Abstract
Introduction and hypothesis
The rs1800255, COL3A1 2209 G>A polymorphism in the alpha 1 chain of collagen type III has been associated with an increased risk of pelvic organ prolapse (POP). In one of our previous studies however, polymerase chain reaction-based restriction fragment length polymorphism (PCR-RFLP) misdiagnosed rs1800255, COL3A1 2209 G>A in 6 % of cases. The high-resolution melting (HRM) analysis on the contrary obtained a 100 % accordance for this specific polymorphism and was used in the present study to validate this risk factor for POP.
Methods
In this case–control study, women with and without symptoms of POP were included and compared. DNA was extracted from blood samples. HRM analysis was used to assess for the presence of the homozygous rs1800255. Groups were compared using the Pearson chi-square, Mann–Whitney, and t tests. The discrepancy between HRM and PCR-RFLP results was investigated using PCR-RFLP results available from our previous study.
Results
The study included 354 women: 272 patients with POP and 82 controls; 18 (7 %) cases versus 3 (4 %) controls had a homozygous rs1800255, COL3A1 2209 G>A polymorphism (odds ratio 1.9, 95 % confidence interval 0.5–6.9, compared to the wild type), and thus no association between POP and the homozygous polymorphism could be demonstrated. A discrepancy between HRM and PCR-RFLP results was found in 8 % of the samples.
Conclusions
The previously found statistically significant association between the rs1800255, COL3A1 2209 G>A polymorphism as measured with PCR-RFLP and POP could no longer be demonstrated. This raises concerns regarding the results of other association studies using PCR-RFLP.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pelvic organ prolapse (POP) is known to have a multifactorial etiology. Over recent years, the importance of genetics in the development of POP has become increasingly evident. Studies among (twin) sisters have shown that there is a high concordance in POP stage between sisters, irrespective of parity [1, 2], and that genetic factors attribute to 40 % of the variation of liability for POP [3]. Until now, a variety of different polymorphisms have been described in relation to POP. Part of these polymorphisms concern genes encoding proteins that are important substances of the extracellular matrix [4–10]. Others concern genes encoding for hormone receptors [11–13].
In 2009, our research group published a report on a polymorphism in the alpha 1 chain of the type III collagen protein-encoding gene (rs1800255, COL3A1 2209 G>A)[5]. In that publication, we presented a case–control study on the presence of polymorphisms in the gene encoding for COL3A1 and their association with POP. To detect these polymorphisms, we used polymerase chain reactions (PCRs) followed by restriction fragment length polymorphism (RFLP) analysis for DNA analysis. We found that the homozygous form of one of these polymorphisms (rs1800255, COL3A1 2209 G>A polymorphism) was associated with POP with an odds ratio (OR) of 5.0 (95 % confidence interval 1.4–17.1) compared to the wild type. Our results were in concordance with data published on the prevalence of this polymorphism in Taiwanese POP patients using the same analysis technique [6].
In order to bring this promising finding within reach for everyday practice, the accuracy of high-resolution melting (HRM) analysis was assessed. The HRM analysis is a low-cost, high-throughput technique to identify DNA polymorphisms by using the differences in melting curves between the different amplicons. However, HRM analysis cannot be used unthinkingly, since the accuracy of the analysis depends on the melting behavior of each specific DNA sequence. This melting behavior is determined by many factors like for instance the length of the amplicon and GC content [14] as well as the use of only a primer pair or in combination with a probe. Therefore, the results obtained differ for each specific case. This makes it sometimes impossible to identify all mutations correctly. For example, in an attempt to use HRM analysis for screening of glucose-6-phosphate dehydrogenase gene mutations in men, all cases were identified as “normal” because hemizygotes and wild types created almost identical melting curve profiles. It was only after improvement of the procedure that it was possible to identify all but one mutation correctly [15]. The same kind of problem occurred in an attempt to use HRM analysis for the identification of different genotypes in sickle cell disease. With the use of HRM, seven of nine genotypes could be distinguished from the wild type without any problem. It was however impossible to differentiate the last two genotypes with use of the HRM analysis method, while only one curve was created for both genotypes [16]. In this case, sequencing remained necessary for detection. Nevertheless, there are also numerous cases in which 100 % accordance between the results of the HRM analysis and sequencing is obtained. For example, in the detection of hemoglobin-beta gene mutation in β-thalassemia [17] and the detection of mutations in the HFE gene in hemochromatosis [18], the results of the HRM screening analysis were 100 % consistent with DNA sequencing. The reason for the 100 % accordance in these cases was the fact that the individual DNA variants all produced melting curves that were clearly distinguishable from each other.
Using DNA sequencing as the gold standard, we performed a study comparing the accuracy of PCR-RFLP and HRM for the detection of the rs1800255, COL3A1 2209 G>A polymorphism in 36 randomly selected DNA samples [19]. That study demonstrated 100 % accordance between HRM analysis and sequencing, with unambiguously distinctive melting profiles for heterozygous, homozygous and wild-type variants. This result was obtained by using an asymmetric amplification in combination with a probe that maximally increased the HRM specificity. However, 6 % of the results obtained by PCR-RFLP were incorrect as compared to HRM and direct sequencing [19].
This inconsistency has resulted in the current study in which the primary aim was to verify the association between the rs1800255, COL3A1 2209 G>A polymorphism and POP by using the HRM analysis method. Our secondary goal was to investigate the discrepancy between HRM and PCR-RFLP results in this cohort.
Materials and methods
Patients
Patients with POP, presenting themselves between January 2007 and September 2009 at the Department of Obstetrics and Gynecology of the Radboud University Medical Center, were asked to participate. As controls, vaginally parous women without symptoms of POP [i.e., most descended edge of the Pelvic Organ Prolapse Quantification (POP-Q) staging system was not beyond 1 cm above the hymen] and without previous POP surgery visiting the department during the same period were included. All women included were Caucasian of Dutch descent. The cases and controls partially overlapped the subjects of our first study [5], who were recruited between January 2007 and August 2008. Exclusion criteria for both groups were hereditary diseases with a known increased risk of POP, such as Ehlers-Danlos syndrome, and problems with the patient’s understanding of the study.
Data on age, parity, and the number of previous POP procedures were collected from all patients. Date at first visit was defined as the date of the first clinic visit. Date of first complaints was defined as the year of onset of the first POP complaints or otherwise the year of the (first) POP treatment. All women underwent gynecologic examination for the assessment of the POP-Q stage. Women were examined in a gynecological chair, in a 45° position, performing maximum Valsalva for all measurements of the POP-Q scale except total vaginal length (TVL), which was assessed during rest. The study was approved by the local Institutional Review Board (IRB) under number CMO 2007/043 and informed consent was obtained from all of the women.
Laboratory analysis
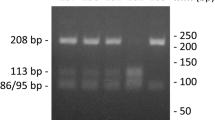
Genomic DNA was isolated from peripheral blood. DNA was isolated using the NucleoSpin Blood Kit (Macherey-Nagel, Düren, Germany) according the supplier’s protocol. PCRs were performed according to standard procedures in a total volume of 25 μl, containing 50 ng DNA (OD260/280 and 260/230 both > 1.8 as measured by nanodrop), 0.4 pmol/μl of each primer (5′-GCC CCA GGA CTT AGA GGT G and 5′-CCT TGC AGA CCA GGA GT, Invitrogen), 2.5 mM MgCl2, 0.2 mM dNTP (Roche, Mannheim, Germany), and 0.012 U/μl ThermoPerfect taq polymerase (Integro, Leuvenheim, The Netherlands). PCR was performed according to the following protocol: 5 min 95 °C, 30 × [1 min 95 °C, 45 s 62 °C, 45 s 72 °C], 5 min 72 °C. Subsequently, the 192 nt amplicon was purified using MultiScreen PCRμ96 plates and a vacuum manifold (Millipore, Billerica, MA, USA) and submitted to the sequence facility at the Radboud University Medical Center for sequence analysis. The forward and reverse sequences were determined separately using the primers described above. Following amplification as described above, presence of the rs1800255 polymorphism was determined subsequently by RFLP [19]. Ten microliters of PCR mix was digested with 2U AluI (New England Biolabs, Frankfurt am Main, Germany) in a final volume of 20 μl 1× Buffer A (New England Biolabs) at 37 °C overnight. Digestion products were resolved on a 1.5 % agarose gel containing ethidium bromide. In the case of homozygous 2209G, the AluI-digested amplicon should reveal bands at 171 and 22 nucleotides. In the case of homozygous 2209A, a single band at 192 nucleotides was expected, and in the case of heterozygosity bands at 192, 171, and 22 were visible. Each DNA sample was analyzed at least in duplicate and results interpreted by two of the authors (LCvK, JRD).
HRM was performed with HPLC purified primers and probes purchased from Qiagen Operon (Cologne, Germany). A 10-μl mixture containing 10 ng genomic DNA, 4 μl of the 2.5× LightScanner Master Mix containing LCGreen Plus® (Idaho Technology, Salt Lake City, UT, USA), 0.1 μM forward primer 5′-TGG TGA ACG TGG ACC TCC TGG ATT G-3′ (position 189572274–189572298 on chromosome 2 of the genomic sequence of rs1800255, COL3A1 2209 G>A polymorphism), 0.5 μM reversed primer 5′-GTT GAA TGG AAT GCT GTG GAG TTA CCT TT-3′ (position 189572352–189572380), and 0.5 μM probe 5′-GGA CTT AGA GGT GGA ACT-3′ GGT CCC-block (position 189572311–189572334) was prepared. The probe contained a C3 spacer (Operon Biotechnologies, Cologne, Germany) that functioned as a 3-foot block on the probe to prevent extension during PCR. The mixture was overlaid with 15 μl mineral oil, and PCR amplification was conducted at initial denaturation for 2 min at 95 °C followed by 50 cycles of denaturation for 30 s at 25 °C, respectively. The use of asymmetric amplification in combination with a probe strongly increases the HRM specificity. This was confirmed by a validation experiment in which sequencing of the single 106 nt amplicon that was generated in this PCR contained the expected sequence. The 96-well LightScanner® System (Idaho Technology) was used to analyze the DNA/dye complex by high-resolution thermal denaturation. Melting curves were generated by ramping between 55 and 95 °C at 0.1 °C/s. The LightScanner software (version 1.5) was used for genotyping analysis. The region of the probe melt was bracketed and fluorescence was normalized. The melting curves were displayed as derivative melting peaks and clustered by auto-grouping as reported previously [19].
Statistics
Missing data were excluded from analysis. The Pearson chi-square test was used to compare categorical variables. All other variables were analyzed using independent samples t test and Mann–Whitney U test. A test for deviation from Hardy–Weinberg equilibrium was performed in the control group; two-sided p values below 0.05 were considered statistically significant. For the power calculation, the assumptions in our previous study were used [5]. All data were analyzed using Statistical Package for the Social Sciences (SPSS) software, version 20.0.
Results
A total of 354 women were included; 272 patients with POP and 82 controls. Using the same assumptions as for our previous study [5], this resulted in a power of 85 % for the present study. The baseline characteristics of the POP and the control groups are presented in Table 1. There were no significant differences between the women with POP and the controls with regard to age at first visit and body mass index (BMI). Parity, however, was significantly different between the two groups, with parity being slightly higher among POP patients. There was no apparent deviation from Hardy-Weinberg equilibrium (p = 0.21).
A total of 354 DNA samples were analyzed with the HRM analysis method. Of those samples, 214 (60 %) had previously been analyzed in the PCR-RLFP analysis on rs1800255, COL3A1 2209 G>A polymorphism [5]. A discrepancy between HRM and PCR-RFLP was found in 17 samples, leading to a reclassification rate of 8 % (95 % confidence interval of 4.9–12.4) (Table 2).
The relation between POP and the COL3A1 2209 G>A polymorphism is presented in Table 1. No association between POP and the homozygous rs1800255, COL3A1 2209 G>A polymorphism could be demonstrated in the present study, with an OR of 1.9 (95 % confidence interval of 0.5–6.9) found. Findings were similar after adjustment for parity.
Discussion
This study was performed to validate the association between POP and the homozygous polymorphism in the alpha 1 chain of collagen type III (rs1800255, COL3A1 2209 G>A polymorphism) after (re)analysis of the DNA samples with the more accurate HRM analysis technique. The results showed that a statistically significant association could no longer be demonstrated.
In our review of the literature describing clinical studies on hereditary factors in POP [20], a discrepancy between the different studies on COL3A1 polymorphism and POP was found. Chen and colleagues [6] reported an association between the homozygous COL3A1 2209 G>A polymorphism as assessed with PCR-RFLP and POP. Jeon et al. [7] found that the GG genotype of this polymorphism, instead of the AA genotype, was significantly associated with POP. Martins et al. [8], however, did not find any association with POP. Our initial explanation for these differences was that all studies were performed in ethnically different populations, i.e., Korean, Dutch, Taiwanese, and Brazilian, with different background risks of POP. However, with the present knowledge, these differences may also be explained by the fact that the methodology to detect this single nucleotide variant (PCR-RFLP) was not sufficiently accurate for the analyses in these studies. The same may hold true for the discrepancy found in the results of the studies on the laminin gamma-1 (LAMC1) polymorphism [rs10911193 (C/T)] performed by Nikolova et al. [4] and Chen et al. [10]. The study group of Nikolova et al. [4] using the accurate sequencing method for their analyses found a statistically significant correlation between the LAMC1 gene polymorphism and POP, whereas Chen et al. [10] using PCR-RFLP analysis found no association. One explanation for the diminished accuracy of PCR-RFLP is the subjective reading of the electrophoresis patterns, as previously reported [19]. Other investigators have similarly described discordant results between PCR-RFLP and direct sequencing. Peters et al. [21] examined the association between three tumor necrosis factor alpha (TNF-α) promoter polymorphisms and severe sepsis after traumatic injury. Of 46 samples, 4 (9 %) were incorrectly classified as heterozygous by RFLP, whereas they were classified as GG-homozygous by sequencing. This error rate is in line with what we have published previously [19] and in this study using a larger sample set (6 and 8 %, respectively).
One limitation of our current study is the fact that 90 of the 304 subjects (30 %) of our previous PCR-RLFP analysis on rs1800255 [5] were not included because the sample was exhausted in our other studies. This hampers the full validation of the previous study results and obviously it cannot be ruled out that this could have influenced the current outcome. However, due to the additional samples, we still had over 80 % power to detect an OR of at least 3.8, the number used in the power calculation of our first study [5]. Nonetheless, to be able to detect an OR of around 2.0 or less, the sample size should have been larger.
Furthermore, it should be noted that we did not use DNA sequencing, the gold standard method for analyzing DNA, in our present study. However, in our previous study [19], the HRM analysis showed a 100 % accordance with DNA sequencing for the analysis of this specific polymorphism, which is in line with other publications [22, 23]. Furthermore, the patterns of the derivate melt curves were unambiguous. For this reason, we decided to use this rapid and inexpensive analysis method.
In conclusion, the previously reported statistically significant association between the rs1800255, COL3A1 2209 G>A polymorphism and POP was not confirmed in this study with more accurate measurement tools. Although a relation between POP status and genetic background has been proven [3, 24] and a relation with collagen status has been suggested [25], this specific polymorphism does not seem to have any additional value in the prediction of POP. However, since POP is likely to be a multigenic disorder, where the relatively small influence of multiple genes adds up to the risk of developing POP, a follow-up study with a larger sample size would be advisable.
Our findings raise concerns regarding studies using PCR-RFLP. Most studies on POP and polymorphisms utilized PCR-RFLP analysis and might therefore be prone to the same inaccuracy. Although we cannot extrapolate the results of our analysis to other studies using PCR-RFLP, we feel that researchers studying the association between diseases and polymorphisms should take this information on accuracy of measurement tools into account.
References
Buchsbaum GM, Duecy EE, Kerr LA et al (2006) Pelvic organ prolapse in nulliparous women and their parous sisters. Obstet Gynecol 108(6):1388–1393
Buchsbaum GM, Duecy EE (2008) Incontinence and pelvic organ prolapse in parous/nulliparous pairs of identical twins. Neurourol Urodyn 27(6):496–498
Altman D, Forsman M, Falconer C et al (2008) Genetic influence on stress urinary incontinence and pelvic organ prolapse. Eur Urol 54(4):918–922
Nikolova G, Lee H, Berkovitz S et al (2007) Sequence variant in the laminin gamma1 (LAMC1) gene associated with familial pelvic organ prolapse. Hum Genet 120(6):847–856
Kluivers KB, Dijkstra JR, Hendriks JC et al (2009) COL3A1 2209 G>A is a predictor of pelvic organ prolapse. Int Urogynecol J Pelvic Floor Dysfunct 20(9):1113–1118
Chen HY, Chung YW, Lin WY et al (2008) Collagen type 3 alpha 1 polymorphism and risk of pelvic organ prolapse. Int J Gynaecol Obstet 103(1):55–58
Jeon MJ, Chung SM, Choi JR et al (2009) The relationship between COL3A1 exon 31 polymorphism and pelvic organ prolapse. J Urol 181(3):1213–1216
Martins KD, de Jármy-DiBella ZI, da Fonseca AM et al (2011) Evaluation of demographic, clinical characteristics, and genetic polymorphism as risk factors for pelvic organ prolapse in Brazilian women. Neurourol Urodyn 30(7):1325–1328
Chen HY, Lin WY, Chen YH et al (2010) Matrix metalloproteinase-9 polymorphism and risk of pelvic organ prolapse in Taiwanese women. Eur J Obstet Gynecol Reprod Biol 149(2):222–224
Chen C, Hill LD, Schubert CM et al (2010) Is laminin gamma-1 a candidate gene for advanced pelvic organ prolapse? Am J Obstet Gynecol 202(5):505.e1–505.e5
Chen HY, Chung YW, Lin WY et al (2008) Estrogen receptor alpha polymorphism is associated with pelvic organ prolapse risk. Int Urogynecol J Pelvic Floor Dysfunct 19(8):1159–1163
Chen HY, Wan L, Chung YW et al (2008) Estrogen receptor beta gene haplotype is associated with pelvic organ prolapse. Eur J Obstet Gynecol Reprod Biol 138(1):105–109
Chen HY, Chung YW, Lin WY et al (2009) Progesterone receptor polymorphism is associated with pelvic organ prolapse risk. Acta Obstet Gynecol Scand 88(7):835–838
Erali M, Voelkerding KV, Wittwer CT (2008) High resolution melting applications for clinical laboratory medicine. Exp Mol Pathol 85(1):50–58
Pan M, Lin M, Yang L et al (2013) Glucose-6-phosphate dehydrogenase (G6PD) gene mutations detection by improved high-resolution DNA melting assay. Mol Biol Rep 40(4):3073–3082
Thomas V, Mazard B, Garcia C et al (2013) UGT1A1 (TA)n genotyping in sickle-cell disease: high resolution melting (HRM) curve analysis or direct sequencing, what is the best way? Clin Chim Acta 424:258–260
Ouragini H, Haddad F, Darragi I et al (2013) Rapid and inexpensive detection of common HBB gene mutations in Tunisian population by high-resolution melting analysis: implication for molecular diagnosis. Hematology 19(2):80–84
Marotta RV, Turri O, Morandi A et al (2011) High resolution melting analysis to genotype the most common variants in the HFE gene. Clin Chem Lab Med 49(9):1453–1457
Lince SL, Kluivers KB, Dijkstra JR et al (2009) Reliable identification of the type III collagen gene polymorphism rs1800255 with the use of high resolution melting analysis. Lab Med 40(10):604–606
Lince SL, van Kempen LC, Vierhout ME et al (2012) A systematic review of clinical studies on hereditary factors in pelvic organ prolapse. Int Urogynecol J 23(10):1327–1336
Peters DL, Barber RC, Flood EM et al (2003) Methodologic quality and genotyping reproducibility in studies of tumor necrosis factor −308G–>A single nucleotide polymorphism and bacterial sepsis: implications for studies of complex traits. Crit Care Med 31(6):1691–1696
Jancik S, Drabek J, Berkovcova J et al (2012) A comparison of direct sequencing, pyrosequencing, high resolution melting analysis, TheraScreen DxS, and the K-ras StripAssay for detecting KRAS mutations in non small cell lung carcinomas. J Exp Clin Cancer Res 31:79
Carbonell P, Turpin MC, Torres-Moreno D et al (2011) Comparison of allelic discrimination by dHPLC, HRM, and TaqMan in the detection of BRAF mutation V600E. J Mol Diagn 13(5):467–473
Allen-Brady K, Cannon-Albright L, Farnham JM et al (2011) Identification of six loci associated with pelvic organ prolapse using genome-wide association analysis. Obstet Gynecol 118(6):1345–1353
Lammers K, Lince SL, Spath MA et al (2012) Pelvic organ prolapse and collagen-associated disorders. Int Urogynecol J 23(3):313–319
Conflicts of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lince, S.L., van Kempen, L.C., Dijkstra, J.R. et al. Collagen type III alpha 1 polymorphism (rs1800255, COL3A1 2209 G>A) assessed with high-resolution melting analysis is not associated with pelvic organ prolapse in the Dutch population. Int Urogynecol J 25, 1237–1242 (2014). https://doi.org/10.1007/s00192-014-2385-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00192-014-2385-y