Abstract
Brain tumor classification and segmentation for different weighted MRIs are among the most tedious tasks for many researchers due to the high variability of tumor tissues based on texture, structure, and position. Our study is divided into two stages: supervised machine learning-based tumor classification and image processing-based region of tumor extraction. For this job, seven methods have been used for texture feature generation. We have experimented with various state-of-the-art supervised machine learning classification algorithms such as support vector machines (SVMs), K-nearest neighbors (KNNs), binary decision trees (BDTs), random forest (RF), and ensemble methods. Then considering texture features into account, we have tried for fuzzy C-means (FCM), K-means, and hybrid image segmentation algorithms for our study. The experimental results achieved a classification accuracy of 94.25%, 87.88%, 89.57%, 96.99%, and 97% with SVM, KNN, BDT, RF, and Ensemble methods, respectively, on FLAIR-, T1C-, and T2-weighted MRI, and the hybrid segmentation attaining 90.16% mean dice score for tumor area segmentation against ground-truth images.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
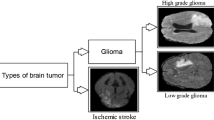
While the brain is the control unit of the human body, brain tumors have become fatal and life-threatening diseases escalating in recent days. It is generally a mass of tissue that originates from an irregular stimulus of tumorous cells in any part of the human brain. It is an activation in a single cell’s genes, which is the source of the act, resulting in the further uncontrollable division of nearby cells. More often than not, a brain tumor emerges inside the brain and its nerves or the brain’s coverings. These tumors are generally classified into malignant tumors or benign tumors [1,2,3]. The non-cancerous cells are termed benign, whereas cancerous cells are called malignant tumors. Benign types do not pervade the brain or its neighboring tissues. However, these can still harm the nearby tissues or vital organs, so they need to be treated in the nick of time. On the other hand, malignant tumors are life-threatening as they invade healthy tissues of the brain and further spread throughout the brain or other regions of the body.
A medical imaging technique, especially magnetic resonance imaging (MRI) [4, 5], plays an essential role in diagnosing and treating brain tumors with positive outcomes. The best parts of MRI, being an important modality of medical imaging [6,7,8], provide automated and precise diagnostic results. One of the most demanding problems while dealing with MR images is to segregate a few distinct cells and tissues from the image. Further, this leads up to the segmentation process. Segmentation of required objects aids physicians in identifying lesions more precisely; hence, it assumes a noteworthy job in computerized medical imaging. While there exist many mainstream segmentation techniques such as thresholding [9], region-based seed growing [10], and graph partitioning [11], still they lack behind in domain application of brain tumor classification due to similarities in intensity between some healthy tissues and brain tumors, which can give rise to uncertainty within the algorithm. This resulted in the usage of multispectral MR sequences [12] for tumor identification by researchers to overcome this problem. Nevertheless, four main difficulties were identified in this approach [13]. First, being the acquisition of such MR sequences is not always attainable owing to the condition as in the severity and urgency of patients. Second, it is an expensive procedure. The third is the presence of redundant information, which further results in the consumption of image processing time, and still, the chances of segmentation errors cannot be ruled out. Finally, the multispectral MRI scans suffer from misalignment and inconsistency, which needs bias correction and image registration in advance before being used in segmentation algorithms [9]. Considering these limitations into consideration, in this research article, we propose a scheme for the classification and segmentation of tumors on two-dimensional single spectral MR sequences. The mainstream contributions of this article are:
-
This is the first article performing classification on the BraTS dataset for the brain tumor as per the best of our knowledge, and we also consider some data from The Cancer Imaging Archive (TCIA).
-
We consider as many as five machine learning algorithms along with multiple hyperparameter conditions.
-
We consider as many as seven texture feature extraction methodologies to generate texture features.
-
Along with tumor classification, we also perform the tumor segmentation method to detect the tumor.
-
Finally, we use a hybrid segmentation approach (hybrid of K-means and fuzzy C-means) rather than any individual approach.
The remaining parts of the article have been presented as follows: Section 2 discusses some noticeable related works in this domain of brain tumor classification and segmentation. Section 3 presents a system overview of the current work. A detail about the dataset and preprocessing tasks is given in sect. 4. Sections 5 and 6 discuss the tumor classification and segmentation process, respectively, with the experimental outcome and discussion. Section 7 is devoted to the conclusion, followed by some selected references.
1 Related works
Many approaches have been recommended for tumor classification and detection on MRI scans. Some of the related works are referred here to further improve our work in that direction.
In [14], Mishra et al. (2020) present brain tumor MRI classification using a support vector machine (SVM) by considering wavelet feature extraction such as DWT, SWT, and DMWT. In [15], Gumei et al. (2019) proposed a regularized extreme learning machine (RELM) brain tumor classification method based on a machine learning approach. They considered hybrid feature extraction on 3064 brain MRI images with max–min preprocessing steps. Basically, GIST and normalized GIST feature descriptors are used in their work.
In [16], Mishra et al. (2019) discussed the classification of the microscopic image to classify normal white cells from infected cells of leukemia. They have considered discrete orthonormal S-transform for feature extraction with linear discriminant analysis for feature reduction and finally Adaboost-based random forest classifier for classification purposes. In [17], Amin et al. (2017) proposed a distinctive approach to detect and classify the brain tumor from an MRI scan. Basically, they go for an SVM classifier with cross-validation after getting the feature by shape, texture, and intensity feature selection procedure to finally classify cancerous MRI from non-cancerous MRI.
In [18], Bahadure et al. (2017), support vector machine (SVM) and Berkeley wavelet transformation (BWT) approaches were investigated for image analysis on MRI. The work on [19] by Joseph et al. (2014) proposed K-means clustering algorithms for segmentation of tumor regions, and in [20], by Alfonse et al. (2016) used SVM for automatic brain tumor classification with accuracy further improved by Fourier transform for feature extraction.
In [21] Ahmadvand et al. (2016), a feature vector of MRI was extracted based on the wavelet transform described as modality fusion vector (MFV). Then for the segmentation of tumorous images, Markov random field model was used. In [22], Atiq Islam et al. (2013), a new texture feature extraction method, MultiFD with Ada Boost classification technique, is used to detect and segment out a brain tumor. In [23], Abbasi et al. (2017), automatic detection of the tumor was performed on 3D images. Histogram-oriented gradients (HOGs) and local binary pattern (LBP) in three orthogonal planes of MRI were used for the random forest classifier for segmenting out the region of interest. The work in [24] by Işın, Ali et al. (2016), emphasizes modern deep learning architectures, mostly on convolutional neural networks (CNNs) for brain tumor classification. CNN takes spatial information within input pixels using convolution and pooling processes. The convolution process extracts features, and round-up operations result in successful classification. Still, it takes a considerably long training period and showcases issues with being able to adhere to a particular solution at the time of the training process [25].
After going through some relevant literature, we have studied that most of the literature is based on either one or two classification algorithms. Similarly, for feature extraction, some literature uses wavelet features, while others use texture features. Furthermore, each of the classification algorithms and feature extraction techniques has its own pros and cons. So, the motivation behind this work is to use multiple classification algorithms and texture features for a better conclusion of the classification of brain tumors. Eventually, by doing so, we can choose the best classification algorithm as per the need of the hour.
In this paper, we have considered texture features and seven techniques of texture feature generation. For classification, we choose SVM, KNN (with Euclidean distance, City-Block distance, and Minkowski distance), binary decision tree, random forest, ensemble method (with Adaboost, Gentleboost, Logitboost, LPboost, Robustboost, RUSboost, and Totalboost). So, as a total, we have considered five mainstream classification methods, even more hyperparameters for some classification algorithms.
2 System overview
Our whole system is partitioned into two parts, i.e., tumor classification and tumor segmentation. In tumor classification, an MR image has been considered as input and classified into whether the MR image is tumorous or not based on the trained classifiers. After that, the tumorous MR image is passed into the tumor segmentation process, where the tumorous region is segmented out to give an idea of the tumor present in the brain. The above-described system overview of the proposed approach is depicted in Fig. 1.
3 Dataset and preprocessing
3.1 Dataset
The datasets used in our system have been gathered from the NCI-MICCAI 2017 and the 2019 Challenge (BraTS-2017 and BraTS-2019) [26,27,28]. These benchmark datasets consist of fully anonymous images from institutions like Bern University, ETH Zurich, Utah University, and Debrecen University. These are the datasets of 3D images of brain tumor medical resonance imaging (MRI), specifically designed for brain tumor segmentation. These datasets consist of multimodal brain MRI scans along with manually annotated tumor regions corresponding to each scan in four different volumes, namely (a) native T1 (T1), (b) contrast-enhanced T1-weighted (T1ce), (c) T2-weighted, (d) fluid attenuated inversion recovery (Flair). Samples of all these modalities of the images are shown in Fig. 2. Also, we have considered data from MR sequences from The Cancer Imaging Archive (TCIA). Overall, the manually inspected central slices of 100 numbers of T1C-, T2- & FLAIR-weighted MR sequences, each of the brain images having high-grade glioma (HGG) along with low-grade glioma (LGG) and 100 number MRI scans of the normal brains are utilized in this study from both the datasets.
3.2 Preprocessing
The raw MRI scans usually contain various artifacts, noise, salts. It also includes uneven intensity distribution as the MRI has been collected from the different scanners and even under multiple situations and positions of the camera. This may reduce the performance of the system. Due to the above reasons, medical images and especially MRI images need mandatory preprocessing steps.
The data of BraTS 2017 and 2019 contain 155 slices for every modality (Flair, T1, T1ce, and T2) of each patient as it is a 3D dataset and is originally provided in .nii (NIfTI) format. Corresponding to each patient, every volume consists of 155 slices of 2D images, each having dimensions of 240 \(\times \) 240, representing the 3D architecture of the brain. We chose five slices out of 155 from each patient’s data. These five slices were chosen manually in a careful way to make sure that they were at least 3 or 4 slices away from each other, and the brain tumor images were present in them. This takes care that different parts of the brain are covered, and the probability of finding a tumor becomes high. Each of the chosen slices was converted to .png (portable network graphic) format as it eased the reading of images for the next preprocessing steps.
For the mandatory preprocessing step, we applied “intensity normalization” and “bias field correction”. The N4ITK method [29] is adopted for the bias field correction job. We have combined the histogram equalization (HE) technique with the fast gray-level grouping (FGLG) technique [30] for intensity normalization of the image. Initially, the histogram obtained from a low-contrast image is further segmented into two sub-histograms with reference to the position of the maximum amplitude of the constituents of the histogram. Contrast enhancement is achieved by leveling the left segments of the histogram constituents with the help of HE and then by the FGLG method to equalize the correct segment of the histogram components of all. Figure 3 shows the result of images from some arbitrary tumorous images after the preprocessing steps.
4 Tumor classification
Automatic brain tumor classification is very important in modern diagnosis science because it sets out prognosis and treatment finding for the patient. For the tumor classification, 100 numbers of MR tumorous images and 100 numbers of MR non-tumorous images are selected of different weights from the dataset. Then we have to consider the texture features into account and apply binary classification techniques along with tenfold cross-validation, as shown in Fig. 4. The various classification algorithms used for this purpose and their corresponding classification accuracy are tabulated in the result section of Table 1. The various texture feature generation techniques are also summarized briefly, as given below.
4.1 Texture features extraction
The texture is an essential characteristic used in identifying regions of interest in an image. The texture of an image region has been determined by how the gray levels are distributed over the pixels in the region. We have extracted texture features using different texture-based feature extraction approaches. Although there is no clear definition of “texture” in the literature, it often describes an image that looks fine or coarse, smooth or irregular, homogeneous or inhomogeneous, etc. Generally, texture denotes characteristics of the surface and appearance of an object based on the structure, size, density, arrangement, and proportion of its fundamental parts. The collections of such features through the process of texture analysis have been described as texture feature extraction. The following texture features have been accounted into the discussion in this study:
4.1.1 First-order statistical feature
The most widely used first-order statistical features that characterize texture for image classification are mean, median, skewness, kurtosis, energy, entropy, average contrast. So, these six features are considered as first-order statistical features used for the feature extraction process [31].
4.1.2 Gray-level co-occurrence matrix (GLCM) feature
The GLCM is one of the essential and traditional methods for texture feature extraction. It is a two-dimensional histogram where the occurrence of pairs of pixels which are parted by a particular distance (i.e., d = 1, 2) as well as an angle (0\(^\circ \), 45\(^\circ \), 90\(^\circ \), and 135\(^\circ \)) is described. The number of gray levels presented in an image influences the required size of the matrix. The statistical features that are extracted from this matrix are energy, entropy, inertia, homogeneity, correlation, absolute value, maximum probability, and inverse difference, i.e., eight features [32]. By taking distances and angles into consideration, finally, there are 64 consolidated features.
4.1.3 Gray-level run length matrix (GLRLM) feature
In GLRLM [33], gray-level run length is a texture primitive, which is regarded as the connected group of pixels of maximum co-linearity with exactly the same gray level. Further, gray-level runs are described based on the direction and length of the run for a specific gray value. For determining GLRLM, different lengths of gray-level runs must be found for certain. The GLR matrices are calculated for angles 0\(^\circ \), 45\(^\circ \), 90\(^\circ \), and 135\(^\circ \).
The extracted features are short-run emphasis (SRE), long-run emphasis (LRE), run percentage (RP), run length non-uniformity (RLN), gray-level non-uniformity (GLN), low gray-level run emphasis (LGRE), high gray-level run emphasis (HGRE), short-run high gray-Level emphasis (SRHGE), short-run low gray-level emphasis (SRLGE), long-run high gray-level emphasis (LRHGE), and long-run low gray-level emphasis (LRLGE), i.e., 11 features. By taking angles into account, there are 44 features.
4.1.4 Histogram-oriented gradient (HOG) feature
The reasoning behind the usage of histogram-oriented gradient features [34] is that it takes the appearance of local object and structure, which can be identified by edge directions and then generalizes. The method locally outlines the gradient orientation of an image. Hence, 80 histogram-oriented gradient features are derived.
4.1.5 Local binary patterns (LBP) feature
Local binary patterns [35] can be used to describe the shape and texture of an image. The image is partitioned into various small regions from which the extraction of features takes place. Binary patterns that describe the neighborhood of pixels in the divided small regions constitute the features. The features obtained from these small regions are sequenced into a single-feature histogram, which creates a portrayal of the image. The resulting histogram will be 256 dimensions. Finally, 256 LBP features are extracted.
4.1.6 Cross-diagonal texture matrix (CDTM) feature
CDTM [36] represents the spatial connection between a pixel and its neighboring pixel at a particular angle and distance. It also finds out texture information around the central pixel by using the eight neighboring pixels. So, a set of six features are extracted from the matrix, i.e., homogeneity, entropy, absolute value, contrast, energy, and inertia difference moment.
4.1.7 Simplified texture spectrum feature
It characterizes local texture information in four directions instead of eight directions, which is used in the original texture spectrum feature. One of the advantages of the texture spectrum approach in image processing is that instead of texture features, it characterizes the texture aspects of an image by the corresponding texture spectrum. The simplified texture spectrum groups the 81 features obtained from the texture spectrum into 15 features [37].
So a total of 6 first-order statistical features, 44 gray-level run length matrix features, 64 gray-level co-occurrence matrix features, 80 histogram-oriented gradient features, 256 local binary pattern features, 6 cross-diagonal texture matrix features, and 15 simplified texture spectrum features, forms a 471-dimensional texture feature vector.
4.2 Feature matrix
All the texture feature vectors obtained from 100 tumorous and 100 non-tumorous images are hence consolidated to form a feature vector matrix of size 200 x 471.
4.3 Feature classification
This section deals with the brain tumor classification from both the tumorous and non-tumorous images using features obtained. Five robust supervised binary classification techniques are applied, and a comparison of results is made. The techniques applied are SVM (linear) [38, 39], KNN (distance = Euclidean, City-Block and Minkowski & k = 1,3,5,7) [40], binary decision trees [41], random forest [42], and ensemble methods [43], i.e., Adaboost, Gentleboost, Logitboost, LPboost, Robustboost, Rusboost, and Totalboost, as shown in Fig. 5. Training–testing samples are chosen randomly. Tenfold cross-validation is applied to ensure the robustness and prevent over-fitting of our models.
The advantages of using the SVM model are more visible in high-dimensional spaces, i.e., more separation margin between classes gives better results and is more effective in the scenario where number samples are less as compared to the number of dimensions [38, 39]. The superiority of using KNN is that it has a straightforward implementation procedure, is robust to noisy data, is effective if the training data are large, and augmentation in training data [40]. The benefits of using a decision tree as a classifier are decision trees required less effort for data preparation during preprocessing compared to other algorithms and do not require normalized data as well as scaling of data [41]. The advantages of using a random forest classifier are handling unlabeled data, robustness to outliers and nonlinear data, quick prediction and training time, and deals with high dimensionality data [42]. The feature that ensemble learning follows is: it combines various machine learning models to improve the final model’s performance. We can call the ensemble technique a meta-algorithm. By following this approach, ensemble learning reduces bias and variance or improves predictions [43]. After the completion of training, the classifiers’ recognition rate on un-tested data is used to indicate the performance algorithm.
4.4 Performance evaluation and experimental setup
The following parameters are used as the performance evaluation metrics for the brain tumor classification system.
-
Accuracy: The texture features consist of many different capabilities for the precise classification of MRI lesions. So we computed its confusion matrix, i.e., a table that is often used to illustrate a trained model’s performance on a given test dataset where the true values are well known. The terms used in a confusion matrix are as follows:
-
True positives (TP): accurately classified +ve samples
-
True negatives (TN): accurately classified -ve samples
-
False negatives (FN): inaccurately classified +ve samples
-
False positives (FP): inaccurately classified -ve samples
We then quantify this capability using accuracy from the following expression:
$$\begin{aligned} Accuracy = (TP+TN)/(TP+TN+FN+FP)\nonumber \\ \end{aligned}$$(1) -
-
Sensitivity: It is the metric that evaluates a model’s ability to predict the true positives of each available category.
$$\begin{aligned} Sensitivity = TP/(TP+FN) \end{aligned}$$(2) -
Specificity: It is the metric that evaluates a model’s ability to predict true negatives of each available category.
$$\begin{aligned} Specificity = TN/(TN+FP) \end{aligned}$$(3) -
Precision: It is the ratio of correctly predicted positive observations to the total predicted positive observations.
$$\begin{aligned} Precision = TP/(TP+FP) \end{aligned}$$(4) -
F1-Score: It is the weighted average of precision and recall (sensitivity).
$$\begin{aligned}&F1-Score =\nonumber \\&2*(Precision*Recall)/(Precision+Recall) \end{aligned}$$(5) -
Experimental Setup: All the source code implementation for preprocessing steps, classification, and segmentation are performed on python 3.6 with the help of standard machine learning libraries. The software systems are compiled on the hardware resource of Windows 10 operating system (64-bit), 8GB memory, Intel(R) Core(TM) i5-10300H CPU @ 2.50GHz with NVIDIA GeForce GTX 1650 Ti graphics.
4.5 Experimental results
We have trained our classifiers for 200 TIC-, T2-, and FLAIR-weighted images of tumorous and non-tumorous MRI scans. The various classifiers used for these image classification tasks are support vector machine (SVM) with the linear model, K-nearest neighbors (KNNs) having k values 1,3,5 and 7 for Euclidean, City-Block and Minkowski distance, binary decision trees (BDTs), random forest (RF), and ensemble methods with Adaboost, Gentleboost, Logitboost, Lpboost, Robustboost, Rusboost and Totalboost as various types of classifiers. The accuracy of the individual classifier is mentioned in Table ztab1.
With reference to Table 1, when we compare among the classifiers for classification accuracy, ensemble algorithms outperform the rest. However, the accuracy of the random forest algorithm has also reached nearby the ensemble. The average accuracy of SVM is 93.33%, KNN is 84.56%, BDT is 89.16%, RF is 96.5%, and ensemble is 96.98% as depicted in Table 2 for BraTS-2017 dataset. We have also performed exploratory analysis using box-plot for T1C, T2, and FLAIR class of tumor to know the shape of the distribution, central value, and variability of classification accuracy, sensitivity, and specificity. Figures 6, 7, and 8 show the box-plot distribution of accuracy, sensitivity, and specificity, respectively, for various classes of tumor. Now, Fig. 9 shows the comparison graph for average classification accuracy, sensitivity, and specificity for various classifiers.
We have also calculated the sensitivity, specificity, precision, and F1-score of respective classifiers as these parameters also hold equal importance to measure the performance of classifiers. Table 3 summarizes the sensitivity of all classification models for all the weights of MRI scans, whereas Table 4 gives the average sensitivity of the classifiers. Similarly, Tables 5 and 6 illustrate the specificity and average specificity of the classifiers for all volumes of brain image, respectively. Now, Table 7 and 8 represents the average precision and F1-score of all the classifiers used in this study. Further, the classification mean CPU time in seconds also depicted in Table 9 shows the performance analysis of various algorithms.
Even if we go through various articles on brain tumor classification and detection, we closely follow some very recent works by Mishra et al. (2020) [14] and Abbasi et al. (2017) [23]. While most of the works follow certain types of methods for feature extraction, then a sole classifier for the classification task. By considering all the pros and cons, we go for seven methods for texture feature extraction, and then, we consider five classifiers (SVM, KNN, BDT, RF, and ensemble) for the classification task. The datasets used in this work are MICCAI’s 2017 and 2019 challenge datasets of 3D brain MRI scans (BraTS-2017 and BraTS-2019). The comparison of our work with some relevant recent works is tabulated in Table 10.
5 Tumor segmentation
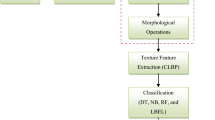
After a successful tumor classification, the region of interest(ROI) must be detected from the tumorous images as the subsequent important work. For the tumor segmentation process, we have taken 100 tumorous images and their corresponding ground-truth images. We have then preprocessed the tumorous images. Then, we have applied a hybrid of K-means and fuzzy C-means clustering techniques to segment tumorous regions followed by morphological operations. Ground-truth images are also preprocessed and applied with morphological operations. Finally, the segmented MR image and ground-truth image are compared by various performance metrics. The above-described image segmentation process is clearly depicted in Fig. 10.
5.1 Segmentation methods
Clustering is a powerful technique that has been reached in image segmentation. We have used clustering techniques for the process of image segmentation of brain tumors as clustering problems are mainly concerned with determining specific pixels in an image belonging together most appropriately. The two clustering techniques which we experimented are as follows:
5.1.1 K-means segmentation
This k-means clustering algorithm [44] is an unsupervised algorithm that helps segment the region of interest from the background of the image. The main principle of this algorithm is: it partitions the given dataset into k number of clusters based on the k-centroids. When we have unlabeled data, we go for this algorithm. Based on a certain similarity in the data present, we find the groups where the number of groups formed is represented by k. The initial requirement of the k-means algorithm is the number of clusters, i.e., k. Then randomly, the centers of k-clusters are selected. By taking various distance matrices available, the distance between each cluster center to each pixel is determined. Then, the pixels are grouped to a specific cluster that has the smallest distance among others, and then, the re-estimation of the centroid is calculated. Again the same approach for other pixels continues until the center converges. Finally, this algorithm aims at minimizing an objective function knows as the squared error function given by Eq. 6:
where \(\left\| x_{i}-v_{j} \right\| \) is the Euclidean distance between \(x_{i}\) and \(v_{j}\) .
k = number of clusters.
n= number of data points in a cluster.
5.1.2 Fuzzy C-means
The case of fuzzy c-means (FCM) [45] is another form of clustering technique, where a single piece of information may belong to more than one cluster. Hence, the pixels of a particular image data will be present in various outcome classes with different degrees of membership. Fuzzy behavior can be studied in fuzzy C-means when data are bound to each cluster employing an objective function. It occurs by the continuous increment of membership function along with cluster centers. The distance of the weighted summation of the data from each cluster center defines the object function. The belongingness of data to the cluster for every cluster is considered as a weight for that data. This membership function is responsible for the bounding of data to each cluster. The weighted mean of data is defined as the center of a specific cluster. The increment is continued until and unless the optimization between iterations exceeds a threshold. Finally, the objective of the fuzzy C-means algorithm is to minimize the objective function given by Eq. 7:
where \(\left\| x_{i}-v_{j} \right\| \) is the Euclidean distance between \(x_{i}\) and \(v_{j}\) .
k = number of clusters.
n= number of data points in a cluster.
\(\mu _{ij}\) = represents the membership of ith data to the jth cluster center.
m = fuzziness index.
5.1.3 Hybrid segmentation
This is the segmentation method, which is the combination of K-means and fuzzy C-means [46]. The K-means clustering method is simple to run and very fast on large datasets, but it fails to determine the specific tumorous regions. On the other hand, fuzzy C-mean methods are mainly used because they help retain the more minute information of the original image for detecting tumor cells precisely compared to the K-means. After experimenting with the algorithms mentioned above, we chose the hybrid segmentation algorithm based on the results. The hybrid segmentation algorithm first performs K-means followed by FCM on the given dataset using the results of K-means. The resulted cluster centers of the K-means algorithm are used as the cluster seeds in FCM algorithms until the termination condition arrives. But, to run the initial iteration of the FCM, the cluster centers and the membership matrix are calculated based on the results of K-means. The remaining iterations continue as in the FCM algorithm. The two output segmented images from the set of tumorous images after applying the hybrid segmentation techniques are shown in Fig. 11.
5.2 Morphological operations on the segmented image
Binary images can have many disfigurements. In specific, after segmentation, the binary regions induced are mainly distorted by texture and noise. Morphological image processing methods are applied for removing such imperfections by considering the structure and form of the image. Firstly, we have applied Erosion operation and then closing operation; after that, holes are filled, and an opening operation is finally applied. The order of operations is totally based on experimental results. The results from the morphological operation on some segmented images are shown in Fig. 12.
5.3 Image preprocessing of ground-truth images
Image preprocessing of ground-truth images is highly required as ground-truth images are in the grayscale format. We have converted them to binary format and then filled the holes to compare ground-truth images with the segmented images. Figure 13 shows the application of preprocessing on a segmented ground-truth image.
5.4 Comparison of segmented image and ground-truth image
For comparison, we have taken three parameters, namely Dice similarity coefficient, Jaccard similarity coefficient, and accuracy.
-
Dice similarity coefficient: The Dice similarity coefficient (DSC) is a performance metric to calculate the accuracy of automatic image segmentation results from MRI scans by comparing it with the ground-truth results. It is basically a statistical measurement metric, providing accuracy in a probabilistic manner. For two sets P and Q, it can be expressed as:
$$\begin{aligned} dice(P,Q)=2*|intersection(P,Q)| / (|P|+|Q|) \end{aligned}$$(8)Where, |P| and |Q| represents the cardinal of sets P and Q, respectively.
-
Jaccard similarity coefficient: The Jaccard similarity index (JSI) compares the members of two sets. The comparison result says that out of the two sets, which members of a set are shared and distinct. It generally measures the similarity of the two sets of data. The probabilistic value of JSI ranges from 0 to 1. The greater the JSI value, the better the similarity between the two sets. For two given sets P and Q, it can be expressed as:
$$\begin{aligned}&Jaccard(P,Q)\nonumber \\&\quad =|intersection(P,Q)| / |union(P,Q)| \end{aligned}$$(9)where |P| and |Q| represent the cardinal of set P and Q, respectively.
-
Accuracy: It is the extent of the result of a measurement or calculation that conforms to be correct. In easier terms, it means how much the resulting data are closer to the original data. Accuracy is determined, as mentioned in Eq. 1.
5.5 Experimental Results
The performance for the comparison between 100 tumorous segmented images and its corresponding ground-truth images is mentioned in Table 11. All these results are calculated for the complete tumor region class, which covers various tumor regions such as enhancing tumor, peritumoral edema, necrosis, and non-enhancing tumor. This indicates that our segmentation process belongs to the binary segmentation class. Hence, we can also say this as the tumor detection procedure. To validate the segmentation results of our proposed system, we compare our segmentation performance with other similar works, which is tabulated in Table 12
Now to have a fair point to discuss, we experimented by superimposing the segmented image on the tumorous MR image. It can be clearly noticed that the tumorous region is correctly superimposed on a tumorous image, and this indicates the correctness of our segmentation task, which is depicted in Fig. 14, for a randomly selected image.
6 Conclusion
This work is toward brain tumor analysis from MRI scans. We have classified T1C-, T2-, and FLAIR-weighted MR images into tumorous and non-tumorous, and further segmentation on the tumorous images. The noteworthy accuracy of the procedure for tumor classification and segmentation on the benchmark datasets signifies our method’s efficacy. The observations prove that the texture features are enough to differentiate between tumor tissues and non-tumorous tissues in the T1C-, T2-, and FLAIR-weighted scans. Moreover, clustering-based segmentation techniques are one of the best ways to segment tumorous regions. By keeping the positive outcomes of our proposed methods, we can claim it on other benchmark datasets of brain tumors and compare the results. Also, we could extend our observation other than texture features and strive to work on multifeatures and diverse datasets.
Availability of Data and Material
Yes.
References
Tandel, Gopal S., Balestrieri, Antonella., Jujaray, Tanay., Khanna, Narender N., Saba, Luca.,Suri, Jasjit S.: Multiclass magnetic resonance imaging brain tumor classification using artificial intelligence paradigm. Computers in Biology and Medicine, 122:103804, (2020)
Padma Nanthagopal, A., Sukanesh Rajamony, R.: Classification of benign and malignant brain tumor ct images using wavelet texture parameters and neural network classifier. J. Vis. 16(1), 19–28 (2013)
Muhammad, S., Salman, K., Khan, M., Wanqing, W., Amin, U., Sung Wook, B.: Multi-grade brain tumor classification using deep cnn with extensive data augmentation. Journal of computational science 30, 174–182 (2019)
Suneetha, B., JhansiRani, A.: A survey on image processing techniques for brain tumor detection using magnetic resonance imaging. In: 2017 International Conference on Innovations in Green Energy and Healthcare Technologies (IGEHT) , pp. 1–6. IEEE (2017)
Pei, L., Bakas, S., Vossough, A., Reza, S.M.S., Davatzikos, C., Iftekharuddin, K.M.: Longitudinal brain tumor segmentation prediction in mri using feature and label fusion. Biomedical Signal Processing and Control 55, 101648 (2020)
Scarapicchia, V., Brown, C., Mayo, C., Gawryluk, J.R.: Functional magnetic resonance imaging and functional near-infrared spectroscopy: insights from combined recording studies. Frontiers in human neuroscience 11, 419 (2017)
Nikolaos P Asimakis, Irene S Karanasiou, PK Gkonis, and Nikolaos K Uzunoglu. Theoretical analysis of a passive acoustic brain monitoring system. Progress in Electromagnetics Research, 23:165–180, 2010
Chaturvedi, C.M., Singh, V.P., Singh, P., Basu, P., Singaravel, M., Shukla, R.K., Dhawan, A., Pati, A.K., Gangwar, R.K., Singh, S.: 2.45 ghz (cw) microwave irradiation alters circadian organization, spatial memory, dna structure in the brain cells and blood cell counts of male mice, mus musculus. Progr. Electromagn. Res. B 29, 23–42 (2011)
Lemieux, L., Hagemann, G., Krakow, K., Woermann, F.G.: Fast, accurate, and reproducible automatic segmentation of the brain in t1-weighted volume mri data. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 42(1), 127–135 (1999)
Tang, H., Wu, E.X., Ma, Q.Y., Gallagher, D., Perera, G.M., Zhuang, T.: MRI brain image segmentation by multi-resolution edge detection and region selection. Comput. Med. Imaging Gr. 24(6), 349–357 (2000)
Chen, V., Ruan, S.: Graph cut based segmentation of brain tumor from mri images. International Journal on Sciences and Techniques of Automatic control & computer engineering 3(2), 1054–1063 (2009)
Jara, H., Sakai, O., Mankal, P., Irving, R.P., Norbash, A.M.: Multispectral quantitative magnetic resonance imaging of brain iron stores: a theoretical perspective. Topics in Magnetic Resonance Imaging 17(1), 19–30 (2006)
Kabir, Y., Dojat, M., Scherrer, B., Forbes, F., Garbay, C.: Multimodal MRI segmentation of ischemic stroke lesions. In: 2007 29th annual international conference of the IEEE engineering in medicine and biology society, pp. 1595–1598 (2007)
Mishra, S.K., Deepthi, VH.: Brain image classification by the combination of different wavelet transforms and support vector machine classification. J. Am. Intell. Human Comput. 12(6), 6741–6749 (2021)
Gumaei, A., Hassan, M.M., Rafiul Hassan, Md., Alelaiwi, A.: Ahybrid feature extractionmethod with regularized extreme learningmachine for brain tumor classification. IEEE Access 7, 36266–36273 (2019)
Mishra, Sonali, Majhi, Banshidhar, Sa, Pankaj Kumar: Texture feature based classification on microscopic blood smear for acute lymphoblastic leukemia detection. Biomed. Signal Process. Control 47, 303–311 (2019)
Amin, J., Sharif, M., Yasmin, M., Fernandes, S.L.: A distinctive approach in brain tumor detection and classification using mri. Pattern Recognition Letters 139, 118–127 (2020)
Bahadure, N.B., Ray, A.K., Thethi, H.P.: Image analysis for MRI based brain tumor detection and feature extraction using biologically inspired BWT and SVM. International journal of biomedical imaging, 2017 (2017)
Joseph, R.P., Singh, C.S., Manikandan, M.: Brain tumor mri image segmentation and detection in image processing. International Journal of Research in Engineering and Technology 3(1), 1–5 (2014)
Marco, A., Salem, A.B.M.: An automatic classification of brain tumors through MRI using support vector machine. Egy. Comp. Sci. J., 40(3), (2016)
Ahmadvand, A., Kabiri, P.: Multispectral MRI image segmentation using Markov random field model. Signal Image Video Process 10(2), 251–258 (2016)
Islam, A., Reza, S.M.S., Iftekharuddin, K.M.: Multifractal texture estimation for detection and segmentation of brain tumors. IEEE transactions on biomedical engineering 60(11), 3204–3215 (2013)
Abbasi, S., Tajeripour, F.: Detection of brain tumor in 3d mri images using local binary patterns and histogram orientation gradient. Neurocomputing 219, 526–535 (2017)
Işın, A., Direkoğlu, C., Şah, M.: Review of MRI-based brain tumor image segmentation using deep learning methods. Procedia Computer Science 102, 317–324 (2016)
Arı, B., Şengür, A., Arı, A.: Local receptive fields extreme learning machine for apricot leaf recognition. In: International Conference on Artificial Intelligence and Data Processing (IDAP16), pp. 17–18 (2016)
Menze, B.H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., Burren, Y., Porz, N., Slotboom, J., Wiest, et al, R.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2014)
Bakas, S., Akbari, H., Sotiras, A., Bilello, M., Rozycki, M., Kirby, J.S., Freymann, J.B., Farahani, K., Davatzikos, C.: Advancing the cancer genome atlas glioma mri collections with expert segmentation labels and radiomic features. Scientific data 4(1), 1–13 (2017)
Spyridon, B., Mauricio, R., Andras, J., Stefan, B., Markus, R., Alessandro, C., Russell, T.S., Christoph, B., Sung, M.H., Martin, R., et al.: Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. arXiv preprintarXiv: 1811.02629 (2018)
Tustison, N.J., Avants, B.B., Cook, P.A., Zheng, Y., Egan, A., Yushkevich, P.A., Gee, J.C.: N4itk: improved n3 bias correction. IEEE transactions on medical imaging 29(6), 1310–1320 (2010)
Humied, I.A., Abou-Chadi, F.E.Z., Rashad, M.Z.: A new combined technique for automatic contrast enhancement of digital images. Egypt. Inf. J. 13(1), 27–37 (2012)
Haralick, R.M., Shanmugam, K., Its Hak, D.I.N.S.T.E.I.N.: Textural features for image classification. IEEE Trans. Syst. Man Cybern. 6, 610–621 (1973)
Qurat-Ul-Ain, G.L., Kazmi, S.B., Jaffar, M.A., Mirza, A.M.: Classification and segmentation of brain tumor using texture analysis. In: Recent advances in artificial intelligence, knowledge engineering and data bases, pp. 147-155 (2010)
Chu, A., Sehgal, C.M., Greenleaf, J.F.: Use of gray value distribution of run lengths for texture analysis. Pattern Recognit. Lett. 11(6), 415–419 (1990)
Tian, S., Bhattacharya, U., Lu, S., Su, B., Wang, Q., Wei, X., Lu, Y., Tan, C.L.: Multilingual scene character recognition with co-occurrence of histogram of oriented gradients. Pattern Recognition 51, 125–134 (2016)
Prakasa, E.: Texture feature extraction by using local binary pattern. INKOM J. 9(2), 45–48 (2016)
Al-Janobi, A.: Performance evaluation of cross-diagonal texture matrix method of texture analysis. Pattern Recognit. 34(1), 171–180 (2001)
He, D.-C., Wang, L.: Simplified texture spectrum for texture analysis. Journal of Communication and Computer 7(8), 44–53 (2010)
Tandel, Gopal S., Biswas, Mainak, Kakde, Omprakash G., Tiwari, Ashish, Suri, Harman S., Turk, Monica, Laird, John R., Asare, Christopher K., Ankrah, Annabel A., Khanna, et al, N.N.: A review on a deep learning perspective in brain cancer classification. Cancers 11(1), 111 (201+9)
Braun, A.C., Weidner, U., Hinz, S.: Classification in high-dimensional feature spaces-assessment using svm, ivm and rvm with focus on simulated enmap data. IEEE J. Sel. Topi. Appl. Earth Obs. Remote Sens. 5(2), 436–443 (2012)
Tan, S.: An effective refinement strategy for KNN text classifier. Exp. Syst. Appl. 30(2), 290–298 (2006)
Tin Kam Ho: A data complexity analysis of comparative advantages of decision forest constructors. Pattern Anal. Appl. 5(2), 102–112 (2002)
Raf Guns and Ronald Rousseau. Recommending research collaborations using link prediction and random forest classifiers. Scientometrics, 101(2), 1461–1473, 2014
Oza, N.C., Tumer, K.: Classifier ensembles: Select real-world applications. Information fusion 9(1), 4–20 (2008)
Dhanalakshmi, P., Kanimozhi, T.: Automatic segmentation of brain tumor using k-means clustering and its area calculation. International Journal of advanced electrical and Electronics Engineering 2(2), 130–134 (2013)
Kalema, K.A., Bukenya, F., Rose, A.A.: A review and analysis of fuzzy-c means clustering techniques. Int J Sci Eng Res 5(11), 1072–7 (2014)
Raja, K.D.: Segmenting images using hybridization of k-means and fuzzy c-means algorithms. In: Introduction to data science and machine learning, IntechOpen (2019)
Sanjay, S., Suraj, S.: Brain tumor segmentation by texture feature extraction with the parallel implementation of fuzzy c-means using CUDA on GPU. In: 2018 5th international conference on Parallel, Distributed and Grid Computing (PDGC), pp. 580–585. IEEE (2018)
Alam, M. S., Rahman, M. M., Hossain, M. A., Islam, M. K., Ahmed, K. M., Ahmed, K. T., Singh BC, Sipon MM,: Automatic human brain tumor detection in MRI image using template-based k means and improved fuzzy c means clustering algorithm. Big Data Cognit Comput 3(2), 27 (2019)
Funding
This study is not funded by any organization or individual.
Author information
Authors and Affiliations
Contributions
First author is the main contributor and supervised by other authors.
Corresponding author
Ethics declarations
Conflicts of interest
All the authors declare that they have no conflict of interest.
Ethics Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to Participate
Yes.
Consent for Publication
Yes.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jena, B., Nayak, G.K. & Saxena, S. An empirical study of different machine learning techniques for brain tumor classification and subsequent segmentation using hybrid texture feature. Machine Vision and Applications 33, 6 (2022). https://doi.org/10.1007/s00138-021-01262-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00138-021-01262-x