Abstract
Summary
We detected disease-causing mutations in 585 of 598 individuals (98 %) with typical features of osteogenesis imperfecta (OI). In mild OI, only collagen type I encoding genes were involved. In moderate to severe OI, mutations in 12 different genes were found; 11 % of these patients had mutations in recessive genes.
Introduction
OI is usually caused by mutations in COL1A1 or COL1A2, the genes encoding collagen type I alpha chains, but mutations in at least 16 other genes have also been associated with OI. It is presently unknown what proportion of individuals with clinical features of OI has a disease-causing mutation in one of these genes.
Methods
DNA sequence analysis was performed on 598 individuals from 487 families who had a typical OI phenotype. OI type I was diagnosed in 43 % of individuals, and 57 % had moderate to severe OI, defined as OI types other than type I.
Results
Disease-causing variants were detected in 97 % of individuals with OI type I and in 99 % of patients with moderate to severe OI. All mutations found in OI type I were dominant and exclusively affected COL1A1 or COL1A2. In moderate to severe OI, dominant mutations were found in COL1A1/COL1A2 (77 %), IFITM5 (9 %), and P4HB (0.6 %). Mutations in one of the recessive OI-associated gene were observed in 12 % of individuals with moderate to severe OI. The genes most frequently involved in recessive OI were SERPINF1 (4.0 % of individuals with moderate to severe OI) and CRTAP (2.9 %).
Conclusions
DNA sequence analysis of currently known OI-associated genes identifies disease-causing variants in almost all individuals with a typical OI phenotype. About 20 % of individuals with moderate to severe OI had mutations in genes other than COL1A1/COL1A2.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteogenesis imperfecta (OI) is a heritable connective tissue disorder that is mainly characterized by bone fragility. The typical clinical picture also includes extraskeletal findings such as blue or gray discoloration of the sclera and tooth abnormalities called dentinogenesis imperfecta [1].
The severity of bone fragility in OI varies widely. This is captured in the traditional classification that distinguishes four phenotypic categories (OI types I to IV) [2]. OI type I represents the mildest phenotype, OI type II is lethal in the neonatal period, OI type III is the most severe form of the disease in individuals surviving the neonatal period, and OI type IV is of moderate severity [3]. Three more phenotypic OI types (V, VI, and VII) have been described, based on specific clinical characteristics [4]. More recently, a large number of additional OI types (OI type VIII and higher) have been proposed on the basis of genetic findings [1] and are listed in the OMIM database. However, the addition of genotypically defined OI types to a phenotypic classification is controversial [5]. The 2015 Nosology and Classification of Genetic Skeletal Disorders exclusively uses phenotypic criteria to classify OI types [3].
It has been known for more than 30 years that OI phenotypes are most often caused by dominant alterations in COL1A1 (MIM 120150) or COL1A2 (MIM 120160), the genes coding for collagen type I alpha chains [1]. The molecular diagnosis of OI was initially performed by examining collagen type I protein from skin fibroblasts [6] and later by Sanger sequencing of COL1A1 and COL1A2. Both approaches have a high diagnostic yield in OI with typical phenotypes. For example, protein analysis in 132 individuals with “typical nonlethal OI” identified biochemical abnormalities in 87 % of cases [7]. In the most detailed Sanger sequencing study to date, pathogenic COL1A1/COL1A2 sequence alterations were detected in 87 % of 142 children with “typical OI” [8]. However, that study also showed that the diagnostic yield varied with the phenotypic group. Pathogenic variants were found in 94 % of individuals with OI type I, in 88 % of patients with OI type III but in only 63 % of children with OI type IV.
Starting in 2006, defects in at least 16 genes other than COL1A1 and COL1A2 have been linked to OI phenotypes [1]. Most of these gene defects lead to recessive forms of OI, even though two genes (IFITM5 [MIM 614757], P4HB [MIM 176790]) are associated with dominant OI, and defects in one gene (PLS3, [MIM 300131]) lead to X-linked bone fragility. Inclusion of these “new” genes in OI sequencing panels can be expected to increase the clinical sensitivity of molecular diagnosis in OI, but it is presently not known what proportion of individuals with a typical OI phenotype have identifiable pathogenic variants in one of the currently known OI genes. It is also not clear how frequently pathogenic variants in each of the newly discovered OI-associated genes cause the disease. Such data are needed to assess the scope for further improvements in the molecular diagnosis of OI and will also be useful for estimating the number of individuals that might benefit from the future development of gene-specific treatment approaches.
In the present study, we therefore assessed the results of molecular testing for the currently known OI genes in 598 individuals who had a typical OI phenotype and who had all been assessed at a single center.
Subjects and methods
Subjects
The study population comprised all individuals with a typical OI phenotype who were evaluated at the Shriners Hospital for Children in Montreal between December 2000 and February 2015 and in whom molecular diagnosis by sequence analysis of genomic DNA had been completed. One of the authors (F.G. or F.R.) assessed each patient clinically and made a diagnosis of OI type I, III, IV, V, VI, VII, or Cole-Carpenter syndrome [4]. Given that the study was performed at a center for pediatric orthopedics, the study population did not include patients with OI type II, which leads to death in the neonatal period.
The 598 individuals reported here were residents of a variety of countries and geographic regions: Canada (n = 370), USA (n = 154), Latin America (n = 44), Europe (n = 16), Middle East (n = 10), and others (n = 4). The ethnic distribution was as follows: Caucasian (n = 457), Hispanic (n = 68), Arab (n = 27), Asian (n = 25), First Nations (n = 13), and others (n = 8).
The spectrum of disease severity varies between OI centers, which makes it difficult to compare the spectrum of molecular diagnosis results between studies. At the same time, the diagnosis of specific OI types is somewhat subjective and can depend on the amount of available clinical information (e.g., the diagnosis of OI type VI requires histological analysis of bone tissue) or on the age of the patient (e.g., hyperplastic callus may not be evident in young children with OI type V). For easier comparison with other studies, we separated the present study cohort into two broad groups—“mild OI” (OI type I) and “moderate to severe OI” (all other OI types). The main distinguishing characteristic between these two categories is the presence of lower extremity deformities in the moderate to severe OI group. The OI type I group only included individuals who had “typical extraskeletal features” of OI. This means that they had either discoloration of sclera or dentinogenesis imperfecta or both. In the absence of such extraskeletal features, it can be difficult to distinguish OI from other causes of increased fracture incidence in children and adolescents. The diagnosis and clinical information were obtained by retrospective chart review.
The study was approved by the Institutional Review Board of McGill University. For individuals in whom a disease-causing mutation had been found as part of the clinical diagnostic workup, the present study was limited to a retrospective chart review and no consent was required. Individuals in whom Sanger sequencing had previously failed to detect a disease-causing variant were invited to have next-generation sequencing (NGS) on a research basis. In these individuals, consent was obtained if they were aged 14 years or older. For individuals below 18 years of age, parental consent was obtained, as well as assent from participants over 8 years of age.
Clinical analyses
Height was measured using a Harpenden stadiometer (Holtain, Crymych, UK). Height and weight were converted to age- and sex-specific Z-scores on the basis of reference data published by the Centers for Disease Control and Prevention [9].
Sequence analysis
Sanger sequencing (Applied Biosystems 3100 DNA sequencer) was used until June 2013 for sequence analyses in genomic DNA after PCR amplification of all exons and exon/intron boundaries of target genes. Until 2007, sequence analysis was limited to those COL1A1 and COL1A2 exons, which were positive on heteroduplex screening [10]; later, all COL1A1 and COL1A2 exons were sequenced without prior screening step. Sequencing of CRTAP (2007), P3H1 (2008), SERPINF1 (2011), and IFITM5 (2012) was added to the Sanger sequencing program, after variants in these genes had been identified as causes of OI.
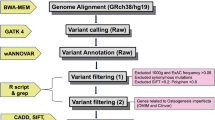
After June 2013, NGS was performed as the primary sequencing methodology, using an Ion Torrent PGM device (Life Technologies), as described [11]. Initially, a metabolic bone gene panel was assessed which included the following OI-related genes: COL1A1 (NM_000088.3), COL1A2 (NM_000089.3), BMP1 (NM_001199.3, NM_006129.4), CRTAP (NM_006371.4), FKBP10 (NM_021939.3), IFITM5 (NM_001025295.2), P3H1 (NM_022356.3), PLOD2 (NM_000935.2), PPIB (NM_000942.4), SERPINF1 (NM_002615.5), SERPINH1 (NM_001235.3), SP7 (NM_152860.1), and TMEM38B (NM_018112.2). Subsequently, the panel was expanded to include additional genes that are associated with OI or bone fragility disorders with a similar phenotype (ALPL (NM_000478.4), CREB3L1 (NM_052854.3), P4HB (NM_000918.3), PLS3 (NM_001136025.4), RUNX2 (NM_001024630.3), SEC24D (NM_014822.2), SPARC (NM_003118.3), and WNT1 (NM_005430.3).
A total of 10 ng DNA/sample were used for target enrichment by multiplex PCR and sequencing on an Ion 316 Chip (Ion PGM Hi-Q Sequencing Kit; Life Technologies), which was performed following the manufacturer’s instructions with the 200-bp single-end run configuration. Data from the sequencing run were processed using Torrent Suite software (version 5.04; Life Technologies) for base calls, read alignments, and variant calling using the reference genomic sequence (hg19) of target genes. Called variants were annotated using Ion Reporter (version 5.0). Variants were filtered out if they were present in the UCSC common SNPs database (http://genome.ucsc.edu), which includes SNPs from dbSNP 138 that have a minor allele frequency equal to or higher than 1 %. Variants were also filtered out if they had been observed in 4 or more of 45 control samples that had previously been sequenced with the same methodology.
The standards and guidelines of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology were used to classify variants as pathogenic or likely pathogenic [12]. Samples where no pathogenic or likely pathogenic variant was found in the initial panel were reanalyzed using the expanded panel. Mutations found by NGS were confirmed by Sanger sequencing.
Statistical analyses
Differences between two groups were tested for significance using the unpaired Student’s t test. Differences between three groups were evaluated by analysis of variance (ANOVA). All tests were two-tailed, and throughout the study, P values <0.05 were considered significant. These calculations were performed using the SPSS Statistics software version 19.0 (SPSS Inc., Chicago, Illinois, USA).
Results
The study included 598 individuals with a clinical diagnosis of OI who were from 487 families. OI type I was diagnosed in 43 % of the study population, and 57 % were classified as having one of the moderate to severe OI types (Table 1). Caucasians made up 86 % of the OI type I cohort and 70 % of the moderate to severe OI group. The family history, where available, was positive for OI in 74 % of individuals with OI type I and in 22 % of individuals with moderate to severe OI.
In 518 individuals, Sanger sequencing had been performed as the initial diagnostic methodology; pathogenic or likely pathogenic variants were found in 483 of these subjects. In 80 individuals, NGS was used as the initial diagnostic methodology. NGS was also performed on the 35 individuals in whom Sanger sequencing had not detected a pathogenic mutation. This resulted in the detection of disease-causing variants in 27 individuals, of whom 6 had disease-causing variants in genes that had not been assessed by Sanger sequencing. The other 21 individuals had mutations in COL1A1 or COL1A2 that had not been identified by previous Sanger sequencing for a variety of reasons, such as presence of a deletion of the entire COL1A1 gene [13] or polymorphisms at the annealing site of Sanger sequencing primers.
Overall, disease-causing variants were detected in 97 % of individuals with OI type I and in 99 % of patients with moderate to severe OI (Table 2). The 13 patients in whom sequence analysis did not reveal a disease-causing variant were from 13 unrelated families. The mean read depth in the samples from these 13 individuals was 744 (range 428 to 1196), similar to the samples where a disease-causing variant was found (799 (range 367 to 1465)). In nine of the samples with negative NGS, coverage of COL1A2 exon 3 was <20. Mutations in this exon were excluded by Sanger sequencing.
All mutations found in OI type I were dominant and exclusively affected COL1A1 or COL1A2 (Fig. 1). COL1A1 haploinsufficiency (i.e., nonsense or frameshift) mutations were found in 126 of these patients, splice site mutations in 55 patients (COL1A1, n = 52; COL1A2, n = 3), triple-helical glycine substitutions in 48 patients (COL1A1, n = 22; COL1A2, n = 26) and other types of COL1A1/COL1A2 mutations in 17 patients.
In moderate to severe OI, dominant mutations were found in COL1A1/COL1A2, in IFITM5 (c.-14C > T in all individuals) [14], and in P4HB (c.1178A > G in two individuals with Cole-Carpenter syndrome) [15]. Mutations in one of the recessive OI-associated gene were observed in 40 individuals, who all had a moderate to severe phenotype (Table 2; Fig. 1). The genes most frequently involved in recessive OI were SERPINF1 and CRTAP. In two individuals with moderate to severe OI, the sequencing result was classified as “inconclusive,” because only one heterozygous mutation was found in a gene associated with recessive OI (SERPINF1, N = 1; SEC24D, N = 1).
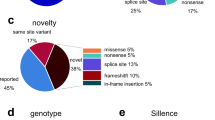
The number of unique mutations per gene was 188 in COL1A1 (66 haploinsufficiency, 52 triple-helical glycine substitutions, 49 splice site, 16 C-propeptide, 5 other), 106 in COL1A2 (92 triple-helical glycine substitutions, 8 splice-site, 6 other), 1 in IFITM5, 1 in P4HB, 3 in CRTAP, 5 in FKBP10, 5 in P3H1, 2 in PLOD2, 2 in PPIB, 7 in SERPINF1, 1 in SPARC, and 4 in WNT1 (Supplemental Tables 1 and 2). Overall, 49 of these mutations were not listed in the OI variant database and were therefore considered novel.
Discussion
In this study, sequence analysis of genomic DNA identified disease-causing mutations in 585 of 598 (98 %) individuals with a typical OI phenotype. The percentage of patients in whom a disease-causing variant could be identified was similar for OI type I and for the moderate to severe OI types, but the mutation spectrum differed between these two groups. In OI type I, we observed only COL1A1 and COL1A2 mutations, whereas in moderate to severe OI, mutations in COL1A1 and COL1A2 explained the disease in less than 80 % of patients, and mutations in 12 different genes were found. Apart from COL1A1/COL1A2, IFITM5 was the most frequently involved gene in moderate to severe OI and was responsible for the disease in 9 % of individuals with moderate to severe OI. Mutations in recessive genes were observed in 12 % of individuals with moderate to severe OI, but were not seen in the context of mild OI.
The present study was conducted in a specialized treatment center for OI, and therefore, the study population comprised a relatively large proportion of individuals with moderate to severe OI (57 %). In recent population-based studies, moderate to severe OI types represented only 31 and 23 %, respectively, of all individuals with OI [8, 16]. To compare mutation findings between studies, it is therefore useful to separately report results for mild OI and for moderate to severe OI. It must be acknowledged, however, that the present “moderate to severe OI” group may be skewed towards the more severe end of the spectrum, given the study setting in a center that receives referrals for specialized care from many countries. Population-based studies would be required to determine the OI mutations spectrum that is not affected by the inherent biases of hospital-based analyses.
It is often assumed that >90 % of individuals with OI have COL1A1 and COL1A2 mutations [17, 18]. While our results in OI type I are consistent with this assumption, we found that COL1A1/COL1A2 mutations explained the disease in only 264 of the 342 individuals (77 %) with moderate to severe OI. Similarly, Lindahl et al. found COL1A1/COL1A2 mutations in 31 of 43 children (72 %) with OI types III and IV [8]. The notion that >90 % of individuals with OI have COL1A1/COL1A2 mutations is mainly based on linkage studies in families with dominant OI [19]. However, the results of linkage studies in dominant OI are not directly applicable to moderate to severe OI, because these individuals typically do not have a positive family history and recessive OI is relatively common in this group.
Overall, we detected mutations in genes other than COL1A1/COL1A2 in about a fifth of individuals with moderate to severe OI. Close to half of the individuals with mutations in these “other” genes had the dominant c.-14C > T mutation in IFITM5, which is the single most prevalent mutation in the present series. This mutation leads to the addition of 5-amino acids to the N-terminus of BRIL, the protein encoded by IFITM5 [20]. No other IFITM5 mutations were found in the present study.
Biallelic mutations in genes linked to recessive OI were present in 12 % of individuals with moderate to severe OI and about 7 % of the entire OI population at our institution. In comparison, about 10 % of all individuals included in the OI variant database have recessive OI (accessed 16 March 2016). However, as observations in newly described genes may be more likely to be reported than variants in well-known genes, the proportion of individuals with recessive OI in the OI variant database is likely an overestimate of the true prevalence of recessive OI. The prevalence of recessive OI may also vary between geographic areas, as recessive disorders may be more prevalent in regions where consanguinity is common.
SERPINF1 and CRTAP mutations were the most common causes of recessive OI in the present study. This may be influenced by the presence of founder effects in the population served by our institution. The SERPINF1 c.295C > T (p.Arg99*) mutation was observed in six individuals from four apparently unrelated families of French Canadian origin [21]. A homozygous intronic c.472-1021C > G CRTAP mutation that introduces a cryptic splice was found in seven members of a community in Northern Quebec who all were diagnosed with OI type VII [22, 23]. Together, these two mutations were responsible for close to one third of all occurrences of recessive OI in the present study.
We detected disease-causing COL1A1/COL1A2 variants in 97 % of individuals with OI type I, which is similar to the results of Lindahl et al. who found COL1A1/COL1A2 mutations in 94 % of such individuals [8]. It is noteworthy that we observed triple-helical glycine substitutions in 19 % of patients with an OI type I phenotype. Even though heterozygous COL1A1 null mutations are sometimes thought to be the sole cause of OI type I [5, 18, 24], our observations highlight the fact that the phenotype caused by some triple-helical glycine substitutions can be clinically indistinguishable from that caused by COL1A1 haploinsufficiency. Similar observations have been made by Lindahl et al. [8].
The mild OI group of the present study included only individuals who had “typical OI type I,” which was defined as having blue/gray sclerae or dentinogenesis imperfecta, or both, in addition to bone fragility. For this reason, previously reported patients with mutations in PLS3 [25] or in the 3′-untranslated region of the short BMP1 transcript [26] were excluded from the present analysis, because these individuals had bone fragility without either scleral discoloration or dentinogenesis imperfecta.
Overall, we failed to detect disease-causing variants in about 2 % of our study population. It is possible that in some of these individuals, OI is caused by variants in the genes that are included in our panel but are located in areas that were not sequenced, such as deep intronic regions. However, it is likely that at least some of these individuals have disease-causing variants in genes that have not yet been associated with OI. In any case, our study suggests that these as yet undiscovered OI-related genes explain the disease in a very small proportion of individuals with OI. Almost all individuals with a typical OI phenotype have pathogenic variants in one of the known OI-associated genes.
References
Forlino A, Marini JC (2016) Osteogenesis imperfecta. Lancet 387:1657–1671
Sillence DO, Senn A, Danks DM (1979) Genetic heterogeneity in osteogenesis imperfecta. J Med Genet 16:101–116
Bonafe L, Cormier-Daire V, Hall C et al (2015) Nosology and classification of genetic skeletal disorders: 2015 revision. Am J Med Genet A 167A:2869–2892
Rauch F, Glorieux FH (2004) Osteogenesis imperfecta. Lancet 363:1377–1385
Van Dijk FS, Sillence DO (2014) Osteogenesis imperfecta: clinical diagnosis, nomenclature and severity assessment. Am J Med Genet A 164A:1470–1481
Bonadio J, Holbrook KA, Gelinas RE, Jacob J, Byers PH (1985) Altered triple helical structure of type I procollagen in lethal perinatal osteogenesis imperfecta. J Biol Chem 260:1734–1742
Wenstrup RJ, Willing MC, Starman BJ, Byers PH (1990) Distinct biochemical phenotypes predict clinical severity in nonlethal variants of osteogenesis imperfecta. Am J Hum Genet 46:975–982
Lindahl K, Astrom E, Rubin CJ, Grigelioniene G, Malmgren B, Ljunggren O, Kindmark A (2015) Genetic epidemiology, prevalence, and genotype-phenotype correlations in the Swedish population with osteogenesis imperfecta. Eur J Hum Genet 23:1042–1050
Ogden CL, Kuczmarski RJ, Flegal KM, Mei Z, Guo S, Wei R, Grummer-Strawn LM, Curtin LR, Roche AF, Johnson CL (2002) Centers for Disease Control and Prevention 2000 growth charts for the United States: improvements to the 1977 National Center for Health Statistics version. Pediatrics 109:45–60
Korkko J, Ala-Kokko L, De Paepe A, Nuytinck L, Earley J, Prockop DJ (1998) Analysis of the COL1A1 and COL1A2 genes by PCR amplification and scanning by conformation-sensitive gel electrophoresis identifies only COL1A1 mutations in 15 patients with osteogenesis imperfecta type I: identification of common sequences of null-allele mutations. Am J Hum Genet 62:98–110
Rauch F, Lalic L, Glorieux FH, Moffatt P, Roughley P (2014) Targeted sequencing of a pediatric metabolic bone gene panel using a desktop semiconductor next-generation sequencer. Calcif Tissue Int 95:323–331
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–424
Bardai G, Lemyre E, Moffatt P, Palomo T, Glorieux FH, Tung J, Ward L, Rauch F (2016) Osteogenesis imperfecta type I caused by COL1A1 deletions. Calcif Tissue Int 98:76–84
Rauch F, Moffatt P, Cheung M, Roughley P, Lalic L, Lund AM, Ramirez N, Fahiminiya S, Majewski J, Glorieux FH (2013) Osteogenesis imperfecta type V: marked phenotypic variability despite the presence of the IFITM5 c.-14C > T mutation in all patients. J Med Genet 50:21–24
Rauch F, Fahiminiya S, Majewski J, Carrot-Zhang J, Boudko S, Glorieux F, Mort JS, Bachinger HP, Moffatt P (2015) Cole-Carpenter syndrome is caused by a heterozygous missense mutation in P4HB. Am J Hum Genet 96:425–431
Wekre LL, Froslie KF, Haugen L, Falch JA (2010) A population-based study of demographical variables and ability to perform activities of daily living in adults with osteogenesis imperfecta. Disabil Rehabil 32:579–587
van Dijk FS, Byers PH, Dalgleish R, Malfait F, Maugeri A, Rohrbach M, Symoens S, Sistermans EA, Pals G (2012) EMQN best practice guidelines for the laboratory diagnosis of osteogenesis imperfecta. Eur J Hum Genet 20:11–19
Pepin MG, Byers PH (2015) What every clinical geneticist should know about testing for osteogenesis imperfecta in suspected child abuse cases. Am J Med Genet C Semin Med Genet 169:307–313
Sykes B, Ogilvie D, Wordsworth P et al (1990) Consistent linkage of dominantly inherited osteogenesis imperfecta to the type I collagen loci: COL1A1 and COL1A2. Am J Hum Genet 46:293–307
Cho TJ, Lee KE, Lee SK et al (2012) A single recurrent mutation in the 5′-UTR of IFITM5 causes osteogenesis imperfecta type V. Am J Hum Genet 91:343–348
Rauch F, Husseini A, Roughley P, Glorieux FH, Moffatt P (2012) Lack of circulating pigment epithelium-derived factor is a marker of osteogenesis imperfecta type VI. J Clin Endocrinol Metab 97:E1550–E1556
Ward LM, Rauch F, Travers R, Chabot G, Azouz EM, Lalic L, Roughley PJ, Glorieux FH (2002) Osteogenesis imperfecta type VII: an autosomal recessive form of brittle bone disease. Bone 31:12–18
Morello R, Bertin TK, Chen Y et al (2006) CRTAP is required for prolyl 3-hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell 127:291–304
Forlino A, Cabral WA, Barnes AM, Marini JC (2011) New perspectives on osteogenesis imperfecta. Nat Rev Endocrinol 7:540–557
Fahiminiya S, Majewski J, Al-Jallad H, Moffatt P, Mort J, Glorieux FH, Roschger P, Klaushofer K, Rauch F (2014) Osteoporosis caused by mutations in PLS3: clinical and bone tissue characteristics. J Bone Miner Res 29:1805–1814
Fahiminiya S, Al-Jallad H, Majewski J, Palomo T, Moffatt P, Roschger P, Klaushofer K, Glorieux FH, Rauch F (2015) A polyadenylation site variant causes transcript-specific BMP1 deficiency and frequent fractures in children. Hum Mol Genet 24:516–524
Acknowledgments
We thank Patty Mason for technical assistance and Mark Lepik for the preparation of the figures. F.R. received salary support from the Chercheur-Boursier Clinicien program of the Fonds de Recherche du Québec-Santé. This study was supported by the Shriners of North America and the Fonds de recherche Québec-Santé.
Roles of the authors
GB performed analyses. FHG contributed patient information. PM revised manuscript content. FR conceptualized the project, contributed patient information, finalized the report, and accepts responsibility for the integrity of the data analysis. All authors have read and approved the final version of the manuscript.
Conflicts of interest
Ghalib Bardai, Pierre Moffatt, Francis H Glorieux, and Frank Rauch declare that they have no conflict of interest.
Web resources
Exome Aggregation Consortium (ExAC) Browser: http://exac.broadinstitute.org/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
Osteogenesis Imperfecta Variant Database: https://oi.gene.le.ac.uk/
UCSC database, version hg19: http://www.genome.ucsc.edu/
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 239 kb)
Rights and permissions
About this article
Cite this article
Bardai, G., Moffatt, P., Glorieux, F.H. et al. DNA sequence analysis in 598 individuals with a clinical diagnosis of osteogenesis imperfecta: diagnostic yield and mutation spectrum. Osteoporos Int 27, 3607–3613 (2016). https://doi.org/10.1007/s00198-016-3709-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00198-016-3709-1