm and Hausner 2007). Essential general transcription factors are the TATA box-binding protein (TBP) (Ouhammouch et al. 2003) and transcription factor B (TFB) (Geiduschek and Ouhammouch 2005). This review is concerned with the transcriptional regulators that control the transcription of genes involved in the uptake and metabolism of maltose and maltodextrins.
TrmB is the trehalose- and maltose-specific repressor for the TM system
In our search for a substrate-specific transcriptional regulator for the TM system, we noticed that in the lateral gene transfer between T. litoralis and P. furiosus harboring the genes encoding the TM system, the inducibility by maltose and trehalose had been conserved. Therefore, we reasoned that the gene encoding the putative regulator must be contained within the transferred DNA segment. We cloned unidentified genes within the TM cluster, expressed them in E. coli and tested the purified proteins for their ability to bind the malE upstream DNA sequence and to shift them in an electrophoretic mobility shift assay (EMSA). In this way, the gene upstream of malK was identified to encode a transcriptional regulator. We named it TrmB (transcriptional regulator for the maltose system). The protein is a homodimer (molecular weight of the monomer is 38,800 Da). Determination of the transcription start point, EMSA tests and footprint analysis in the presence and absence of maltose and scanning mutagenesis revealed the perfectly palindromic sequence ATACTTTTAGTAT as the TrmB binding site overlapping the putative BRE/TATA box of the malE gene (Fig. 1c). In vitro transcription assays at 80°C with purified RNA polymerase, TBP and TFB showed that TrmB was able to block malE transcription, and the presence of maltose, but not maltotriose, allowed again transcription in the presence of TrmB (Fig. 2a). Similarly, the presence of trehalose allowed transcription in the presence of TrmB (not shown). These results demonstrated that TrmB acts as transcriptional repressor for malE and that trehalose and maltose function as inducers (Lee et al. 2003). Purified TrmB binds maltose in a positive cooperative fashion (half maximal binding occurs at 20 μM) indicating conformational changes upon binding maltose. The affinity of TrmB for trehalose is at least 20 lower than for maltose and no indication for sigmoidality in the binding behavior can be detected. The difference in binding affinity for the two inducers finds its explanation in the transport and metabolism of both sugars. Trehalose and maltose are transported with equal rate and equal high affinity (Km about 20 nM). However, in vivo trehalose can only slowly be metabolized since it accumulates to high internal concentrations when trehalose is present in the medium as shown for T. litoralis (Lamosa et al. 1998), whereas maltose is fast metabolized. Significantly, other sugars are binding substrates for TrmB. Thus, maltose, glucose, sucrose, maltotriose and trehalose are bound by TrmB (listed in decreasing affinities), but only maltose and trehalose act as inducers on the TM promoter, as tested by EMSA or in vitro transcription assays.
In vitro transcription of the TM (a) and the MD (b) system. malE (a) and mdxE (b) transcription was performed at 80°C using the basic transcriptional components of P. furiosus. As template, linearized DNA containing the malE or the mdxE promoter was used. First lane (a), control assay with DNA for P. furiosus glutamate dehydrogenase. Lanes 2 and 3 malE and mdxE transcription in the presence of maltose or maltotriose, but in the absence of TrmB. Lanes 4–6, malE or mdxE transcription in the presence of 0.8–3.2 μg TrmB. Lanes 7–14, transcription in the presence of 3.2 μg TrmB and 25, 100, 250 μM, 1, 2.5, 10, 25, 100 mM maltose or maltotriose. Taken from Lee et al. (2003), with permission from the authors and the publisher
TrmB is the maltodextrin and sucrose-specific repressor for the MD system
The gene cluster encoding the MD system is shown in Fig. 1b. The order of genes (as well as their sequence) encoding the transport machinery is very similar to those of the TM system. However, upstream of mdxK, the gene encoding the ABC subunit of the system, PF1935 is located encoding an annotated amylopullulanase. A gene encoding an obvious operon-specific regulator such as trmB in the TM operon is missing. However, TrmB the regulator of the TM system was found to also control the MD system. The transcriptional start point was determined, and EMSA and footprint analysis were done. The data are summarized in Fig. 1c. TrmB binds to the upstream region of mdxE. But in contrast to the binding site in the TM promoter (that overlaps the BRE/TATA box), TrmB binding overlaps the transcriptional start site of the MD operon downstream of its putative BRE/TATA box. The sequence that is protected by TrmB from DNAse I digestion (ATCGATGATACTAACATGGGA…) contains the first half of the palindrome seen in the TM promoter, but it is different otherwise. In vitro transcription assays of the mdxE gene were done in the presence and absence of TrmB and in the presence of different sugars (Fig. 2b). TrmB is able to block mdxE transcription, but maltose and trehalose, the sugars that are able to relieve repression on the TM promoter, are unable to do so on the MD promoter. However, surprisingly, maltotriose, maltotetraose, maltopentaose as well as sucrose are able to counteract repression of the MD operon by TrmB (only the data for maltotriose are shown in Fig. 2b) (Lee et al. 2005). We conclude that TrmB exhibits dual promoter specificity and is controlled by different sugars depending on the promoter where TrmB is bound to. To our knowledge, this has not been observed with any procaryotic transcriptional regulator.
Glucose binding by TrmB exhibits features that are reminiscent of catabolite repression in bacteria
Glucose does not relieve repression by TrmB of the TM nor the MD promoter, even though glucose is bound quite well by TrmB. In contrast, when the repression by TrmB on either the TM operon or the MD operon was relieved by maltose (in case of the TM promoter), or by maltotriose (in case of the MD promoter) repression was again established by the additional presence of glucose (Fig. 3a, b). The effect of glucose is not simply a competition to remove the inducer maltose or maltotriose from TrmB, but in addition it increases the repression of both operons by TrmB. This can be seen in Fig. 4 where during EMSA the band shifting of the TM promoter by nonsaturating amounts of TrmB is increased by glucose. Also, TM promoter DNA increases the affinity of TrmB to bind glucose from 73 to 27 μM. These data show that TrmB can integrate signals that arise from different sugars in the cytoplasm. Maltose and trehalose act as inducers for the TM system, whereas maltodextrins and sucrose act as inducers for the MD system. Glucose functions as corepressor for both, the TM as well as the MD system (Lee et al. 2005, 2007a). The effect of glucose on TrmB activity is reminiscent of catabolite repression in bacteria, where the utilization (transport) of glucose interferes with the transcription of genes encoding transporters for alternate sugar carbon sources (Postma et al. 1996). There, the PEP-dependent phosphotransferase system (PTS) mediates catabolite repression. The PTS has not been found in archaea, but the exclusion of alternate carbon sources by glucose has been observed (Lubelska et al. 2006). P. furiosus does not transport glucose, but metabolizes glucose derived from cytoplasmic dextrin metabolism. Metabolic overflow of glucose will repress both the MD and the TM system, even in the presence of inducer, to curb the uptake of glucose producing sugars. The properties of TrmB offer a simple explanation for this phenomenon in the absence of PTS (Lee et al. 2007a).
The effect of glucose on the induced in vitro transcription of the TM and the MD system. a C-terminally His-tagged TrmB (0.2 μM) was used during in vitro transcription at 80°C of a TM operon fragment in the presence of the inducer maltose and the co-repressor glucose. Lane 1, transcription of the operon fragment in the absence of TrmB; lane 2, transcription in the presence of TrmB; lanes 3–8, transcription in the presence of 500 μM maltose and increasing glucose concentrations. b C-terminally His-tagged TrmB (1.6 μM) was used during in vitro transcription at 80°C of an MD operon fragment in the presence of the inducer maltotriose and the co-repressor glucose. Lane 1, transcription of the operon fragment in the absence of TrmB; lane 2, transcription in the presence of TrmB; lanes 3–6, transcription in the presence of 1 mM maltotriose and increasing glucose concentrations Taken from Lee et al. (2007a), with permission from the authors and the publisher
Glucose as corepressor of TrmB. EMSA of TM promoter DNA by nonsaturating concentrations of TrmB and increasing amounts of glucose (Lee et al. 2007a), with permission from the authors and the publisher
The crystal structure of the sugar-binding domain of TrmB
The unfavorable biochemical properties (precipitation at high protein concentration) of full length TrmB prevented its crystallization so far. However, an N-terminal truncated TrmB (∆2-109) could be purified and crystallized. In contrast to the full-length protein, it appears to be a monomer in solution. The protein lacks the DNA-binding domain, but retains the sugar-binding site. The binding affinity of the truncated protein for maltose, glucose sucrose and maltotriose was determined to be 6.8, 25, 34, and 160 μM KD, significantly higher than the affinity of full-length protein. Cooperative binding of maltose as seen with the full-length protein was no longer observed with the truncated version. This may indicate that the full-length protein is under conformational restraints that is altered (relieved) by promoter DNA binding. The 3D structure was solved at 1.5 Å with maltose bound to it (Krug et al. 2006). Maltose is in close contact with seven amino acids of which six are in contact with the nonreducing glucosyl residue of maltose Fig. 5. The structure harboring the sugar-binding site consists of two distinctive domains (N and C-domains). The N-domain (to which in the full length protein the DNA-binding domain would be attached) forms an 8-stranded β-sheet flanked by two large helices on one side and one large helix crossing the β-sheet on the other side. Because the latter helix provides the only maltose-binding residues of the N-domain, we designate it the “sugar-binding helix”. The C-domain forms a strand, a helix and an irregular flattened 7-stranded β-barrel with its axis roughly parallel to the strains of the N-domain. All residues binding the nonreducing glucosyl residue of maltose are contributed by two neighboring loops of the C-domain. Based on the observed mode of binding for maltose, it appears likely that all substrates are bound with their nonreducing α-glucosyl moiety to the same six amino acid residues of the C-domain. Obviously, the fixation of the common non-reducing glucosyl residue and the varying interaction with the remaining portion of the sugars on the sugar-binding helix are the basis for the surprisingly large range of different substrates and the differential effects of the different sugars on promoter selection.
Crystal structure of the sugar binding domain of TrmB. The protein and the bound maltose are indicated as ribbon, ball and stick representation, respectively. Maltose is bound with its non-reducing glucosyl residue towards the squashed barrel domain. The N (blue) and C (orange) termini are indicated. With modifications taken from Krug et al. (2006) with permission of the authors and the publisher
The maltose binding of the structure appears to be a novel sugar-binding fold quite different from the binding fold of well-characterized prokaryotic transcriptional regulators and binding proteins of ABC transporters (Lewis et al. 1996; Hars et al. 1998; Borths et al. 2002; Quiocho and Ledvina 1996). The bound maltose in TrmB∆2-109 sticks to the surface-exposed edge of the cleft between the N- and the C-domain oriented with its axis roughly perpendicular to the cleft (Fig. 5). The buried surface between the two domains is rich in phenylalanines and other hydrophobic residues rendering a large movement of these domains (typical for periplasmic binding proteins) in response to the sugar binding rather unlikely. The DNA-binding domain of the full-length TrmB is in proximity to the N-terminal end of the sugar-binding helix suggesting transduction of differential induced-fit movements upon sugar binding via this sliding helix. This must be the basis for the differential sugar and promoter recognition that is so typical for TrmB. Major efforts are under way to determine the crystal structure of full-length protein in the presence of diferent sugars and different promotors to unravel the surprising versatility of TrmB. The binding site for TrmB in front of the TM operon is palindromic, necessitating a dimer of TrmB to be recognized. Its binding site in front of the MD operon only harbors one-half of this palindrom suggesting a different structure of the DNA-binding domain of TrmB to be involved. This is supported by mutational analysis of the DNA-binding domain of TrmB that can differentiate promoter recognition (unpublished).
The TrmB-like protein TrmBL1 is a global regulator for genes encoding maltodextrin transport, as well as for glycolytic and for gluconeogenic enzymes in P. furiosus recognizing the TGM motif
Recently, van de Werken et al. (2006)) reported the occurrence of a conserved sequence motif TGM (for Thermococcales–Glycolytic-Motif), upstream of genes encoding glycolytic as well a gluconeogenic enzymes in P. furiosus, indicating common transcriptional control of the respective genes. This motif was also conserved in another representative of the Thermococcales, Thermococcus kodakaraensis, but not in Pyrococcus abyssi or Pyrococcus horikoshii. Since TrmB is not found in T. kodakaraensis, it was unlikely that TrmB would be identical with this unknown global regulator rcognizing TGM. Therefore, we searched for TrmB-like proteins in the genomic sequence of P. furiosus and other Thermococcales. Table 1 gives a survey of the occurrence of TrmB and the TrmB-like proteins in the sequenced members of Thermococcales. The sequence identity of one orthologue between the different organisms is more than 70%. The percentage identity among the different members is between 22 and 30%. Surprisingly, not all TrmB-like proteins occur in all species. Figure 6 compares the amino acid sequences of TrmBL1 (PF0124) with TrmB and the TrmBL1 paralog TK1769 from T. kodakaraensis, as well as with TrmBL2 from P. furiosus. TrmB and TrmBL1 of P. furiosus showed 29% amino acid sequence identity over the entire length of the protein. Especially, the N-terminus, which in TrmB of P. furiosus contains the DNA-binding domain (amino acids 1–90) is highly conserved (45% sequence identity). The percentage sequence identity between TrmBL1 of P. furiosus (PF0124) and of T. kodakaraensis (TK1769) was 67%. TrmBL1 and TrmBL2 appeared as likely candidates for the global regulator recognizing TGM, since both occurred in P. furiosus as well as in T. kodakaraensis, the two organisms that carried the TGM motif. However, as seen in Fig. 6 TrmBL2, even though highly conserved among the Thermococcales, lacks part of the extended C-terminal domain, which in the crystal structure of TrmB harbors the sugar-binding domain. Therefore, only TrmBL1 was the likely candidate for the postulated global regulator.
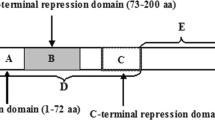
Sequence comparison between the TrmB paralogues. Amino acid sequence alignment were done between TrmB (PF1743), TrmBL1 (PF1204) from P. furiosus, the TrmBL1-paralog (TK1769) from T. kodakaraensis and TrmBL2 (PF0496) from P. furiosus. The position of the helix–turn–helix motif in the N-terminal region is indicated. Boxed are two amino acids in the recognition helix that are essential for target recognition. Strictly conserved residues between all three sequences are indicated by an asterisk, and highly and lowly conserved residues are indicated by colon and dots, respectively. Seven amino acid residues of the maltose-binding site at the C-terminal region of TrmB are indicated by black boxes. Sequence alignments were done by the ClustalW program. Taken from Lee et al. (2007b), with permission of the authors and the publisher
TrmBL1 of P. furiosus has a molecular mass of 39 kDa (by SDS-PAGE). By molecular sieve chromatography, TrmBL1 appears in equilibrium of a tetrameric and an octameric form that is shifted to the octamer in the presence of maltose and more effectively of maltotriose) (Lee et al. 2007b). By EMSA TrmBL1 was shown to recognize the MD (K d about 0.2 μM) (that does carry the TGM sequence) as well as the TM promoter DNA (K d about 1.25 μM) that does not carry the TGM motif (Fig. 7). To determine the TrmBL1 binding site in the MD promoter, TrmBL1 protection analysis against DNaseI digestion was performed. TrmBL1 interacted with a region downstream of the BRE-TATA box overlapping the transcriptional start site of the mdxE operon (Fig. 7b). The TrmBL1 binding site was 5′-TATGTATCACTATCGATGATACTAACATGGG-3′ (bold letters indicating the TGM sequence). This TrmBL1 binding sequence also partly overlaps the TrmB binding site in the MD promoter (Fig. 7). Figure 8 shows EMSA with TrmBL1 of promoter regions upstream of three gene clusters encoding glycolytic enzymes that carry the TGM sequence. These enzymes are: ADP-dependent glucokinase, (PF0312), glyceraldehyde-3-phosphate ferredoxin oxidoreductase, (PF0464), and ADP-dependent phosphofructokinase (PF1784). Intrerestingly, the promoter region of the gene encoding TrmBL1 itself, PF0124 that does not contain the TGM sequence, is also recognized by TrmBL1. The first three promoter regions were bound to TrmBL1 with similar high affinity. Complete shifting of the DNA occurred below 1.25 μM of TrmBL1 (K d about 0.6 μM). The promoter of trmBL1 itself also bound TrmBL1, although with a lower affinity (K d around 1 μM).
a EMSA of the TM and the MD promoter DNA in the presence of TrmBL1. The concentration of TrmBL1 for TM was 0.375, 0.625, 1, 1.25 μM and 0.125, 0.25, 0.5, 1 μM for the MD promoter. Thus, TrmB showed higher binding affinity for the MD promoter than for the TM promoter. B, The position of the TGM sequence in front of three different promoters (underlined). The TGM motif was proposed by van de Werken et al. (2006). The dashed line represents the TrmB binding site. The start of the open reading frame (ATG) is in italics. Taken from Lee et al. (2007b), with permission of the authors and the publisher
EMSA with TrmBL1 on the promoter regions containing TGM sequence. The promoters of genes encoding glycolytic enzymes: ADP-glucokinase (PF0312), glyceraldehyde-3-phosphate ferredoxin oxidoreductase (PF0464) and ADP-phosphofructokinase (PF1784). These promoters contain a palindromic sequence, called TGM. The promoter of PF0124 encoding TrmBL1 does not harbor the TGM sequence. The concentration of TrmBL1 was in each test 0.63, 1.25, 2.5, and 3.75 μM, respectively. Taken from (Lee et al. 2007b) with permission of the authors and the publisher
As demonstrated by molecular sieve chromatography, TrmBL1 does respond to maltose and more so to maltotriose by shifting from a tetrameric to octameric quarternary structure. Thus, it should bind maltotriose and maltose. Maltose, maltotriose and fructose relieve repression by TrmBL1 in the in vitro transcription assay driven by the promoter upstream of the gene encoding PF1784 (phosphofructokinase), as shown for maltose in Fig. 9. TrmBL1 inhibits transcription; the three sugars do rescue transcription to some extent but not in a monotonous fashion. With maltose, induction (rescue from repression) reached two peak value of expression between 250 μM and 1 mM maltose, before vanishing above 2.5 mM. All three sugars showed this phenomenon, though not at the same sugar concentration. Possibly, this phenomenon is due to two different binding affinities of the tetrameric and octameric forms of TrmBL1 to promoter DNA. High DNA-binding affinity would be exhibited by the tetramer in the absence of sugar that can be released by low sugar concentration. At high sugar concentration, the octamer would form with high affinity for promoter DNA, again preventing transcription even in the presence of maltose.
The effect of maltose on the in vitro transcriptional repression by TrmBL1 on the operon encoding phosphofructokinase. The test was done under optimized conditions in the presence of 600 mM potassium glutamate. Taken from (Lee et al. 2007b), with permission of the authors and the publisher
During in vitro transcription assays using the TGM-containing promoter of the gene encoding PF1784, we found that both, TrmBL1 as well as TrmB, prevented transcription. Yet, whereas maltose, maltotriose and fructose lifted repression by TrmBL1 (shown for maltose in Fig. 11), these sugars were unable to lift repression by TrmB. Nevertheless, it is clear that TrmB shows binding towards these sugars with affinities that largely exceed the binding affinity of TrmBL1 for these sugars. This reflects the unique properties of TrmB to act as a repressor that is inactivated by an inducer or as a repressor that is activated by a corepressor depending on the operator that it is bound to (Lee et al. 2007a).
By in vitro transcription assays it was shown that TrmBL1 not only acts as repressor for the MD operon and genes encoding glycolytic enzymes (for instance phosphofructokinase), but it also acts +1 as transcriptional activator for the genes encoding enzymes of the gluconeogenic pathway such as fructose 1.6-bisphosphatase (PF0613) (Fig. 10). In this case, the TGM motif is located upstream of the BRE/TATA box.
The dual role of a TrmBL1 homologue, Tgr (for Thermococcales glycolytic regulator) (TK1769, see Table 1), to act as a repressor for genes encoding glycolytic enzymes and as activator for genes encoding gluconeogenetic enzymes has recently also been shown in T. kodakaraensis (Kanai et al. 2007). The authors were able to construct a deletion in the gene encoding tgr. Micoarray analysis of wild type and mutant strain showed indeed that the expression of a large number of genes encoding glycolytic enzymes was elevated in the mutant, whereas genes encoding gluconeogenic enzymes were reduced. Figure 11 summarizes the dual function of TrmBL1 (Tgr) in the regulation of carbohydrate metabolism.
Cross-regulation by TrmB and TrmB-like proteins
As shown above, TrmBL1/Tgr acts as global regulator at the crosspoint of glycolysis and gluconeogenesis. The TGM sequence, either downstream or upstream of the BRE/TATA, box allows binding of TrmBL1 and determines repression or activation of the respective gene. Yet, neither is the TGM motif exclusively used as TrmBL1 binding site nor does the TGM sequence prevent the binding (and subsequent regulation) by TrmB (for instance: MD, PF0464, or PF1784). All TGM-containing promoters are recognized very well by TrmBL1. However, also the gene encoding TrmBL1 (PF0124) itself whose promoter does not harbor the TGM sequence is recognized by TrmBL1 and appears to be autoregulated (repressed). In vitro transcription assays with this promoter DNA in the presence of TrmBL1 showed strong inhibition that was relieved by as little as 50 μM glucose. Note that glucose does not relieve repression by TrmBL1 when acting on TGM containing promoters. Thus, curiously, TrmBL1 in the absence of sugar represses its own expression, but repression is relieved by glucose! In contrast maltose and maltotriose, sugars that relieve repression on TGM harboring promoters, have no effect on TrmBL1 when acting on the PF0124 promoter. Extrapolated to the in vivo situation, this would mean that excess glucose stimulates the synthesis of TrmBL1 and therefore would in turn stimulate the expression of genes encoding gluconeogenic enzymes, but represses those encoding glycolytic enzymes. In addition, excess glucose would via TrmB further repress the genes encoding the TM and the MD system as discussed above. This emphasizes the important role of cytoplasmic glucose in controlling transport, glycolysis and gluconeogenesis.
Considering the evolutionary aspects of maltose/maltodextrin ABC transporters (Noll et al. 2008), one would speculate that TrmBL1 is the “older” regulator evolved for the control of glycolytic and gluconeognic systems (even though it is not present in all sequenced Thermococcales), whereas TrmB would represent a more recent aquisition (possibly from bacterial origin) initially geared for the specific control of the TM system. Yet, obvious homologues of TrmB in bacteria have not been recognized so far.
The third TrmB-like protein is TrmBL2. Only preliminary testing has been done with this interesting regulator. As shown in Fig. 6 it lacks a large C-terminal domain, which in the TrmB crystal structure was shown to contain the sugar-binding site. Therefore, TrmBL2 should not be able to bind sugar. Nevertheless, it is apparently the evolutionarily the most conserved regulator and is found in all sequenced Thermococcales, unlike the other TrmB-like proteins (Table 1). TrmBL2 recognizes the MD promoter very well, but no obvious pattern in respect to other TGM-containing promoters can be deduced (Lee et al. 2007b). Of course, there is the question of sugar specificity of TrmBL2. In the absence of a sugar-binding site, we speculate on a mechanism that has been observed in the E. coli maltose system. There, MalT the central activator of all mal genes can be controlled by the regulatory domain of MalK, the ABC subunit of the binding protein-dependent transporter (Böhm and Boos 2004). The same “regulatory” domain has been observed in the ABC subunit of the TM and the MD transporter of P. furiosus (Diederichs et al. 2000). TrmBL2 could follow the same mechanism and obtain its specificity by interacting with the regulatory domain of the two ABC transporters.
The surprising conclusion that arises from these studies is the multiplicity of regulatory input acting on the different promoters. In the case of the MD promoter, no less than three regulators are participating, two of which are shown to be substrate controlled. The dissection of the role of each regulator acting in vivo will be challenge for the future.
References
Borths EL, Locher KP, Lee AT, Rees DC (2002) The structure of Escherichia coli BtuF and binding to its cognate ATP binding cassette transporter. Proc Natl Acad Sci USA 99:16642–16647
Böhm A, Boos W (2004) Gene regulation in prokaryotes by subcellular relocalization of transcription factors. Curr Opin Microbiol 7:151–156
Costantino HR, Brown SH, Kelly RM (1990) Purification and characterization of an α-glucosidase from a hyperthermophilic archaebacterium, Pyrococcus furiosus, exhibiting a temperature optimum of 105 to 115°C. J Bacteriol 172:3654–3660
Diederichs K et al (2000) Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon Thermococcus litoralis. EMBO J 19:5951–5961
Diez J, Diederichs K, Greller G, Horlacher R, Boos W, Welte W (2001) The crystal structure of a liganded trehalose/maltose-binding protein from the hyperthermophilic archaeon Thermococcus litoralis at 1.85 Å. J Mol Biol 305:905–915
DiRuggiero J et al (2000) Evidence of recent lateral gene transfer among hyperthermophilic Archaea. Mol Microbiol 38:684–693
Evdokimov AG, Anderson DE, Routzahn KM, Waugh DS (2001) Structural basis for oligosaccharide recognition by Pyrococcus furiosus maltodextrin-binding protein. J Mol Biol 305:891–904
Geiduschek EP, Ouhammouch M (2005) Archaeal transcription and its regulators. Mol Microbiol 56:1397–1407
Greller G, Horlacher R, DiRuggiero J, Boos W (1999) Molecular and biochemical analysis of MalK, the ATP-hydrolyzing subunit of the trehalose maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J Biol Chem 274:20259–20264
Hars U, Horlacher R, Boos W, Welte W, Diederichs K (1998) Crystal structure of the effector-binding domain of the trehalose-repressor of Escherichia coli, a member of the LacI family, in its complexes with inducer trehalose-6-phosphate and noninducer trehalose. Protein Sci 7:2511–2521
Horlacher R, Xavier KB, Santos H, DiRuggiero J, Kossmann M, Boos W (1998) Archaeal binding protein-dependent ABC transporter: molecular and biochemical analysis of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J Bacteriol 180:680–689
Kanai T et al (2007) A global transcriptional regulator in Thermococcus kodakaraensis controls the expression levels of both glycolytic and gluconeogenic enzyme-encoding genes. J Biol Chem 282:33659–33670
Kengen SWM, Tuininga JE, de Bok FAM, Stams AJM, de Vos WM (1995) Purification and characterization of a novel ADP-dependent glucokinase from the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem 270:30453–30457
Koning SM, Konings WN, Driessen AJ (2002) Biochemical evidence for the presence of two alpha-glucoside ABC-transport systems in the hyperthermophilic archaeon Pyrococcus furiosus. Archaea 1:19–25
Krug M, Lee S-J, Diederichs K, Boos W, Welte W (2006) Crystal structure of the sugar binding domain of the archaeal transcriptional regulator TrmB. J Biol Chem 281:10976–10982
Lamosa P, Martins LO, DaCosta MS, Santos H (1998) Effects of temperature, salinity, and medium composition on compatible solute accumulation by Thermococcus spp. Appl Environ Microbiol 64:3591–3598
Lee S-J et al (2003) TrmB, a sugar-specific transcriptional regulator of the trehalose/maltose ABC transporter from the hyperthermophilic archaeon Thermococcus litoralis. J Biol Chem 278:983–990
Lee S-J, Moulakakis C, Koning SM, Hausner W, Thomm M, Boos W (2005) TrmB, a sugar sensing regulator for ABC transporter genes in Pyrococcus furiosus exhibits dual promoter specificity and is controlled by different inducers. Mol Microbiol 57:1797–1807
Lee S-J, Seitz S, Surma M, Hausner W, Thomm M, Boos W (2007a) Differential signal transduction via TrmB, a sugar sensing transcriptional repressor of Pyrococcus furiosus. Mol Microbiol 64:1499–1505
Lee SJ, Surma M, Seitz S, Hausner W, Thomm M, Boos W (2007b) Characterization of the TrmB-like protein, PF0124, a TGM-recognizing transcriptional regulator of the hyperthermophilic archaeon Pyrococcus furiosus. Mol Microbiol 65:305–318
Lewis M et al (1996) Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science 271:1247–1254
Lubelska JM, Jonuscheit M, Schleper C, Albers SV, Driessen AJM (2006) Regulation of expression of the arabinose and glucose transporter genes in the thermophilic archaeon Sulfolobus solfataricus. Extremophiles 10:383–391
Noll KM, Lapierre P, Gogarten JP, Nanavati DM (2008) Evolution of mal ABC transporter operons in the Thermococcales and Thermotogales. BMC Evol Biol 8:7
Ouhammouch M, Dewhurst RE, Hausner W, Thomm M, Geiduschek EP (2003) Activation of archaeal transcription by recruitment of the TATA-binding protein. Proc Natl Acad Sci USA 100(9):5097–5102
Postma PW, Lengeler JW, Jacobson GR (1996) Phosphoenolpyruvate:carbohydrate phosphotransferase system. In: Neidhardt FC et al (eds) Escherichia coli and Salmonella typhimurium; cellular and molecular biology. American Society of Microbiology, Washington, pp 1149–1174
Qu Q, Lee S-J, Boos W (2004) TreT, a novel trehalose glycosyltransferring synthase of the hyperthermophilic archaeon Thermococcus litoralis. J Biol Chem 279:47890–47897
Quiocho FA, Ledvina PS (1996) Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol Microbiol 20:17–25
Schut GJ, Brehm SD, Datta S, Adams MWW (2003) Whole-genome DNA microarray analysis of a hyperthermophile and an archaeon: Pyrococcus furiosus grown on carbohydrates or peptides. J Bacteriol 185:3935–3947
Schäfer T, Schönheit P (1993) Gluconeogenesis from pyruvate in the hyperthermophilic archaeon Pyrococcus furiosus: involvement of reactions of the Embden–Meyerhof pathway. Arch Microbiol 159:354–363
Shockley KR, Ward DE, Chhabra SR, Conners SB, Montero CI, Kelly RM (2003) Heat shock response by the hyperthermophilic archaeon Pyrococcus furiosus. Appl Environ Microbiol 69:2365–2371
Siebers B, Schonheit P (2005) Unusual pathways and enzymes of central carbohydrate metabolism in Archaea. Curr Opin Microbiol 8:695–705
Thomm M, Hausner W (2007) Transcriptional Mechanisms. In: Garrett R, Klenk H-P (eds) Archaea Evolution, Physiology and Molecular Biology. Blackwell Publishing, U.K
van de Werken HJ, Verhees CH, Akerboom J, de Vos WM, van der Oost J (2006) Identification of a glycolytic regulon in the archaea Pyrococcus and Thermococcus. FEMS Microbiol Lett 260:69–76
Verhees CH et al (2003) The unique features of glycolytic pathways in Archaea. Biochem J 375:231–246
Xavier KB, Martins LO, Peist R, Kossmann M, Boos W, Santos H (1996) High-affinity maltose/trehalose transport system in the hyperthermophilic archaeon Thermococcus litoralis. J Bacteriol 178:4773–4777
Xavier KB, Peist R, Kossmann M, Boos W, Santos H (1999) Maltose metabolism in the hyperthermophilic archaeon Thermococcus litoralis: purification and characterization of key enzymes. J Bacteriol 181:3358–3367
Acknowledgments
The work of the authors cited in this review was supported by the Deutsche Forschungsgemeinschaft (SPP1112 “Genome function and gene regulation in Archaea”)
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Harald Huber.
Rights and permissions
About this article
Cite this article
Lee, SJ., Surma, M., Hausner, W. et al. The role of TrmB and TrmB-like transcriptional regulators for sugar transport and metabolism in the hyperthermophilic archaeon Pyrococcus furiosus . Arch Microbiol 190, 247–256 (2008). https://doi.org/10.1007/s00203-008-0378-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-008-0378-2