Abstract
The organization of all genes required for the anaerobic conversion of toluene to benzoyl-CoA was investigated in denitrifying Azoarcus-like strain EbN1. All of these genes are clustered within 25.3 kb of contiguous DNA sequence, which includes only a few intervening sequences. The toluene-catabolic genes are organized in two apparent operons. One contains the genes (bssCAB) for the three subunits of benzylsuccinate synthase, which initiates anaerobic toluene degradation by converting toluene to (R)-benzylsuccinate. The BssCAB proteins of strain EbN1 are most similar to those of Thauera aromatica strain K172. The bssCAB genes are part of a larger putative operon (bssDCABEFGH), which contains the gene bssD, encoding the activase for benzylsuccinate synthase, and four genes (bssEFGH) encoding proteins of unknown function. RT-PCR experiments showing continuation of transcription over the three largest intergenic regions of the bss operon support the assumed structure. Moreover, BssG was identified as toluene-induced protein. Downstream of the bss genes, another large putative operon (bbsA–H) was identified that contains all genes required for β-oxidation of benzylsuccinate to benzoyl-CoA, e.g. bbsEF, encoding succinyl-CoA:(R)-benzylsuccinate CoA-transferase. Immediately upstream of the bss operon, genes for a two-component regulatory system were identified; their products may sense toluene and induce the expression of both catabolic operons. The order and sequences of the bss and bbs genes are highly similar among toluene-degrading denitrifiers. The bss and bbs genes of the FeIII-reducing Geobacter metallireducens display less sequence similarity and are organized differently. The genes between the bss and bbs operons and in the flanking regions differ between strain EbN1 and the other strains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oxygen-independent degradation pathways of hydrocarbons have been demonstrated as novel metabolic capacities in bacteria during the last decade (for overview see Heider et al. 1999; Spormann and Widdel 2000; Widdel and Rabus 2001; Widdel et al. 2003). While aerobic bacteria initialize the degradation of hydrocarbons exclusively by O2-dependent mono- or dioxygenase reactions, anaerobic bacteria must employ fundamentally different activation mechanisms. The best understood and apparently most widespread of these anaerobic mechanisms is the radical-catalyzed addition of hydrocarbons to fumarate, yielding substituted succinate derivatives. This reaction has been recognized for the activation of several alkyl-substituted benzenes as well as for n-alkanes (for overview, see Heider et al. 1999; Spormann and Widdel 2000; Widdel and Rabus 2001; Widdel et al. 2003).
Our understanding of fumarate-dependent hydrocarbon activation and the consecutive degradation pathways is most advanced in the case of toluene (Fig. 1). Formation of (R)-benzylsuccinate from toluene and fumarate is catalyzed by the glycyl-radical enzyme benzylsuccinate synthase, a heterohexameric enzyme of α2β2γ2 composition whose subunits are encoded by the bssCAB genes (Beller and Spormann 1998; Coschigano et al. 1998; Leuthner et al. 1998; Achong et al. 2001; Kane et al. 2002). The glycyl radical present in activated benzylsuccinate synthase (Krieger et al. 2001; Duboc-Toia et al. 2003) is generated by an S-adenosylmethionine (SAM)-dependent activating enzyme, as known for pyruvate formate-lyase and anaerobic ribonucleotide reductase (for overview, see Sawers and Watson 1998). The gene for the putative activating enzyme, bssD, is encoded immediately upstream of the bssCAB genes. Expression of the bss genes is probably controlled by a two-component regulatory system and the respective genes (tdiSR) are mostly located in close proximity to the bss operon (Coschigano and Young 1997; Leuthner and Heider 1998; Achong et al. 2001). It should be noted that, although the genes involved in anaerobic toluene degradation in Thauera aromatica strain T1 were originally designated tutEFDGH (Coschigano 2000), we will use the bss nomenclature to avoid confusion. Further degradation of (R)-benzylsuccinate to benzoyl-CoA in Thauera aromatica strain K172 follows a modified β-oxidation pathway (Leuthner and Heider 2000; Leutwein and Heider 2001, 2002), which is initiated by activation of (R)-benzylsuccinate to the CoA-thioester. Benzylsuccinyl-CoA is oxidized to benzoylsuccinyl-CoA and cleaved to benzoyl-CoA and succinyl-CoA. The latter is used for the activation of benzylsuccinate in a CoA-transfer reaction, thus releasing succinate for the regeneration of fumarate via succinate dehydrogenase. All enzymes required for β-oxidation of benzylsuccinate are encoded in the bbs operon. The sizes and genetic compositions of the regions between the bss and bbs operons are currently unknown. Further degradation of benzoyl-CoA proceeds via reductive dearomatization, hydrolytic ring cleavage, β-oxidation to acetyl-CoA units, and terminal oxidation to CO2 (Harwood et al. 1999; Boll et al. 2002).
Proposed reaction sequence for the anaerobic degradation of toluene in denitrifying strain EbN1 to the level of benzoyl-CoA (modified from Boll et al. 2002; Leuthner and Heider 2000). The fumarate cosubstrate of benzylsuccinate synthase is recycled during activation of benzylsuccinate and subsequent β-oxidation to benzoyl-CoA. The latter is further oxidized via ring cleavage to carbon dioxide (not shown). Reducing equivalents ([H]) are used for the reduction of nitrate to dinitrogen. Enzyme names of shown (bold) gene products are as follows: BssABC, benzylsuccinate synthase; BbsEF, succinyl-CoA:(R)-benzylsuccinate CoA-transferase; BbsG, (R)-benzylsuccinyl-CoA dehydrogenase; BbsH, phenylitaconyl-CoA hydratase; BbsCD, 2-[hydroxy(phenyl)methyl]-succinyl-CoA dehydrogenase; BbsAB, benzoylsuccinyl-CoA thiolase. For coding genes of these enzymes, see Fig. 2
Among the known bacteria that degrade toluene anaerobically, Azoarcus-like strain EbN1 is unique in utilizing ethylbenzene as an alternative hydrocarbon substrate (Rabus and Widdel 1995). Despite chemical and structural similarities between the two hydrocarbons, their anaerobic degradation pathways differ completely. Whereas toluene catabolism follows the common route (Fig. 1), ethylbenzene is anaerobically hydroxylated and dehydrogenated to acetophenone, which is then carboxylated and converted to benzoyl-CoA as the first common intermediate of the two pathways (Rabus and Heider 1998; Kniemeyer and Heider 2001). Since both pathways are regulated independently (Rabus and Heider 1998; Champion et al. 1999), strain EbN1 is a useful study organism to gain insights into the largely unexplored principles of specific substrate sensing and gene activation in anaerobic hydrocarbon metabolism. The genes for the ethylbenzene degradation pathway, including those for putative regulators, have recently been identified on a 56-kb contig from strain EbN1 (Rabus et al. 2002a). Here, we describe the organization of the genes needed for anaerobic toluene catabolism, which were identified on a different contig from this bacterium.
Materials and methods
Bacterial strain, growth conditions, and isolation of genomic DNA
The denitrifying bacterium Azoarcus-like strain EbN1 (β-Proteobacteria) was isolated from anoxic freshwater mud sampled in Bremen, Germany (Rabus and Widdel 1995). Strain EbN1 was cultivated and cells were harvested as previously described (Rabus and Widdel 1995). The procedure for isolation of genomic DNA was modified (Rabus et al. 2002a) from methods reported by others (Ausubel et al. 1992; Zhou et al. 1996).
Construction of shotgun libraries, DNA sequencing, and sequence assembly
Two shotgun libraries with average insert sizes of 1.5 and 3.5 kb were generated for DNA sequencing. The obtained sequences were assembled. Regions of weak quality within the analyzed contig were improved by resequencing and primer walking. Final sequence quality was based on three independent reads and sequencing of both strands. This procedure was recently described in more detail (Rabus et al. 2002a). The nucleotide sequence has been deposited at EMBL under the accession number BX682953.
Gene prediction, functional assignment, and data management
The program ORPHEUS (Frishman et al. 1998) was used for gene prediction. The program was adjusted, falsely predicted ORFs were removed, and ORFs refined as previously described (Rabus et al. 2002a).
Similarity searches were carried out by the BLAST programs (Altschul et al. 1997) and screening of the amino acid sequences of the predicted ORFs against the non-redundant protein database and the translated nucleotide database sequences. The predicted ORFs were functionally assigned with the INTERPRO system (Apweiler et al. 2001) and screening against the Clusters of Orthologous Groups of proteins (COGs; Tatusov et al. 2001). To evaluate the reliability of gene annotation (i.e. functional assignment), an additive scoring tool was used. More details about this procedure are given in Rabus et al. (2002a). Results of the automated ORF prediction and functional assignment were manually controlled for the entire contig (36.3 kb). Multiple alignments were generated with ClustalW (Thompson et al. 1994), and used to determine identities of amino acids with GAP (Wisconsin Package Version 10.2, Genetics Computer Group (GCG), Madison, Wis., USA).
The methods used for functional assignment (BLAST, INTERPRO and the additive scoring tool) are all implemented in the annotation platform HTGA (High Throughput Genome Annotation; Rabus et al. 2002a). More details about the design of the HTGA system are provided on the Web page http://www.micro-genomes.mpg.de/ebn1/.
The genomic sequence of Geobacter metallireducens is currently being determined by the DOE Joint Genome Institute (http://www.jgi.doe.gov). DNA sequence data from the draft version (November 7, 2002) are accessible at NCBI under accession number NZ_AAAS01000001 (http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/genom_table.cgi). A sequence region of about 60 kb of the G. metallireducens genome was used to compare bss and bbs genes among different toluene-degrading bacteria.
RT-PCR experiments
Total RNA was prepared from toluene-grown cells of strain EbN1 by the hot-phenol method as described by Aiba et al. (1981). The dried RNA was dissolved in water, and potential contaminating DNA was removed by treating the samples with RNase-free DNase (Promega, Mannheim, Germany) according to the manufacturer’s instructions. The quality of RNA preparations was controlled using the Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany), and cDNA was synthesized with gene-specific reverse primers by H Minus M-MuLV reverse transcriptase (MBI Fermentas, St. Leon Roth, Germany) according to the manufacturer’s protocol. PCR was carried out on a 50-µl scale under standard conditions with REDTaq Polymerase (Sigma-Aldrich, Munich, Germany) and the synthesized cDNAs as templates (2 µl individual reverse transcription products). To prove the absence of DNA in the RNA preparations, controls (cDNA synthesis) were carried out in the absence of reverse transcriptase. To cover the intergenic region between bssA and bssB, reverse primer bssBrev (TTACACGTGGTCGCGGAA) and forward primer bssAfor (GACCTGATCGTGCGGGTATC) were used (expected product of 470 bp). Likewise, primers bssErev (GGCTCAGGGTCTCGGTATTCA) and bssBfor (GGATACCCATCATGAGCGCA) were used for the intergenic region between bssB and bssE (expected product of 435 bp), and primers bssFrev (GATGATGTCGCCGGTGTTG) and bssEfor (ACGTCGGTCTCGGCAAGAT) for that between bssE and bssF (expected product of 285 bp). The sizes of the obtained RT-PCR products were analyzed by agarose gel electrophoresis.
Two-dimensional gel electrophoresis and analysis by mass spectrometry
Differential analysis of protein patterns by two-dimensional gel electrophoresis was based on cells of strain EbN1 anaerobically grown on toluene or benzoate. Samples were prepared and separated by two-dimensional gel electrophoresis as recently described (Rabus et al. 2002b; Gade et al. 2003). Gels were loaded with 50 and 500 µg protein, respectively, and stained with silver and colloidal Coomassie brilliant blue, respectively.
Toluene-specific protein spots were selected and excised manually using a cutting tool with a 1.5-mm needle. Proteolytic digests were carried out on a PROTEINEER dp digest and sample preparation robot (Bruker Daltonik, Bremen) using a commercial digestion kit (DP 96 Kit; Bruker) containing all necessary buffers, porcine trypsin as proteolytic enzyme, and α-cyano-4-hydroxycinnamic acid as MALDI matrix. The PROTEINEER dp run consisted of several washing steps, incubation for 4 h with trypsin, extraction, and thin-layer sample preparation on an AnchorChip600 MALDI target (Bruker).
MS fingerprint and MS/MS fragment spectra were acquired with an ultraflex TOF/TOF instrument (Bruker), equipped with a gridless reflectron, Lift cell, and Scout MTP ion source in positive ionization mode. In a single automated run, MS spectra were acquired and, based on the resulting peak lists, up to ten precursor ions were selected and submitted to Lift MS/MS without manual interference. The combined information of the fingerprint and fragment spectra was submitted to a protein database search (Mascot search engine; Matrix Science, London) carried out against a database containing the genetic information described in this study. The MS-tolerance was set to 30 ppm for external calibration.
Results and discussion
The present study describes a shotgun DNA sequencing approach to identify genes of strain EbN1 (related to Azoarcus, β-Proteobacteria) that are involved in the anaerobic conversion of toluene to benzoyl-CoA. The general pathway of anaerobic toluene catabolism is depicted in Fig. 1. DNA fragments from shotgun sequencing were screened for genes related to toluene metabolism by similarity of the gene products to known protein sequences of T. aromatica strains K172 (Leuthner et al. 1998; Leuthner and Heider 2000) and T1 (Coschigano et al. 1998), and Azoarcus sp. strain T (Achong et al. 2001). By this approach, a 36,333-bp contig was assembled from a total of 831 sequence reads. Based on the criteria described in the Materials and methods section and in a recent study (Rabus et al. 2002a), 35 ORFs were finally predicted on the contig (Fig. 2). INTERPRO/COG references, BLASTP hits, and assigned functions for each ORF are listed in Table 1. Bioinformatical annotation of gene function was complemented by a combined physiological/RT-PCR/proteomic approach. Identified genes correlated to anaerobic toluene degradation are indicated at each reaction step of the pathway shown in Fig. 1.
Scale model of gene organization in the investigated contig from denitrifying strain EbN1. Putative functions of the depicted ORFs are listed in Table 1. Color coding: blue genes related to anaerobic oxidation of toluene to benzoyl-CoA (hues indicate initial reaction and β-oxidation-like reaction sequence as explained in the text); orange regulatory proteins, gray ORFs with assigned putative functions; white hypothetical proteins. The scale indicates nucleotide position on the DNA fragment
Genes of the benzylsuccinate synthase operon bssDCABEFGH
Benzylsuccinate synthase was previously purified from T. aromatica strain K172 and demonstrated to catalyze the formation of (R)-benzylsuccinate from toluene and fumarate (Leuthner et al. 1998). Three genes, bssA, bssB and bssC, were identified that apparently code for the α- (BssA), β- (BssB) and γ-subunits (BssC) of the heterohexameric enzyme. Identification was based on similarity to translated gene sequences from T. aromatica strains K172 (Leuthner et al. 1998) and T1 (Coschigano et al. 1998), and Azoarcus sp. strain T (Achong et al. 2001). Active benzylsuccinate synthase of strain EbN1 is assumed to carry a radical on Gly-825 of the α-subunit (BssA). The Gly-825 probably functions in storing the radical and in generating a transient catalytically active thiyl residue on Cys-489. As the first step in catalysis, the thiyl radical may then abstract a hydrogen atom from the methyl group of toluene (Heider et al. 1999; Himo 2002). A similar radical-based mechanism has previously been demonstrated for pyruvate formate-lyase (Becker et al. 1999) and anaerobic ribonucleotide reductase (Eklund and Fontecave 1999). Amino acid sequences of the subunits of all currently known benzylsuccinate synthases display a high degree of similarity. The bss gene products of Azoarcus-like strain EbN1 are most similar to those of T. aromatica strain K172 (>94% identity), and slightly less similar to those of Azoarcus strain T, T. aromatica strain T1, and Thauera sp. strain DNT-1 (around 80% identity). The sequences from the latter three denitrifying strains are again >95% identical to each other, whereas the bss gene products of the phylogenetically more remote G. metallireducens are only 71–73% identical to those of any of the denitrifying strains (Fig. 3).
Phylogenetic tree of BssA subunits of benzylsuccinate synthases from different strains of bacteria. Sequences were retrieved from the databases and trimmed to the first aligned amino acid. Accession numbers: AY032676, Azoarcus-like strain T; AJ001848, Thauera aromatica strain K172; AF113168, T. aromatica strain T1; AB066263, Thauera sp. strain DNT-1; NZ_AAAS01000001, Geobacter metallireducens
Directly upstream of bssC, bssD, coding for an activating enzyme required for glycyl radical generation, was detected as part of the bss operon. As shown for bssD from T. aromatica strain K172 and Azoarcus sp. strain T, the start codon of bssD appears to be GTG, rather than an in-frame ATG located 34 codons further upstream. This is evident from the lack of similarity of the translated sequence between the ATG and GTG codons to those of other activating enzymes and from the location of the predicted promoter (see below). The BssD protein sequence from Azoarcus-like strain EbN1 is again most similar to the ortholog from T. aromatica strain K172 (79% identity) and less similar to those of Azoarcus sp. strain T, T. aromatica strain T1, Thauera sp. DNT-1, and G.metallireducens (50–63% identity). All BssD proteins contain three conserved Cys motifs in the N-terminal part of the proteins. The first motif (position 29–36 in strain EbN1) has a C-x3-C-x2-C structure characteristic of the emerging class of SAM-dependent radical generators (Sofia et al. 2001), whereas the other two (positions 55–65 and 89–99 in strain EbN1) correspond to the consensus sequences (C-x2-C-x2-C-x3-C) of typical [Fe8S8] ferredoxins. Sequence similarity with the pyruvate formate-lyase (PFL)-activating enzyme from Escherichia coli suggests that BssD is involved in introducing the glycyl radical into BssA. The C-x3-C-x2-C motif of PFL-activase was demonstrated to coordinate an atypical [Fe4S4] cluster that is required for the activity of the enzyme (Külzer et al. 1998) and for direct binding and reduction of SAM to methionine and an adenosyl radical (Walsby et al. 2002).
Downstream of the bssDCAB genes, at least four further genes of a putative continued operon are located, which we designate bssEFGH. The derived bssE product contains a Walker-type ATP/GTP binding-site motif and is similar to an emerging class of chaperone-like ATPases required for assembly, operation, and disassembly of protein complexes (Neuwald et al. 1999). Moreover, orthologs of bssE were independently shown to be part of the bss operons of three other bacterial strains (Coschigano 2000; Achong et al. 2001; Hermuth et al. 2002). bssF was predicted to be part of the operon, because genes coding for highly similar proteins are found directly downstream of bssE in all known and sufficiently far sequenced bss operons (intergenic distances ranging from 0 to 47 bases). The derived gene products do not show similarity to other known proteins, precluding any prediction of possible function. An extended transcript of the bss operon containing bssE and bssF was recently shown for Azoarcus sp. strain T, and the same study revealed an additional RNA 5′-end in front of bssF. To date, it is unknown whether this 5′-end reflects transcription initiation within the coding sequence of bssE or RNase processing of the extended bss transcript (Achong et al. 2001). The next gene encoded in the DNA sequence, bssG, is very closely spaced to bssF with an intergenic region of only nine bases. A gene coding for an orthologous protein is also present immediately downstream of bssF in T. aromatica strain K172, but not in the operon of the more distantly related FeIII-reducing G. metallireducens. The bss operons of other denitrifying toluene degraders are not sequenced sufficiently far to detect possible orthologs. Finally, a further gene, bssH, is located immediately downstream of bssG. Although no bssH orthologs are known from other strains due to insufficient sequence information, the observed one-base-overlap of the GTG start codon of bssH with the TGA stop codon of bssG strongly suggests the presence of a common transcript. The bssH gene product displays sequence similarity to members of the major facilitator superfamily (MFS) and the Bcr/CflA subfamily of drug resistance transporters. Some members of these transporter families are involved in uptake/efflux of aromatic compounds (Saier 2000). Because a hydrophobic compound like toluene is expected to diffuse freely across the cytoplasmic membrane, one may speculate that BssH functions in export of toxic levels of toluene from the cytosol rather than in toluene uptake. An alternative function in specific transport of benzylsuccinate appears unlikely, since strain EbN1 does not grow with benzylsuccinate.
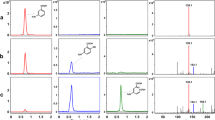
In strain EbN1, the largest intergenic regions of the putative bssDCABEFGH operon are between bssA and bssB (97 bp), between bssB and bssE (119 bp), and between bssE and bssF (44 bp). These intergenic regions may be considered long enough to cause rho-dependent transcriptional termination, and the first two even contain putative RNA secondary structures resembling rho-independent termination signals (Fig. 4B). Therefore, we tested for continuation of transcription over these intergenic gaps by RT-PCR. Using primer pairs covering the intergenic regions, RT-PCR products of the expected sizes were obtained from total RNA of toluene-grown cells of strain EbN1, which were absent when the reverse transcriptase reaction was omitted (Fig. 4A). The last two genes of the predicted operon (bssGH) are so closely spaced to their preceding genes (intergenic distances 9 and 0 bases) that termination of transcription downstream of bssF or bssG is not plausible. In support of the suggested operon organization, the BssG protein in toluene-grown cells was specifically detected by two-dimensional gel electrophoresis (data not shown) and identified unambiguously by mass spectrometry of tryptic peptides (Fig. 5). The assumed ATG codon of bssG is the only reasonable start codon preceding the coding sequence of the first identified peptide (starting at amino acid 22). In contrast to this ATG start codon, two alternative GTG codons (codon positions 9 and 20) are not associated with possible ribosome binding sites.
Transcription of intergenic regions in the bss operon. A RT-PCR analysis. Lane 1 Size-marker, lane 2 RT-PCR reaction product of the intergenic region between bssA and bssB (expected product of 470 bp), lane 4 RT-PCR reaction product of the intergenic region between bssB and bssE (expected product of 435 bp), lane 6 RT-PCR reaction product of the intergenic region between bssE and bssF (expected product of 285 bp). Lanes 3, 5, 7 Corresponding controls in which reverse transcriptase was absent from the reaction mixtures. B Predicted RNA secondary structures in the intergenic regions. Analogous, but not identical RNA structures are also present in the corresponding intergenic regions from T. aromatica strain K172 (Hermuth et al. 2002). N changed base,△ deleted base, N inserted base
Identification of the bssG gene product by mass spectrometry. Tryptic peptides of the protein separated by 2D-electrophoresis were analyzed by MALDI MS and MS/MS (ultraflex TOF/TOF). The bssG gene product was identified unambiguously. A MALDI MS-spectrum. MS-peaks are assigned with the BssG sequence position (top) and the peptide mass (bottom). B BssG sequence map. Bars Peptides confirmed by mass spectrometry; positions in the BssG sequence are indicated. The sequence coverage is 65%. C MALDI MS/MS spectrum of peptide TVQLYYENVAR (position 142–152). MS/MS-peaks are assigned with the ion-type (top) and the peptide mass (bottom). Cleavage occurs most frequently at the peptide bond resulting in b-ions (fragments starting from N-terminus) and y-ions (fragments starting from C-terminus)
At present it cannot be determined whether the bss operon continues beyond bssH. One of two possible translational start codons of the gene following bssH (c2B001; see Fig. 2 and Table 1) overlaps with the bssH stop codon, indicating transcriptional read-through (if used). The other one is located 336 bases downstream, allowing for enough space for transcription termination of the bss operon and expression of the downstream genes as an independent operon. In any case, the three genes (c2A200, c2A203 and c2A204; see Fig. 2 and Table 1) following c2B001 are expected to be part of the same transcription unit, based on the short intergenic regions between the genes (0–22 bases). Thus, the bss operon of strain EbN1 is predicted to consist of eight genes and might even contain up to 12 genes (corresponding to 8.7 or 12.8 kb).
Operon encoding enzymes for β-oxidation of benzylsuccinate: bbsA-H
Further degradation of (R)-benzylsuccinate proceeds via β-oxidation to benzoyl-CoA and succinyl-CoA (see Fig. 1). To date, this reaction sequence has only been studied with T. aromatica strain K172. Based on N-terminal sequences of toluene-induced, electrophoretically separated proteins, nine genes (bbsA–I) were identified that form the bbs operon in this strain (Leuthner and Heider 2000). The bbsEF genes code for the two subunits of succinyl-CoA:(R)-benzylsuccinate CoA-transferase, bbsG for the subunit of 2-(R)-benzylsuccinyl-CoA dehydrogenase, as shown with the purified and characterized enzymes (Leutwein and Heider 2001, 2002). The next three enzymes of the pathway are encoded by bbsH (phenylitaconyl-CoA hydratase), bbsCD (two subunits of an alcohol dehydrogenase), and bbsAB (two subunits of a thiolase), respectively (C. Feil, K. Hermuth and J. Heider, unpublished data).
Using the bbs gene sequences from T. aromatica strain K172, orthologs (2cA309-316) of all bbs genes (except for bbsI, the only gene unaccounted for in the operon of T. aromatica strain K172) were also identified in Azoarcus-like strain EbN1. Therefore, the functions of all eight conserved bbs genes of strain EbN1 can be annotated. The bbs gene products of both strains displayed high sequence similarity (shared identical residues ranging from 82.7 to 95.0%), and strong similarity is also retained on the DNA level (89.1% identity for the overall operon). The bbs operons of Azoarcus-like strain EbN1 and T. aromatica strain K172 mainly differ in the length of the intergenic region between bbsF and bbsG. Closer inspection of this gap revealed the presence of another putative gene in the bbs operon of strain EbN1 (termed bbsJ) which is lacking in strain K172. bbsJ codes for a protein of 9.8 kDa that contains a C-x-x-C and a C-R-C motif, but does not show apparent similarity to entries in the current databases. Still, it must be regarded as an expressed gene of the operon, based on its good ribosome binding sequence and its short intergenic distances to the preceding and the following genes. Most interestingly, DNA alignment of the bbs operons from strains EbN1 and K172 shows that there is recognizable nucleotide similarity between the rather large bbsFG intergenic gap of strain K172 and the last quarter of bbsJ from strain EbN1. One possible interpretation of this finding would be that the bbs operon of T. aromatica strain K172 has evolved from a bbsJ-containing operon by deletion of most of that gene and some subsequent deterioration of the remaining “scar” sequence. Genes coding for bbsA-H orthologs were also detected in the current draft version of the G. metallireducens genome. However, the degree of similarity between corresponding bbs genes of G. metallireducens and T. aromatica strain K172 was lower (derived amino acid sequence identities ranging from 39.1 to 75.5%). Moreover, three additional genes are inserted into the bbs operon of G. metallireducens. Two of these clearly code for the subunits of an electron-transferring flavoprotein (ETF), which is expected to serve as physiological electron acceptor for benzylsuccinyl-CoA dehydrogenase (Leutwein and Heider 2002); the third codes for a protein similar to a domain of heterodisulfide reductases, thiol:fumarate oxidoreductases or succinate dehydrogenases and may therefore also be involved in electron transfer from benzylsuccinyl-CoA to the respiratory chain. The organization of the bbs operon in Azoarcus-like strain EbN1, T. aromatica strain K172, and G. metallireducens is shown in Fig. 6.
Conserved promoter structures of the bss and bbs operons
The 5′-flanking DNA sequences of the bss and bbs operons of strain EbN1 were analyzed for similarity to the characterized promoter regions of the orthologous operons from T. aromatica strain K172 (Leuthner and Heider 2000; Hermuth et al. 2002) and Azoarcus sp. strain T (Achong et al. 2001), as well as to the upstream sequences of other sequenced bss operons. As shown in Fig. 7, several conserved sequence motifs can be identified upstream of the bss and bbs operons of strain EbN1 and all other known bss and bbs operons from denitrifying bacteria. Two of these motifs are similar to typical −10 and −35 boxes of E. coli and are underlined in Fig. 7. The spacing of these two sequence motifs varies by one to two bases in the different operons, but would be consistent with RNA polymerase binding in all cases. Further conserved motifs of all sequences are located in the regions between bases −40 to −50 and around base −65, relative to the mapped (or presumed) transcriptional starts. The strong conservation of these sites and their location just upstream of the RNA polymerase binding site identifies these motifs as prime candidates for binding of regulatory protein(s) that may be involved in induction of gene expression in response to anaerobiosis and/or toluene availability. It should be noted that the putative promoter region suggested for T. aromatica strain T1 in Fig. 7 differs from a previously mapped promoter 305 bases upstream of the assumed GTG start codon of the corresponding bssD ortholog (“tutE”; Coschigano 2000). The transcriptional start point of bssD proposed in Fig. 7 would not have been detected under the experimental conditions employed. The occurrence of internal 5′-ends of RNA species in the bss operon, as detected upstream of bssC in T. aromatica strains K172 and T1 (Hermuth et al. 2002; Coschigano 2000) and upstream of bssF in Azoarcus sp. strain T (Achong et al. 2001), cannot be predicted for strain EbN1 because the respective DNA sites are not sufficiently conserved.
Promoter consensus of bss and bbs operons from denitrifying bacteria. Bacterial strains depicted are Azoarcus-like strains EbN1 and T, as well as Thauera strains K172, T1 and DNT-1. Mapped transcription starts are labeled in bold, the distances to the respective translation start codons are indicated by numbers. Sequences similar to known Escherichia coli −10 and −35 boxes are underlined. Conserved sequences that may either be involved in regulator or RNA polymerase binding are indicated by shading
Regulatory proteins for the bss and bbs operons
Directly upstream of the bss operon of Azoarcus-like strain EbN1, two adjacent genes, tdiR and tdiS, code for a two-component regulatory system. The gene organization and the corresponding gene products most closely resemble the TdiSR system of Azoarcus sp. strain T (Achong et al. 2001; 79 and 81% identity). The next similar proteins in the database are orthologous two-component systems from T. aromatica strain K172 and T1 (Leuthner and Heider 1998; Coschigano and Young 1997; 68 and 69% identity). The sensor component consists of two sensory PAS domains and a histidine kinase domain, each occupying about a third of the protein (Leuthner and Heider 1998). PAS domains are implicated in monitoring light, redox, or hydrocarbon stimuli in diverse sensory proteins (Taylor and Zhulin 1999) and are obviously well suited to serve the regulatory requirements in the present case. The regulator consists of domains for a response regulator and a helix-turn-helix motif (Leuthner and Heider 1998). All known TdiSR-like systems are encoded in direct proximity of the respective bss/tut genes and are suggested to be involved in transcriptional control of the toluene catabolic genes. Thus, it appears likely that the TdiSR system also regulates transcription of the bss and bbs operons in Azoarcus-like strain EbN1. The involvement of two-component regulatory systems in transcriptional control of (aerobic) toluene metabolism was first demonstrated for the TodST-system of Pseudomonas putida F1 (Lau et al. 1997). The sensor and regulator components of the predicted anaerobic regulatory systems even show significant similarity with their aerobic counterparts (Leuthner and Heider 1998). Recently, we reported the presence of another TdiSR-like two-component regulatory system in strain EbN1, whose genes are in close proximity to those coding for ethylbenzene dehydrogenase (Rabus et al. 2002a). The components of this putative ethylbenzene-responsive regulatory system are 35–40% identical to those of the known TdiSR systems and share the same domain organization. Remarkably, the first (PAS) and the last (His kinase) domains of the putative ethylbenzene sensor are much more similar (41–43% identity) to those of the known toluene sensors than the second (PAS) domain (16% identity). Thus, strain EbN1 possesses two TdiSR-like two-component regulatory systems, which may be able to discriminate (possibly via the second PAS domain) between toluene and ethylbenzene, thereby allowing a finely tuned, substrate-dependent regulation of the respective degradation pathways.
Other genes
The genes c2A174, c2A173 and c2A172 probably encode enzymes involved in β-oxidation. The ORF c2A174 codes for an enoyl-CoA hydratase-type enzyme of unknown function, and c2A173 for an alcohol dehydrogenase that is apparently a fusion protein of two subdomains, each similar to “short chain” alcohol dehydrogenases (Jörnvall et al. 1995). Similar fusions are known in other operons containing genes for β-oxidation enzymes, e.g. in the operons for aerobic phenylacetate catabolism (Luengo et al. 2001; Mohamed et al. 2002). Finally, c2A172 codes for a standard thiolase of unknown function. The divergently transcribed gene c2A175 apparently codes for a transcriptional regulator, which may be involved in regulation of the β-oxidation-related genes. It belongs to the IclR family of bacterial regulatory proteins (INTERPRO entry IPR005471). Members of this family have been implicated in regulation of organic acids catabolism: acetate utilization via the glyoxylate bypass in E. coli (Sunnarborg et al. 1990) or protocatechuate degradation in Acinetobacter sp. strain ADP1 (Popp et al. 2002).
The distance to the next gene (c2A178) is long enough (93 bases) to expect an independent promoter for it. The gene product of this gene is clearly an electron transferring flavoprotein ETF: ubiquinone oxidoreductase, which is required for channeling the redox equivalents derived from acyl-CoA dehydrogenase reactions (as reduced ETF) into the respiratory chain. The gene is located within 20 kb of the next gene coding for an ETF-reducing enzyme, namely benzylsuccinyl-CoA dehydrogenase (“BbsG”). The gene product of c2A179 is similar to the caiE and paaY gene products whose genes are correlated to the operons involved in carnitine metabolism and aerobic phenylacetate catabolism, respectively.
Insertion sequence: ISE1
Upstream of the bbs operon an insertion element (ISE1) was detected that contains the genes istA and istB, whose products are highly similar to known transposases/cointegrases and correlated helper proteins, respectively. Sequence similarities of istA with IS1162 of Pseudomonas fluorescens (Solinas et al. 1995) and istB with IS1474 of P. alcaligenes (Yeo and Poh 1997) indicate affiliation to the IS21 family. The coding region (istAB) is flanked by 13-base imperfectly (underlined) matching inverted repeats (32914-TGCGGATTCCGAC/GTCGGAATGCGCA-35670). They are highly similar to conserved terminal parts of inverted repeats in other members of the IS21 family (http://www-is.biotoul.fr/), such as IS408 (TGCG T/G ATT C/T C) and IS1162 (TGCG T/G ATTTTC). The occurrence of a conserved mismatch in the fifth position from the start/end of the terminal region of the inverted repeat is striking. The short inverted repeats in isE1 are possibly the result of a deletion. Introduction of a single gap in the right inverted repeat allows the inverted repeats to be enlarged (TGCGGATTCCGACCCAACGTGACCGC/GCGGTCACGTT.CGTCGGAATGCGCA) to 26 bases. These longer inverted repeats possibly represent the original sequence also containing a conserved region (AACGTGA) of the inverted repeats of IS408 (http://www-is.biotoul.fr/). The inverted repeats are directly flanked by 7 bases of direct repeats (GGCTGTG), which probably originated from duplication of the target site. Such an insertion element appears to be absent in either flanking region of the bbs operons in G. metallireducens or T. aromatica strain K172.
Genetic organization
The order of the first six genes of the bss operon (bssDCABEF) is identical in all currently known examples (except for T. aromatica strain T1, where only partial sequence information on bssF is available). The five operons from denitrifying strains can be arranged into two subgroups differing in the length of the intergenic distance between bssB and bssE. In the operons of Azoarcus-like strain EbN1 and T. aromatica strain K172, these genes are separated by a transcribed intergenic region of 122 bases which may contain a stable RNA structure (Hermuth et al. 2002). This distance is much shorter in the operons from Azoarcus sp. strain T, T. aromatica strain T1, and Thauera sp. strain DNT-1 (47 bases). The different operon organizations are correlated with the respective benzylsuccinate synthases belonging to different similarity subgroups (see preceding section on the bss operon), and therefore allow for a facile PCR-based discrimination of the type of bss operon in denitrifying toluene-degrading strains (S. Zorn, K. Verfürth, J. Heider, unpublished). In the bss operon of the phylogenetically remote G. metallireducens, bssG and bssH are lacking, as well as the following genes found in strain EbN1.
The gene organization of the bbs operons in strains EbN1 and K172 differ in the presence of bbsJ between bbsF and bbsG in strain EbN1 and the presence of bbsI gene as the last gene of the operon of strain K172. The function of the bbsI and bbsJ gene products in β-oxidation of benzylsuccinate is not known; the two proteins share no similarity. Interestingly, the preliminary sequence of the bbs operon of G. metallireducens only codes for orthologs of BbsA–H, but does not contain genes for a BbsI- or BbsJ-like protein. Moreover, this operon starts with the bbsEFGH genes, followed by three inserted genes (see above section on bbs operon) not present in the other bbs operons, and ends with the bbsABCD genes. The different bbs operon organizations of Azoarcus-like strain EbN1, T. aromatica strain K172, and G. metallireducens are shown in Fig. 6.
In strain EbN1, five genes coding for proteins of unknown function are predicted to be located between the predicted bssDCABEFGH and bbsABCDJEFGH operons. Four of these genes may be cotranscribed with the bss operon, but their relevance for anaerobic toluene catabolism is unknown. In G. metallireducens, the intercalating sequence between the bss and bbs operons is about 4.4 kb and codes for six predicted genes. The ORF following bssF in G. metallireducens codes for a TodX-like protein, which is part of the toluene catabolic operon and implicated in toluene transport in Pseudomonas sp. (Wang et al. 1995). Even though this TodX-like protein does not display pronounced similarity with BssH from Azoarcus-like strain EbN1, the presence of genes coding for potential toluene transporters in (or next to) the bss operons in both organisms is remarkable. The other five genes of G. metallireducens are apparently not involved in toluene degradation and are disparate from the corresponding genes in strain EbN1. Moreover, the known flanking genes of the bbs operons (both flanks) and the bss/tdiSR operons (only 5′-flank) of T. aromatica strain K172 also differ from those of strain EbN1 and G. metallireducens. Therefore, the operons involved in anaerobic toluene metabolism seem to be embedded in quite different genomic contexts in varying bacterial species.
Similar to the genes involved in anaerobic toluene metabolism, those of anaerobic ethylbenzene metabolism of strain EbN1 are organized in two apparent operons, (ebdABCDped and apc1–5bal; Rabus et al. 2002a). The intercalating sequence (about 16 kb) between these operons contains genes coding for two different two-component regulatory systems, which are probably involved in sequential regulation of the upper (ebdABCDped) and lower part (apc1–5bal) of the degradation pathway. Strain EbN1 grows with either ethylbenzene or acetophenone, which is in accordance with a sequential regulation of these two operons (Rabus et al. 2002a). In contrast, strain EbN1 grows with toluene, but not with benzylsuccinate, which may at least partially be explained by a strictly coordinated regulation of the bss and bbs operons (mediated by the tdiSR gene products and toluene as inducer).
Conclusions
A unique property of strain EbN1 is its capacity to degrade toluene and ethylbenzene anaerobically via two completely different pathways. The identification of the corresponding genes will enable more detailed investigations into their substrate-dependent regulation. Based on the observed expression of “bssG” in toluene-metabolizing cells, the bss operon can be predicted to include this gene. This demonstrates the benefit of proteomic approaches for the annotation of hypothetical proteins. Considering the high degree of conservation among benzylsuccinate synthases, it will be interesting to compare them to the so far unknown subunit sequences of (1-methylpentyl)succinate synthase, the proposed n-hexane activating enzyme of Azoarcus-like strain HxN1 (Rabus et al. 2001). These two types of enzymes might differ on the sequence level, since they activate chemically different hydrocarbons and form products with different stereochemical properties. The high sequence similarity of the bss and bbs operons of different species may be utilized for environmental studies to monitor expression of genes involved in anaerobic degradation of aromatic hydrocarbons. In fact, expression of bssA has only recently been determined in fuel-contaminated groundwater by real-time RT-PCR (Beller et al. 2002). Such in situ expression studies together with a determination of key metabolites, e.g. benzylsuccinates (Beller 2000; Elshahed et al. 2001), could substantially advance our analytical capabilities to assess biodegradation of hydrocarbons in oil reservoirs or bioremediation efforts at contaminated sites.
References
Achong GR, Rodriguez AM, Spormann AM (2001) Benzylsuccinate synthase of Azoarcus sp. strain T: cloning, sequencing, transcriptional organization, and its role in anaerobic toluene and m-xylene mineralization. J Bacteriol 183:6763–6770
Aiba H, Adhya S, de Combrugghe B (1981) Evidence for two functional gal promotors in intact Escherichia coli. J Biol Chem 256:11905–11910
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MDR, Durbin R, Falquet L, Fleischmann W, Gouzy J, Hermjakob H, Hulo N, Jonassen I, Kahn D, Kanapin A, Karavidopoulou Y, Lopez R, Marx B, Mulder NJ, Oinn TM, Pagni M, Servant F, Sigrist CJA, Zdobnov EM (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res 29:37–40
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1992) Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York
Bairoch A, Apweiler R (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res 28:45–48
Becker A, Fritz-Wolf K, Kabsch W, Knappe J, Schultz S, Wagner AFV (1999) Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase. Nat Struct Biol 6:969–975
Beller HR (2000) Metabolic indicators for detecting in situ anaerobic alkylbenzene degradation. Biodegradation 11:125–139
Beller HR, Spormann AM (1998) Analysis of the novel benzylsuccinate synthase reaction for anaerobic toluene activation based on structural studies of the product. J Bacteriol 180:5454–5457
Beller HR, Kane SR, Legler TC, Alvarez PJJ (2002) A real-time polymerase chain reaction method for monitoring anaerobic, hydrocarbon degrading bacteria on a catabolic gene. Environ Sci Technol 36:3977–3984
Boll M, Fuchs G, Heider J (2002) Anaerobic oxidation of aromatic compounds and hydrocarbons. Curr Opin Chem Biol 6:604–611
Champion KM, Zengler K, Rabus R (1999) Anaerobic degradation of ethylbenzene and toluene in denitrifying strain EbN1 proceeds via independent substrate-induced pathways. J Mol Microbiol Biotechnol 1:157–164
Coschigano PW (2000) Transcriptional analysis of the tutE tutFDGH gene cluster from Thauera aromatica strain T1. Appl Environ Microbiol 66:1147–1151
Coschigano PW, Young LY (1997) Identification and sequence analysis of two regulatory genes involved in anaerobic toluene metabolism by strain T1. Appl Environ Microbiol 63:652–660
Coschigano PW, Wehrman TS, Young LY (1998) Identification and analysis of genes involved in anaerobic toluene metabolism by strain T1: putative role of a glycine free radical. Appl Environ Microbiol 64:1650–1656
Duboc-Toia C, Hassan AK, Mulliez E, Ollagnier-de Choudens S, Fontecave M, Leutwein C, Heider J (2003) Very high-field EPR study of glycyl radical enzymes. J Am Chem Soc 125:38–39
Eklund H, Fontecave M (1999) Glycyl radical enzymes: a conservative structural basis for radicals. Structure 7:R257-R262
Elshahed MS, Gieg LM, McInerney MJ, Suflita JM (2001) Signature metabolites attesting to the in situ attenuation of alkylbenzenes in anaerobic environments. Environ Sci Technol 35:682–689
Frishman D, Mironov A, Mewes H-W, Gelfand M (1998) Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res 26:2941–2947
Gade D, Thiermann J, Markowsky D, Rabus R (2003) Evaluation of the 2D DIGE method for protein profiling. Soluble proteins of the marine bacterium Pirellula sp. strain 1. J Mol Microbiol Biotechnol 5:240–251
Harwood CS, Burchardt G, Hermann H, Fuchs G (1999) Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol Rev 22:439–458
Heider J, Spormann AM, Beller HR, Widdel F (1999) Anaerobic bacterial metabolism of hydrocarbons. FEMS Microbiol Rev 22:459–473
Hermuth K, Leuthner B, Heider J (2002) Operon structure and expression of the genes for benzylsuccinate synthase in Thauera aromatica strain K172. Arch Microbiol 177:132–138
Himo T (2002) Catalytic mechanism of benzylsuccinate synthase, a theoretical study. J Phys Chem B 106:7688–7692
Jörnvall H, Persson B, Krook M, Atrian S, Gonzàlez-Duarte R, Jeffrey J, Ghosh D (1995) Short-chain dehydrogenases/reductases (SDR) Biochemistry 34:6003–6013
Kane SR, Beller HR, Legler TC, Anderson RT (2002) Biochemical and genetic evidence of benzylsuccinate synthase in toluene-degrading, ferric iron-reducing Geobacter metallireducens. Biodegradation 13:149–154
Kniemeyer O, Heider J (2001) Ethylbenzene dehydrogenase, a novel hydrocarbon-oxidizing molybdenum/iron-sulfur/heme enzyme. J Biol Chem 276:21381–21386
Krieger CJ, Roseboom W, Albracht SPJ, Spormann AM (2001) A stable organic free radical in anaerobic benzylsuccinate synthase from Azoarcus sp. strain T. J Biol Chem 276:12924–12927
Külzer R, Pils T, Kappl R, Hüttermann J, Knappe J (1998) Reconstitution and characterization of the polynuclear iron-sulfur cluster in pyruvate formate-lyase-activating enzyme. Molecular properties of the holoenzyme form. J Biol Chem 273:4897–4903
Lau PCK, Wang Y, Patel A, Labbé D, Bergeron H, Brousseau R, Konishi Y, Rawlings M (1997) A bacterial basic region leucine zipper histidine kinase regulating toluene degradation. Proc Natl Acad Sci USA 94:1453–1458
Leuthner B, Heider J (1998) A two-component system involved in regulation of anaerobic toluene metabolism in Thauera aromatica. FEMS Microbiol Lett 166:35–41
Leuthner B, Heider J (2000) Anaerobic toluene catabolism of Thauera aromatica: the bbs operon codes for enzymes of β-oxidation of the intermediate benzylsuccinate. J Bacteriol 182:272–277
Leuthner B, Leutwein C, Schulz H, Hörth P, Haehnel W, Schiltz E, Schägger H, Heider J (1998) Biochemical and genetic characterization of benzylsuccinate synthase from Thauera aromatica: a new glycyl radical enzyme catalysing the first step in anaerobic toluene metabolism. Mol Microbiol 28:615–628
Leutwein C, Heider J (2001) Succinyl-CoA:(R)-benzylsuccinate CoA-transferase: an enzyme of the anaerobic toluene catabolic pathway in denitrifying bacteria. J Bacteriol 183:4288–4295
Leutwein C, Heider J (2002) (R)-Benzylsuccinyl-CoA dehydrogenase of Thauera aromatica, an enzyme of the anaerobic toluene catabolic pathway. Arch Microbiol 178:517–524
Luengo JM, Garcia JL, Olivera ER (2001) The phenylacetyl-CoA catabolon: a complex catabolic unit with broad biotechnological applications. Mol Microbiol 39:1434–1442
Mohamed MES, Ismail W, Heider J, Fuchs G (2002) Aerobic metabolism of phenylacetic acids in Azoarcus evansii. Arch Microbiol 178:180–192
Neuwald AF, Aravind L, Spouge JL, Koonin EV (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res 9:27–43
Popp R, Kohl T, Patz P, Trautwein G, Gerischer U (2002) Differential DNA-binding of transcriptional regulator PcaU from Acinetobacter sp. strain ADP1. J Bacteriol 184:1988–1997
Rabus R, Widdel F (1995) Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch Microbiol 163:96–103
Rabus R, Heider J (1998) Initial reactions of anaerobic metabolism of alkylbenzenes in denitrifying and sulfate-reducing bacteria. Arch Microbiol 170:377–384
Rabus R, Wilkes H, Behrends A, Armstroff A, Fischer T, Pierik AJ, Widdel F (2001) Anaerobic initial reaction of n-alkanes in a denitrifying bacterium: evidence for (1-methylpentyl)succinate as initial product and for involvement of an organic radical in n-hexane metabolism. J Bacteriol 183:1707–1715
Rabus R, Kube M, Beck A, Widdel F, Reinhardt R (2002a) Genes involved in the anaerobic degradation of ethylbenzene in a denitrifying bacterium, strain EbN1. Arch Microbiol 178:506–516
Rabus R, Gade D, Helbig R, Bauer M, Glöckner FO, Kube M, Schlesner H, Reinhardt R, Amann R (2002b) Analysis of N-acetylglucosamine metabolism in the marine bacterium Pirellula sp. strain 1 by a proteomic approach. Proteomics 2:649–655
Saier Jr MH (2000) A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev 64:354–411
Sawers G, Watson G (1998) A glycyl radical solution: oxygen-dependent interconversion of pyruvate formate-lyase. Mol Microbiol 29:945–954
Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE (2001) Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res 29:1097–1106
Solinas F, Marconi AM, Ruzzi M, Zennaro E (1995) Characterization and sequence of a novel insertion sequence, IS1162, from Pseudomonas fluorescens. Gene 155:77–82
Spormann AM, Widdel F (2000) Metabolism of alkylbenzenes, alkanes, and other hydrocarbons in anaerobic bacteria. Biodegradation 11:85–105
Sunnarborg A, Klumpp D, Chung T, LaPorte DC (1990) Regulation of the glyoxylate bypass operon: cloning and characterization of iclR. J Bacteriol 172:2642–2649
Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29:22–28
Taylor BL, Zhulin IB (1999) PAS domains: internal sensors of oxygen, redox potential and light. Microbiol Mol Biol Rev 63:479–506
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Walsby CJ, Hong W, Broderick WE, Cheek J, Ortillo D, Broderick JB, Hoffman BM (2002) Electron-nuclear double resonance spectroscopic evidence that S-adenosylmethionine binds in contact with the catalytically active [4Fe-4S]+ cluster of pyruvate formate-lyase activating enzyme. J Am Chem Soc 124:3143–3151
Wang Y, Rawlings M, Gibson DT, Bergeron H, Brousseau R, Lau PC (1995) Identification of a membrane protein and a truncated LysR-type regulator associated with the toluene degradation pathway in Pseudomonas putida F1. Mol Gen Genet 246:570–579
Widdel F, Rabus R (2001) Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr Opin Biotechnol 12:259–276
Widdel F, Boetius A, Rabus R (2003) Anaerobic biodegradation of hydrocarbons including methane, In: Balows A, Trüper HG, Dworkin W, Harder W, Schleifer KH (eds), The prokaryotes: an evolving electronic resource for the microbiological community. Heidelberg, Springer Science Online (http://www.prokaryotes.com)
Yeo CC, Poh CL (1997) Characterization of IS1474, an insertion sequence of the IS21 family isolated from Pseudomonas alcaligenes NCIB 9897. FEMS Microbiol Lett 149:257–263
Zhou J, Bruns MA, Tiedje JM (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322
Acknowledgements
We wish to thank Katja Heitmann (MPI Berlin) for finishing the DNA sequence, Ulrike Schweiger-Hufnagel and Markus Lubeck (Bruker Daltonik) for help with the mass spectrometry analysis, Lars Wöhlbrandt (MPI Bremen) for providing total RNA, and Friedrich Widdel (MPI Bremen) for general support of genomic/proteomic work at our institute and critical reading of the manuscript. This work was supported by the Max-Planck-Society, the Deutsche Forschungsgemeinschaft and Bruker Daltonik GmbH.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kube, M., Heider, J., Amann, J. et al. Genes involved in the anaerobic degradation of toluene in a denitrifying bacterium, strain EbN1. Arch Microbiol 181, 182–194 (2004). https://doi.org/10.1007/s00203-003-0627-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-003-0627-3