Abstract
Simocyclinone D8 is a potent inhibitor of bacterial gyrase, produced by Streptomyces antibioticus Tü 6040. It contains an aminocoumarin moiety, similar to that of novobiocin, which is linked by an amide bond to a structurally complex acyl moiety, consisting of an aromatic angucycline polyketide nucleus, the deoxysugar olivose and a tetraene dicarboxylic acid. We have now investigated the enzyme SimL, responsible for the formation of the amide bond of simocyclinone. The gene was cloned, expressed in S. lividans T7, and the protein was purified to near homogeneity, and characterized. The 60 kDa protein catalyzed both the ATP-dependent activation of the acyl component as well as its transfer to the amino group of the aminocoumarin ring, with no requirement for a 4′-phosphopantetheinyl cofactor. Besides its natural substrate, simocyclinone C4, SimL also accepted a range of cinnamic and benzoic acid derivatives and several other, structurally very diverse acids. These findings make SimL a possible tool for the creation of new aminocoumarin antibiotics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aminocoumarin antibiotics are characterized by a 3-amino-4,7-dihydroxycoumarin moiety and are relatively rare in nature. The best-known representatives of this class are novobiocin, clorobiocin and coumermycin A1 (Fig. 1), which are potent inhibitors of gyrase and are produced by different Streptomyces strains. In the last years, we have identified the biosynthetic gene clusters of these three antibiotics, and the function of most genes contained therein has been elucidated (Li and Heide 2004). Several genes encoding novel, unusual enzymes were discovered in these clusters (Pojer et al. 2003a, 2003b), including the unusual amide synthetases NovL, CloL and CouL (Steffensky et al. 2000; Schmutz et al. 2003; Galm et al. 2004a). The latter enzymes catalyze the formation of an amide bond between the aminocoumarin moiety of the antibiotic and an acyl moiety (Fig. 1). In contrast to the well-studied non-ribosomal peptide synthetases, which have been investigated in detail in the biosynthetic pathways of many peptide antibiotics (Marahiel et al. 1997), the amide synthetases of aminocoumarin antibiotic biosynthesis do not contain a 4′-phosphopantetheinyl cofactor and do not form a covalent bond between the substrate and the enzyme. Rather, these monomeric enzymes of approx. 60 kDa catalyze the activation of the acyl moiety in form of an acyl adenylate and its subsequent, direct transfer to the amino group of the aminocoumarin ring (Steffensky et al. 2000).
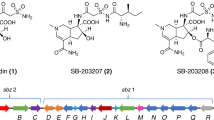
Structures of the aminocoumarin antibiotics novobiocin, clorobiocin, coumermycin A1, simocyclinone D8 and rubradirin. The amide bond generated by SimL/NovL/CloL/CouL is highlighted in grey, and the names of the amide synthetases catalyzing the formation of these amide bonds are given. Within the formulae of simocyclinone D8 and novobiocin, the simocyclinone C4 moiety and the substituted aminocoumarin moiety (Ring B), respectively, are indicated by a box
We recently generated new aminocoumarin antibiotics by mutasynthesis experiments, feeding synthetic analogues of the genuine acyl moiety of clorobiocin, which is 3-dimethylallyl-4-hydroxybenzoic acid (3-DMA-4HB), to a mutant strain of the clorobiocin producer (Galm et al. 2004a, 2004b). The product yields of these experiments were limited by the substrate specificity of the amide synthetase CloL: product yields obtained from feeding different acyl moieties in vivo correlated directly with the conversion rates of the acyl moieties by the amide synthetase CloL in vitro (Galm et al. 2004a). In order to gain access to further new aminocoumarin antibiotics by this approach, we were therefore interested in expanding the substrate specificity of the amide synthetase reaction. This prompted us to search for additional amide synthetases involved in aminocoumarin antibiotic biosynthesis.
Besides novobiocin, clorobiocin and coumermycin A1, two other classes of aminocoumarin antibiotics have been reported in nature, i.e. the simocyclinones and rubradirin. Their acyl components are entirely different from those of the aforementioned antibiotics (Fig. 1). In the biosynthetic gene cluster of rubradirin (Sohng et al. 1997), several genes for putative adenylate-forming enzymes were identified, but is it unknown which of those may be responsible for the acylation of the aminocoumarin ring. Also the nature of the acyl substrate of this reaction is unknown. In the simocyclinone biosynthetic gene cluster (Galm et al. 2002; Trefzer et al. 2002), however, a candidate gene for the amide synthetase was identified and named simL. It has been suggested that the structurally complex acyl moiety (Fig. 1), called simocyclinone C4, is assembled before this acyl moiety is transferred to the aminocoumarin moiety (Schimana et al. 2000, 2001; Holzenkämpfer et al. 2002). Simocyclinone has recently been identified as a potent inhibitor of bacterial gyrase with a novel mechanism of action (Flatman et al. 2005).
In the present study, we have provided experimental evidence that SimL indeed catalyzes the amide bond formation between the acyl component simocyclinone C4 and a 3-amino-4,7-dihydroxycoumarin moiety. The enzyme, which was overexpressed and purified to apparent homogeneity, showed surprisingly high substrate tolerance, making it an attractive tool for the generation of new aminocoumarin antibiotics.
Materials and methods
Bacterial strains, plasmids and culture conditions
For standard cloning procedures Escherichia coli XL1 Blue MRF′ (Stratagene) and standard protocols as described by Sambrook and Russell (2001) were used. Streptomyces lividans TK23 (Kieser et al. 2000) was used as host strain for the isolation of the Streptomyces vector pGM9 (Muth et al. 1989) and was grown in YMG medium (Kieser et al. 2000) at 28°C for 2–3 days. Protoplast preparation and transformation of S. lividans T7 (Heinzelmann et al. 2001) for protein expression was carried out according to standard methods (Kieser et al. 2000). E. coli transformants with pGM9 fusion plasmids were selected with kanamycin (50 mg·ml−1); transformants with pRSET B were selected with carbenicillin (50 mg·ml−1). For selection of transformants of S. lividans T7, kanamycin (50 mg·ml−1) was used.
Inactivation of simL
For the inactivation of simL two fragments of 1,546 bp and 1,493 bp were amplified by PCR using the primers ΔsimL1_for (5′-TAGCCG TCTAGACTTTCACC-3′), Δ simL1_rev (5′-TGTTCAG GATATCCCGGACG-3′), ΔsimL2_for (5′-GAGGCCACC GATATCGAATGAG-3′) and Δ simL2_rev (5′-CGGGGTCGAT CTGCAGGAATCC-3′). Restriction sites introduced in the sequence are underlined in the primer sequences. PCR was performed using Pfu polymerase (Amersham Biosciences, Freiburg, Germany). The conditions for the PCR were one cycle at 96°C (5 min) followed by 25 cycles at 96°C (60 s), 55°C (60 s) and 72°C (3.5 min), and finally one cycle at 72°C (5 min). The amplified PCR fragment ΔsimL1 was digested with XbaI and EcoRV and cloned into the corresponding sites of the cloning vector pcDNA2.1 (Invitrogen, La Jolla, Calif., USA), resulting in pTLΔsimL-1. The PCR fragment ΔsimL2 was digested with EcoRV and PstI and ligated into the same sites of pTLΔsimL-1 to give pTLΔsimL-2, containing an in-frame deletion of 1,521 bp within simL. The fusion product of Δ simL1 and ΔsimL2 was excised from pTLΔsimL-2 as a 3.0-kb XbaI-it/PstI fragment and cloned into the same sites of pKC1132 (Bierman et al. 1992), a suicide vector containing the apramycin resistance gene, resulting in pTLΔsimL-4. S. antibioticus Tü 6040 was transformed with pTLΔsimL-4 as described by Kieser et al. (2000) using 4 mg lysozyme ml−1. The regeneration took place on R2YE plates for 16 h at 30°C.
Selection of recombinant mutants
After the transformation of S. antibioticus Tü 6040 protoplasts, resistant colonies were selected using 100 μg·ml−1 apramycin. The single crossover mutant SASCO4 was grown in the absence of apramycin, allowed to sporulate and then examined for loss of resistance to apramycin due to double-crossover events. Two mutants, SADCO2312 and SADCO2414, were examined further. Chromosomal DNA from wild-type S. antibioticus Tü 6040, as well as from mutants SASCO4, SADCO2312 and SADCO2414, was digested with SalI and hybridized with a probe containing parts of simK, which is located upstream of simL. The signal of the wild-type was expected at 3.2 kb, that of the simL− mutant at 1.7 kb.
Culturing of S. antibioticus and analysis of simocyclinone formation
For the production of simocyclinone D8, wild-type and mutant strains of S. antibioticus Tü 6040 were grown 5–8 days at 28°C in a medium consisting of 2.0% soybean meal and 2.0% glycerol in baffled shake-flasks. For analysis of secondary metabolites the medium was centrifuged (5,000 rpm, 4°C) and analysed as described below using a Nucleosil RP120-5 column (5 μM, 250×2 mm, Macherey-Nagel, Düren, Germany).
Cloning of simL
The gene encoding SimL was PCR amplified from cosmid VII-8g isolated from S. antibioticus Tü 6040 (Galm et al. 2002). The PCR amplification was accomplished using the forward primer simLN_for (5′-GGAGGATGTGCTGCAGAAGGCAACGAG-3′) and the reverse primer simLN_rev (5′-TCACAAGCTTCTCATTCGCCATGGGTG-3′). These primers introduced a PstI and a HindIII restriction site (highlighted in boldface). The PCR was performed using Pfu polymerase (Amersham Biosciences), and the fragment was cloned in the vector pBluescript SK(-) (Stratagene) after digestion of insert and vector with PstI and HindIII and gel purification. The conditions for the PCR were one cycle at 96°C (5 min) followed by 30 cycles at 96°C (90 s), 65°C (90 s) and 72°C (5 min), and finally one cycle at 72°C (10 min). The resulting vector pTLsimLN11 was digested with PstI and HindIII and gel-purified. The fragment was ligated into linearized pRSET B (Invitrogen) to give the N-terminal (His)6-tagged product pTLsimLN12. The construct was linearized with HindIII and fused with HindIII-digested Streptomyces expression vector pGM9 resulting in the vector pTL2.
Protein expression and purification
S. lividans T7 strains harbouring the expression constructs were cultured in YEME medium (Kieser et al. 2000) supplemented with kanamycin (10 mg·ml−1) for 2 days at 28°C in 300 ml baffled flasks. Each flask containing 50 ml YEME medium, supplemented with kanamycin (10 mg·ml−1) and thiostreptone (25 mg·ml−1), was inoculated with 1 ml of the preculture. After 24 h cultivation at 28°C (170 rpm in baffled flasks with steel springs) cells were harvested by centrifugation (5,000 g, 4°C, 10 min) and frozen overnight at −70°C. For lysis the cells were thawed and resuspended in 1 ml lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 8 mg lysozyme·ml−1) per gram (wet wt.) and incubated on ice for 30 min. Cells were broken by sonication (Branson Sonifier 250) for 10 min in 2-min intervals. The lysate was cleared by centrifugation (17,000 g, 4°C, 30 min). The His-tagged proteins were purified by nickel affinity chromatography by using Ni-nitrilotriacetic acid resin (Qiagen, Valencia, Calif., USA) to obtain nearly homogeneous protein fractions. The storage buffer contained 50 mM KH2PO4, pH 8.0, 5 mM DTT, 50 μM PMSF and 10% (v/v) glycerol.
The collected fractions were analysed by SDS-PAGE according to the method of Laemmli (1970) in 10% gels.
Isolation of simocyclinone C
For the production of simocyclinone C4, S. antibioticus Tü 6040 was cultivated in 300 ml baffled flasks containing 1.0 g NaCl, 1.0 g KH2PO4, 0.5 g MgSO4·7 H2O, 2 ml trace elements solution, 25 g soluble starch and 1.46 g L-glutamine per litre as described by Schimana et al. (2001). The components were dissolved in bidistilled H2O, adjusted to pH 7.3 and sterilized. The trace elements solution contained 1.0 g FeSO4·7 H2O, 0.1 g CuSO4·5H2O, 0.1 g MnSO4·H2O and 0.1 g ZnSO4·7 H2O per litre bidistilled H2O. After growth of these cultures for 48 h at 28°C and 120 rpm, 5 ml of these cultures was inoculated into 500 ml baffled flasks containing 250 ml media. After inoculation the cells were cultured at 28°C and 120 rpm for 6 days. The medium was acidified with 1 M HCl to pH 4.0 and extracted twice with an equal volume of ethyl acetate. After evaporation of the solvent the residue was dissolved in 40 ml methanol, passed through a Sephadex LH-20 column (100 cm × 5 cm, Amersham Biosciences) and eluted with methanol. The fractions from the Sephadex LH-20 column were analysed with HPLC using the method described below. Fractions containing simocyclinone C4 were pooled and further purified on a preparative HPLC column (Multosphere 120 RP18-5, 5 μm, 250 mm × 20 mm, C&S Chromatographie Service, Düren, Germany) using the same gradient as described below but with a flow of 3 ml·min−1 and 1% aqueous formic acid instead of the phosphoric acid. A 7.4-mg simocyclinone C4 sample was obtained. The structure of simocyclinone C4 was confirmed by 1H-NMR measured on an AMX 400 spectrometer (Bruker, Karlsruhe, Germany) using CD3OD as solvent.
Enzyme activity assay
For the characterization of SimL the assay established for NovL (Steffensky et al. 2000) was slightly modified. The assay was carried out in buffer containing a final concentration of 50 mM Tris-HCl (pH 8.0), 5 mM ATP and 5 mM MnCl2. The concentration of 3-amino-4,7-dihydroxy-8-methylcoumarin (ring B) was 200 μM and 1 mM of the acid component was used. Reactions were started by adding 2 μg purified SimL-N-His. Incubation was carried out at 30°C and stopped at specific time points with 5 μl 1.5 M trichloroacetic acid. The quenched reactions were centrifuged to remove precipitated protein (5 min, 15,000 rpm) and extracted with 1 ml ethyl acetate. An amount of 900 μl of the extract was transferred into a new tube and evaporated. The residue was dissolved in 30 μl methanol for HPLC analysis. For each reaction a control with heat-inactivated SimL was incubated.
HPLC analysis of enzymatic assay
Assay products were analyzed by using a Nucleosil RP120-5 column (5 μM, 250×2 mm, Macherey-Nagel) with a linear gradient from 30% to 100% acetonitrile in 0.01% aqueous phosphoric acid. Flow rate was 0.2 ml·min−1 and UV absorption was recorded at 340 nm. Authentic novobiocic acid (Pfizer, Kalamazoo, Mich., USA) and simocyclinone D-Met (Galm et al. 2002) were used as standard.
For the quantification of the different acyl components tested with SimL 1 mM 3-amino-4,7-dihydroxy-8-methylcoumarin (ring B) was incubated with 1 mM acyl component under conditions described above. The relative activity was calculated by using novobiocic acid as standard for UV absorption.
Liquid chromatography–mass spectrometry analysis of newly generated amincoumarins
Liquid chromatography–mass spectrometry (LC-MS) analysis was used to confirm the existence of newly generated aminocoumarins. Negative electrospray ionization mass spectra were obtained from a ThermoFinnigan TSQ Quantum instrument (electrospray voltage, 3 kV; heated capillary temperature, 300°C; sheath and auxiliary gas, nitrogen) equipped with a Multosphere RP18-5-column (5 μm, 4 mm × 250 mm, Macherey-Nagel) under basic conditions achieved by adding ammonia (15 μl·min−1). The solvents used were solvent A (99.9% H2O, 0.1% HCOOH) and solvent B (99.9% acetonitrile, 0.1% HCOOH). The profile for separation was a linear gradient from 70% A/30% B to 100% B in 14 min, 100% B for 15 min and then equilibration time with 70% A/30% B for 11 min. The flow rate was 0.5 ml·min−1. The collision-induced dissociation spectra during HPLC run were recorded with collision energy of +20 eV, collision gas argon and collision pressure 1.0×10−3 torr (133 mPa).
Results
Sequence analysis of SimL
Figure 2 shows an alignment of the predicted gene product of simL with the established amide synthetases NovL, CloL and CouL. Typical motifs of adenylate-forming enzymes, including BoxI, BoxII, motif A8 and motif A10 (Stuible et al. 2000; Marahiel et al. 1997) are highlighted. None of these proteins shows a 4′-phosphopantetheinyl attachment site, in clear contrast to the non-ribosomal peptide synthetases (Marahiel et al. 1997). Whereas NovL, CloL and CouL share, on average, 89% identity with each other on the amino acid level, the predicted SimL shows only 35% identity to these enzymes. Especially motif A8, which was shown to be involved in the coordination of pyrophosphate release in adenylate-forming enzymes (Stuible et al. 2000) is poorly conserved in SimL. Therefore, the function of SimL had to be confirmed experimentally.
Inactivation of simL
The gene simL, which codes for a protein of 519 amino acids, was inactivated in the simocyclinone producer, S. antibioticus Tü 6040, by an in-frame deletion of 1521 nucleotides (507 amino acids) within the coding sequence. An inactivation construct containing the shortened gene flanked by approx. 1.5 kb of homologous sequences on both sides was introduced into the S. antibioticus genome by protoplast transformation and homologous recombination (Kieser et al. 2000). Mutants resulting from single crossover events were selected, and subsequent selection for double crossover events yielded the desired gene replacement mutant. The correct simL− genotype was confirmed by Southern blotting, in comparison to the wild type, single crossover mutants and revertants to the wild type (Fig. 3). The simL− mutant and the wild-type strain, were cultured in production medium, and antibiotic formation was analyzed by HPLC. Whereas the wild type clearly produced the complete antibiotic, simocyclinone D8 (30 mg·l−1), no antibiotic formation was detected in the simL− mutants, suggesting that simL is essential for simocyclinone D8 formation. However, our attempts to unequivocally show the accumulation of the presumed substrates of the SimL reaction, i.e. the aminocoumarin ring and simocyclinone C4, remained unsuccessful. The aminocoumarin moiety is chemically unstable and may therefore have escaped detection. However, explanations for the lack of simocyclinone C4 accumulation remain speculative and therefore the experiment did not give definite proof of the function of simL. To obtain such proof, we decided to overexpress and purify the SimL protein and to investigate its reaction in vitro.
Overexpression and purification of recombinant SimL
The protein SimL was expressed as a histidine fusion protein for purification by metal affinity chromatography. A (His)6-tag was fused to the N-terminus of the protein by use of the pRSET B expression vector (Invitrogen). This construct was subsequently ligated into the Streptomyces expression vector pGM9 (Muth et al. 1989) and transformed into S. lividans T7. After induction with thiostreptone the formation of a protein of about 60 kDa (calculated mass, 59.338 kDa) could be clearly detected by SDS/PAGE (Fig. 4). Metal affinity chromatography with Ni-NTA resin yielded the protein in near homogeneity after this one step procedure, in a yield of 0.6 mg·l−1 culture.
Demonstration of the enzymatic activity of SimL
The presumed acyl substrate of SimL, i.e. simocyclinone C4 (Fig. 1), was isolated from cultures of S. antibioticus Tü 6040 and purified by column chromatography on Sephadex LH-20 followed by preparative reversed-phase HPLC. The structure of the isolated compound was confirmed by 1H NMR in comparison to the literature data (Holzenkämpfer et al. 2002).
The presumed amino substrate of SimL, i.e. 3-amino-4,7-dihydroxy-8-chlorocoumarin (Fig. 1) was not available as such, but the very similar 3-amino-4,7-dihydroxy-8-methylcoumarin was kindly provided by Pfizer. A previous mutasynthesis experiment had established that this compound was readily accepted by the amide synthetase of simocyclinone biosynthesis in vivo (Galm et al. 2002).
When these two compounds were incubated with SimL in the presence of ATP and Mn2+, a very efficient conversion of simocyclinone C4 into a new product was detected by HPLC (Fig. 5). Formation of this product was dependent on active SimL protein. The enzymatic product cochromatographed with the product of the aforementioned mutasynthesis experiment (Fig. 5), i.e. simocylinone D-met, which differs from simocyclinone D8 by carrying a methyl group (instead of a chlorine) in position 8 of the aminocoumarin ring. LS-MS analysis confirmed the expected molecular mass of this compound ([M-H]−=910), and the UV–Vis spectrum was identical to that of the previously identified simocyclinone D-met.
HPLC analysis of the product of the amide synthetase SimL. a SimL assay with heat-inactivated SimL. b Assay with active SimL. c Simocyclinone C4 and D-met from a feeding experiment. Simocyclinone C4 always showed two peaks in HPLC, which gave identical molecular ions in mass spectrometry and which, after individual isolation, showed rapid interconversion into each other. Therefore, they may represent structural isomers
Optimal product formation was obtained at pH 8.0, with ATP and MnCl2 concentrations of 5 mM each. Under these conditions, specific activity was 600 pkat·mg−1 protein which corresponds to a turn-over rate of 2.5 min−1. Aminocoumarin concentrations larger than 200 μM inhibited the reaction. The reaction did not require the presence of coenzyme A as cosubstrate, nor a previous transfer of a 4′-phosphopantetheinyl cofactor to the protein.
Kinetic constants were determined by holding the aminocoumarin concentration constant at 200 μM and varying the concentration of the acyl substrate, and vice versa by holding the simocyclinone C4 concentration constant at 1 mM and varying the concentration of the amino substrate. This resulted in apparent KM values of 20.5 μM for the aminocoumarin substrate and 20.4 μM for simocyclinone C4, calculated by the Hanes–Woolf method. These results are in good agreement with previous data obtained for the amide synthetases NovL and CouL (Steffensky et al. 2000; Schmutz et al. 2003).
Substrate specificity of SimL
Because we were interested in the potential of SimL for mutasynthesis experiments (Galm et al. 2004a), we focussed our attention on the specificity of SimL for different acyl substrates. The genuine acyl substrate of SimL, simocyclinone C4, is structurally very different from the acyl substrates of the previously investigated amide synthetases NovL, CloL and CouL (Fig. 1). Incubation of the substrate of CouL, 3-methylpyrrole-2,4-dicarboxylic acid, in the SimL assay did not result in a product formation detectable by HPLC with UV detection. However, the substrate of NovL and CloL, i.e. 3-DMA-4HB was clearly accepted by SimL, prompting us to investigate the kinetic parameters for this substrate. The KM for this compound was determined as 253 μM, with Vmax resulting as 1.7 nkat·mg−1 protein. Therefore, though 3-DMA-4HB was less well accepted than the genuine acyl substrate, this experiment proved that SimL could tolerate substrates similar to the genuine acyl moiety of novobiocin and clorobiocin, which may be useful for mutasynthesis experiments. Conversion of 3-DMA-4HB was achieved at a very reasonable rate at concentrations, which may easily be reached in mutasynthesis experiments.
Subsequently, we tested a range of benzoic and cinnamic acid derivatives (Table 1). Many of these compounds were accepted by SimL. Highest conversion rates were observed for cinnamic acid, 4-hydroxycinnamic acid and 4-hydroxybenzoic acid. Benzoic acids with different substituents in position 4, or with additional substituents in position 3, were accepted as well, whereas 3,5-disubstiuted aromatic compounds yielded no readily detectable product formation. Likewise, product formation was below detection limit (UV detection) for structurally different substrates like DL-tyrosine or fatty acids. Muconic acid, which may be regarded as an analogue of the tetraene dicarboxylic acid attached directly to the aminocoumarin moiety in simocyclinone D8 (Fig. 1), was likewise not readily accepted.
The identity of all products was confirmed by LC-MS analysis. During this analysis, it became apparent that even those acyl substrates, which had not resulted in a UV-detectable product formation all showed traces of product formation in the SimL assay, which could clearly be identified in the highly sensitive LC-MS analysis.
Unfortunately, novenamine, which is the glycoside of novobiocic acid with 3-carbamoyl-noviose (i.e. novobiocin lacking the 3-DMA-4HB moiety), was not accepted by SimL, which shows a specificity of SimL for its amino substrate.
Discussion
In the present study, we provided experimental evidence that the gene simL of the simocyclinone biosynthetic gene cluster codes for an amide synthetase, which links the amino group of an aminocoumarin moiety to an acyl moiety. Our study provides biochemical support to the hypothesis, derived previously from the nature of the metabolites accumulated in S. antibioticus Tü 6040 (Holzenkämpfer et al. 2002; Schimana et al. 2001), that the structurally complex acyl component of the simocyclinones, consisting of an aromatic angucycline polyketide nucleus, the deoxysugar olivose and a tetraene dicarboxylic acid, is assembled before it is connected with the aminocoumarin moiety during simocyclinone D8 biosynthesis.
The substrate tolerance of SimL is clearly greater than that of the previously examined amide synthetases NovL, CloL and CouL, making SimL an attractive tool for the generation of new aminocoumarin antibiotics, both by chemoenzymatic synthesis (Freel Meyers et al. 2004) and by mutasynthesis (Galm et al. 2004a).
The substrate range for such experiments may be even further expanded by use of the corresponding amide synthetase involved in the biosynthesis of rubradirin (Fig. 1). Expression experiments with the gene rub11 (Sohng et al. 1997) resulted in soluble, purified protein (data not shown), but this did not show enzyme activity in our assays. This may be due to the fact that the genuine acyl substrate of the amide synthetase of rubradirin biosynthesis is still unknown. Further experiments to identify the amide synthetase of rubradirin biosynthesis are in progress.
References
Bierman M, Logan R, O’Brien K, Seno ET, Rao RN, Schoner BE (1992) Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49
Flatman RH, Howells AJ, Heide L, Fiedler H-P, Maxwell A (2005) Simocyclinone D8: an inhibitor of DNA gyrase with a novel mode of action. Antimicrob Agents Chemother 49:1093–1100
Freel Meyers CL, Oberthuer M, Xu H, Heide L, Kahne D, Walsh CT (2004) Characterization of NovP and NovN: completion of novobiocin biosynthesis by sequential tailoring of the noviosyl ring. Angew Chem Int Ed 43:67–70
Galm U, Schimana J, Fiedler H-P, Schmidt J, Li S-M, Heide L (2002) Cloning and analysis of the simocyclinone biosynthetic gene cluster of Streptomyces antibioticus Tü 6040. Arch Microbiol 178:102–114
Galm U, Dessoy MA, Schmidt J, Wessjohann LA, Heide L (2004a) In vitro and in vivo production of new aminocoumarins by a combined biochemical, genetic and synthetic approach. Chem Biol 11:173–183
Galm U, Heller S, Shapiro S, Page M, Li S-M, Heide L (2004b) Antimicrobial and DNA gyrase-inhibitory activities of novel clorobiocin derivatives produced by mutasynthesis. Antimicrob Agents Chemother 48:1307–1312
Heinzelmann E, Kienzlen G, Kaspar S, Recktenwald J, Wohlleben W, Schwartz D (2001) The phosphinomethylmalate isomerase gene pmi, encoding an aconitase-like enzyme, is involved in the synthesis of phosphinothricin tripeptide in Streptomyces viridochromogenes. Appl Environ Microbiol 67:3603–3609
Holzenkämpfer M, Walker M, Zeeck A, Schimana J, Fiedler H-P (2002) Simocyclinones, novel cytostatic angucyclinone antibiotics produced by Streptomyces antibioticus Tü 6040–II. Structure elucidation and biosynthesis. J Antibiot 55:301–307
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces Genetics, 2nd edn. John Innes Foundation, Norwich, UK
Laemmli, UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Li S-M, Heide L (2004) Functional analysis of biosynthetic genes of aminocoumarins and production of hybrid antibiotics. Curr Med Chem: Anti-Infect Agents 3:279–295
Marahiel MA, Stachelhaus T, Mootz HD (1997) Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev (Wash) 97:2651–2674
Muth G, Nussbaumer B, Wohlleben W, Puehler A (1989) A vector system with temperature-sensitive replication for gene disruption and mutational cloning in streptomycetes. Mol Gen Genet 219:341–348
Pojer F, Kahlich R, Kammerer B, Li S-M, Heide L (2003a) CloR, a bifunctional non-heme iron oxygenase involved in clorobiocin biosynthesis. J Biol Chem 278:30661–30668
Pojer F, Wemakor E, Kammerer B, Chen H, Walsh CT, Li S-M, Heide L (2003b) CloQ, a prenyltransferase involved in clorobiocin biosynthesis. Proc Natl Acad Sci USA 100:2316–2321
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schimana J, Fiedler H-P, Groth I, Süssmuth R, Beil W, Walker M, Zeeck A (2000) Simocyclinones, novel cytostatic angucyclinone antibiotics produced by Streptomyces antibioticus Tü 6040. I. Taxonomy, fermentation, isolation and biological activities. J Antibiot 53:779–787
Schimana J, Walker M, Zeeck A, Fiedler H-P (2001) Simocyclinones: diversity of metabolites is dependent on fermentation conditions. J Ind Microbiol Biotechnol 27:144–148
Schmutz E, Steffensky M, Schmidt J, Porzel A, Li S-M, Heide L (2003) An unusual amide synthetase (CouL) from the coumermycin A1 biosynthetic gene cluster from Streptomyces rishiriensis DSM 40489. Eur J Biochem 270:4413–4419
Sohng JK, Oh TJ, Lee JJ, Kim CG (1997) Identification of a gene cluster of biosynthetic genes of rubradirin substructures in S. achromogenes var. rubradiris NRRL3061. Mol Cell 7:674–681
Steffensky M, Li S-M, Heide L (2000) Cloning, overexpression, and purification of novobiocic acid synthetase from Streptomyces spheroides NCIMB 11891. J Biol Chem 275:21754–21760
Stuible H, Buttner D, Ehlting J, Hahlbrock K, Kombrink E (2000) Mutational analysis of 4-coumarate: CoA ligase identifies functionally important amino acids and verifies its close relationship to other adenylate-forming enzymes. FEBS Lett 467:117–122
Trefzer A, Pelzer S, Schimana J, Stöckert S, Bihlmaier C, Fiedler H-P, Welzel K, Vente A, Bechthold A (2002) Biosynthetic gene cluster of simocyclinone, a natural multihybrid antibiotic. Antimicrob Agents Chemother 46:1174–1182
Acknowledgements
We thank Dr. H-P. Fiedler for providing us with strain S. antibioticus Tü6040 and reference substance. We thank J.K. Sohng and C.G. Kim for providing cosmids containing the rubradirin biosynthetic gene cluster. This work was supported by a joint grant from the Deutsche Forschungsgemeinschaft (DFG) and KOSEF.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Luft, T., Li, SM., Scheible, H. et al. Overexpression, purification and characterization of SimL, an amide synthetase involved in simocyclinone biosynthesis. Arch Microbiol 183, 277–285 (2005). https://doi.org/10.1007/s00203-005-0770-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-005-0770-0