Abstract
This paper proposes an automatic method for cell segmentation and classification of erythrocytes in thin blood smears with hemolytic anemia. First, to remove the background and noises in the blood images, the proposed method detects a series of changes on the edges and analyzes the edge changes by using the 8-connection chain codes technique to recognize isolated erythrocytes. For segmenting the overlapping erythrocytes, the 8-connection chain codes technique obtains the edge direction of the cells to effectively figure out the points of high concavity. Then, the adapted high concavity information is used to separate overlapping erythrocytes and to extract features from each segmented erythrocyte. After segmenting, all the erythrocytes can be treated equally and the differences between adjacent chain codes of each erythrocyte can be calculated. Furthermore, the proposed method extracts the variation of eight directions from each individual erythrocyte as their features for classifying into four main hemolytic anemia types. Finally, classification process identifies abnormal erythrocytes and the types of hemolytic anemia by using a trained bank of classifiers, utilizing the proposed method to calculate the quantity of erythrocytes and recognize the types of hemolytic anemia effectively.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Medical images have become major tools and techniques used for many clinical diagnosis and clinical trials [1, 2]. Digital image processing has been widely applied in the medical domain, but the majority of image processing techniques still requires human work. Feature analysis and automatic image segmentation systems allow doctors to diagnose diseases faster and more accurately, increasing efficiency of treatment by doctors. Especially for blood smears, the analysis of blood cells suffer from the drawback of time-consuming judgement and the inspection results vary depending on the clinician’s experience. Hence, researchers have been encouraged to develop a computerized medical decision support system focusing on automatic image segmentation and classification that can analyze different types of medical images and extract useful information for medical clinicians [3–5].
Blood cells form in bone marrow. A blood cell can be categorized into four types including platelets, erythrocytes, white blood cells (WBCs), and plasma [5]. Cells in blood smears may overlap each other, and there is great variation in shape, texture, color, size, and morphology of the nucleus and cytoplasm. Hemolytic anemia is the abnormal breakdown of erythrocytes either in the blood vessels or elsewhere in the body and the erythrocyte shape is an important criterion for the diagnosis. The cells are broken down at a faster rate than the bone marrow can produce new cells. Hemolytic anemia is classified as either inherited or acquired. Moreover, there are four subtypes of hemolytic anemia: hereditary elliptocytosis, sickle cell anemia, thalassemia and glucose-6-phosphate dehydrogenase [6]. Hence, this study focuses on an automatic method for segmentation and classification of hemolytic anemia in blood smears.
Image segmentation is the process to partitioning a digital image into multiple regions or objects to simplify further analysis and classification [7, 8]. Furthermore, the techniques of image segmentation can be categorized as supervised or unsupervised classification [5]. In general, to solve the image segmentation problem effectively, techniques need to combine domain knowledge [9]. For example, due to illumination inconsistencies or the image capture conditions of blood cells, this uncertainty makes blood cell image segmentation a challenging task [10–13]. Many segmentation methods for blood cells have been proposed and most of them are threshold-based, region-based, edge-based or clustering schemes [5]. Recent works in this area have mainly focused on segmenting WBCs, erythrocytes, and platelets. Based on this segmentation, some studies have performed blood cell counting and classified abnormalities in cells. Mohamed and Far [14] presented gray scale contrast enhancement and filtering techniques to segment the blood cell nuclei of WBCs. Furthermore, Hiremath et al. [15] proposed histogram equalization, thresholding and edge detection techniques to segment, identify and classify WBCs. In addition, Fatichah et al. [16] proposed that WBC classification can be achieved by optimizing the parameters of local fuzzy patterns based on the fuzzy contrast measure to extract the nucleus and cytoplasm region of the WBC texture. The optimal parameter values of the nucleus and cytoplasm region of a WBC image and the best accuracy rate of WBC classification can therefore be achieved. Sharif et al. [17] used the combination of \(\hbox {YC}_\mathrm{b}\hbox {C}_\mathrm{r}\) color conversion and a morphological operator as a mask to remove the WBC and carry out erythrocyte segmentation by passing erythrocytes through a marker-controlled watershed algorithm which handles overlapping cells. Khan et al. [18] proposed a statistical method for detection of mitotic cells in breast cancer histopathology image that models the pixel intensities in mitotic and non-mitotic regions by a Gamma–Gaussian mixture model and employs a context aware post-processing to reduce false positives. Hahn et al. [19] proposed an ellipse detection scheme using curve segments for classifying megakaryocytes. This major method uses the ellipse function to detect a curve and apply the curve information to segment the object. However, this process is quite complex and takes too much time to calculate in an ellipse equation. Jung et al. presented an unsupervised Bayesian classification scheme for separating overlapped nuclei [20]. The major method segments the overlapped nuclei by applying the prior knowledge about the regular shape of clumped nuclei. However, this process constructs a complex model for analyzing as to how many nuclei the overlapped ones should be separated from.
Moreover, the chain codes technique [21, 22] was proposed for representing directions of adjacent pixels as line drawings and planar curves. This technique can be used to observe the shape of objects. Shape representation is an important topic in many image processing applications, including computer vision, compression of cartography and medical image analysis. The chain codes technique moves along a digital curve or a sequence of border pixels based on 8-connection. The direction of each movement is encoded using a numbering scheme from 0 to 7 \(\{i \vert i = 0, 1, 2,\ldots 7\}\) in a counter-clockwise direction and the encoded symbol represents the angular direction of \(i \times 45^{\circ }\). These codes represent the direction of the neighboring pixels connected in a \(3\times 3\) window, as shown in Fig. 1a. In the 8-connected chain codes, we must move from one contour pixel to the next pixel and each code can be considered as the angular direction in multiples of \(45^{\circ }\). Figure 1b shows an example of chain codes using 8-connected path.
Hence, to improve upon the above issues, an effective method is proposed to apply in the segmentation and classification of hemolytic anemia in blood smears. The proposed technique recognizes normal and abnormal erythrocytes using directional information from chain codes. With the chain codes technique [21, 22] applied in the proposed method, using the edge directions of the object, we do not need to use mathematics computing to find the appropriate points of the curve to segment the object effectively. Furthermore, the scheme splits overlapping erythrocytes in blood smears using a procedure based on 8-connection chain codes. Finally, the classification of hemolytic anemia into its four subtypes is carried out by incorporating three novel features including the differential value of chain codes, irregularity of erythrocytes and variation of eight directions. The above three features are used to classify the hemolytic anemia in the Bayes classifier, logistic model trees, and rules classifier. According to these characteristics, hemolytic anemia can then be classified into four subtypes: hereditary elliptocytosis, sickle cell anemia, thalassemia and glucose-6-phosphate dehydrogenase.
The remainder of this paper is organized as follows. Section 2 describes the proposed method in detail. Section 3 presents the results and discussion. Finally, Sect. 4 concludes the paper.
2 The proposed method
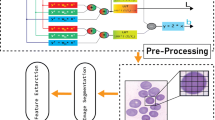
A general overview of the procedure for estimating erythrocytes in blood images is shown in Fig. 2. Image analysis and recognition include four main phases: (1) image preprocessing; (2) recognition of isolated erythrocytes; (3) segmentation of overlapping erythrocytes; (4) feature extraction. Briefly, the image preprocessing applies Otsu’s method [23, 24] and mathematical morphology [12, 13, 25, 26] to remove background images and noises automatically.
The edge changes of erythrocytes are analyzed using the 8-connection chain codes technique to distinguish isolated erythrocytes. The next step segments the overlapping erythrocytes, also using the chain code technique to detect high concavity in the edges of erythrocytes and separate each overlapping erythrocytes. First, the differences between each continuous chain code can be calculated. Second, extract features from the variation of individual erythrocyte of eight directions for classifying four different types of hemolytic anemia. The final step of the classification process identifies abnormal erythrocytes and types of hemolytic anemia by applying a trained bank of classifiers.
2.1 Image preprocessing
The preprocessing phase includes three steps for image analysis and segmentation. First, Otsu’s [23, 24] method is a high-speed and effective thresholding approach for image binarization. It is mainly exploited to discriminate the background and objects on a gray level histogram. However, in a bi-level image, the contour of each object is rubbed. The proposed technique adopts mathematical morphology [12, 13, 25, 26] to remove noises of small objects and smooth the edges of the object. First, in the background, both overlapping and isolated erythrocytes are segmented with an automatic threshold. In the different magnification ratios of thin blood smears, the automatic threshold can be used to recognize erythrocytes as either overlapping or isolated. We calculate mean and standard deviation of object areas, and then set the threshold as mean plus standard deviation in that the standard deviation expresses the differences in the number of degree from the object areas. Meanwhile, we determine if each isolated erythrocyte has an area less than the threshold, and the overlapping erythrocytes have an area greater than the threshold. After that, isolated erythrocytes and overlapping erythrocytes are extracted, respectively.
2.2 Recognition of isolated erythrocyte
First, each isolated erythrocyte is extracted with an automatic threshold from the background in the blood image. The shape of a healthy erythrocyte is a complete circle, and the chain code technique represents the shape of each isolated erythrocyte. Therefore, we analyze edge characteristics using the 8-connection chain codes technique to detect the pathological changes of cells in the blood image. We use the cycle of eight directions from the chain codes where the shape of normal erythrocytes is round. The proposed method defines four blocks for analyzing a circle; each block consists of a sequence of chain codes selected from the eight directions. The scanning model is counter-clockwise. If the erythrocyte is normal, the sequences of the four blocks will be Block1–2–3–4 as shown in Fig. 3. Take Fig. 1b as an example, the sequence of chain codes is 667700011223344455 and we can combine the duplicate codes. Hence, the result of the combined sequence of chain codes is 67012345. With the four blocks we can observe the variation of chain codes, and the sequences of the four blocks are round from Block 1 [6, 7, 0] to Block 4 [4, 5, 6]. Nevertheless, the abnormal erythrocyte does not follow this round of blocks; therefore, in this step, we can observe the variation of the contours of the isolated erythrocytes. Through the sequences of the four blocks, the abnormal erythrocyte can be identified.
2.3 Segmentation of overlapping erythrocytes
The overlapping erythrocytes are located based on the obtained erythrocyte area range. Meanwhile, the chain code technique is also applied to represent the shapes of objects. Since the high concavity of overlapping erythrocytes possesses a symmetric feature, after carrying out the process stated above, this section defines two types with the four blocks according to the directions in a circle. As shown in Fig. 4a, Type 1 contains two pairs: Block 2–Block 1 (denoted as No. 1), and Block 4–Block 3 (denoted as No. 2). On the other hand, Type 2 is composed of two pairs, including Block 1–Block 4 (denoted as No. 3), and Block 3–Block 4 (denoted as No. 4). Because of the coexistence of both Numbers 1 and 2, this is categorized as a Type 1 condition and then we segment Numbers 3 and 4. Similarly, Fig. 4b holds a Type 2 condition because both Numbers 3 and 4 exist. The detected high concavity information obtained by applying Type 1 and Type 2 can determine how many erythrocytes should be split in each object for segmenting each of the overlapped erythrocytes, as shown in Fig. 4c. Note that, in most cases of overlapped nuclei, the erythrocytes are dispersed overlapped and can be found with symmetrical concavity types as shown in Fig. 4. Even with three or more nuclei dispersed overlapping, the proposed chain codes technique can be applied to get the high concavities. For example, if there are three nuclei overlapped, we can find six concavities with three symmetrical concavity types which may or may not be the same type. However, if the nuclei are tightly overlapped, this kind of nuclei could not be provided as the medical judgment of hemolytic anemia. Hence, the proposed technique does not consider this type of overlapped nuclei.
2.4 Feature extraction
The feature extraction process identifies and extracts relevant information from the blood images allowing the filtering of unhealthy erythrocytes from the isolated erythrocytes and the overlapping erythrocytes. The criteria of classifying four main types of hemolytic anemia are based on the following three characteristics: differential value of chain codes, erythrocyte irregularity, and variation of eight directions. According to these characteristics, hemolytic anemia can be classified into four subtypes: hereditary elliptocytosis, sickle cell anemia, thalassemia and glucose-6-phosphate dehydrogenase, as shown in Fig. 5a–d. Afterwards, 14 features are extracted from hemolytic anemia images based on these three characteristics for classification.
First, in step 1: differential value of chain codes. Chain codes represent the shape of an erythrocyte, and therefore, this step calculates differences between the adjacent chain codes. We use an example to illustrate our feature extraction method. An individual erythrocyte using the 8-connection chain codes technique can be represented by the following series: \(A=\{6,6,6,7,7,7,5,5,5,4,4,3,3,0,0\}\), and pairwise subtracted sequence of chain codes can be shown as follows: series \(A'\!=\!\{0,0,-1,0,0,2,0,0,1,0,1,0,3,0,-6\}\). Afterwards, the series of \(A'\) can then generate the mean and standard deviation through calculation.
In step 2: erythrocyte irregularity. First is to use the differential value of radius to represent the degree of circularity of the erythrocyte. Euclidean distances are calculated based on the distance from the center pixel to each pixel of the boundaries of each individual erythrocyte. Thus, the maximum value of Euclidean distance is set as the maximum radius \(r_{i}^\mathrm{max}\) and the minimum value of Euclidean distance is set as the minimum radius \(r_{i}^\mathrm{min}\). The center pixel is found out by the crossing of x- and y-axis with most pixels. The maximum covering area is the circle with \(r_{i}^\mathrm{max}\), because normal erythrocyte should be similar to a circular shape. By subtracting minimum radius \(r_{i}^\mathrm{min}\) from the maximum radius \(r_{i}^\mathrm{max}\), we obtain the differential value. The smaller the difference is, the closer the erythrocyte is to being a circle. Hence, the ratio of radius \(S_{i}\) is shown in following Eq. (1):
Though the radius of the erythrocyte can represent the degree of circularity, this feature does not provide enough information to recognize the shape of the erythrocyte as an ellipse or circle. To better capture the shape information, we extract area irregularity based on region. The ratio of area irregularity \(A_{i}\) is shown in the following Eq. (2):
where \(\pi b_i^{r_i^{\mathrm{max}^{2}}}\) is the square of the maximum radius multiplying \(\pi \) in the object that can represent the area of largest possible circle, and \(b_i^\mathrm{org}\) is the original area of the object.
In step 3: variation of eight directions, the 8-connection chain code better describes the shape of the erythrocytes, and we extract the variation of eight directions from the individual erythrocyte as feature. We, hereby, provide an example to illustrate our feature extraction method. The individual erythrocyte using the 8-connection chain codes techniques results in the following series: \(A=\) {6, 6, 7, 7, 7, 0, 0, 0, 0, 1, 1, 1, 2, 2, 3, 3, 3, 4, 4, 4, 4, 5, 5, 6, 6}, and we construct an \(8\times 8\) matrix to record the variation of eight directions as shown in Fig. 6. The matrix of rows and columns is the direction of the chain code, which is used to record the direction from one pixel to another. In the sequence of series \(A\), the first code is 6 and the next is also 6, therefore the variation of eight directions is described as (6, 6) in the matrix. Similarly, the second and third code can be described as (6, 7). Eventually, all the variations of direction will be accumulated in the matrix. The results of calculation for series \(A\) are as shown in Fig. 6. Afterward, we transform the matrix from two to one dimensional so that matrix contains 64 vectors, and obtains the largest 5 numbers and the smallest 5 numbers from the matrix as features to observe a regular pattern of shape. Based on the approach described above, we can extract 14 features. An example from a glucose-6-phosphate dehydrogenase blood smear is shown in Table 1.
3 Results and discussion
The experiments in this study, including the segmentation algorithm and extraction features, are implemented with Matlab 9.0. The erythrocyte classification is processed using Weka [27]. We used 24 microscopic images of thin blood smears to test the performance of the proposed segmentation algorithm as shown in Fig. 7. Then, we input the abnormal erythrocytes from the segmentation results to conduct the classification of hemolytic anemia. The proposed segmentation method is compared with manual segmentation and the performance of our automatic hemolytic anemia classification system is evaluated by using three classifiers: the J48 tree classifier, Bayes classifier, and DTNB rule classifier. The proposed scheme can recognize the types of hemolytic anemia effectively.
3.1 Comparisons of the proposed method with manual recognition feature extraction
The proposed method uses 24 blood images to generate segmentation results. The number and size of erythrocytes are different in each image, yet our method can successfully segment each abnormal erythrocyte in the images. This section identifies three types of erythrocytes. The first type is isolated abnormal erythrocytes, the second type is isolated normal erythrocytes, and the third type is overlapping erythrocytes. The images with erythrocytes require manual annotations and numbering to serve as ground truth. After image segmentation, the erythrocytes are segmented and numbered to corresponding erythrocytes. Then, the classifiers use 14 extracted features of the erythrocytes to train or test the cell’s recognition rate.
The segmentation results are evaluated using the true positive (TP), true negative (TN), false positive (FP) and false negative (FN) metrics. The formulas of TP, TN, FP and FN metrics are defined in the following Eqs. (3)–(6):
where TP represents the overlapping erythrocytes and abnormal erythrocytes correctly identified in the image, TN describes the normal erythrocytes correctly identified as standard in the image, and FP describes the normal erythrocyte incorrectly identified as abnormal in the image, and FN describes the overlapping erythrocytes and abnormal erythrocytes in the image misidentified as standard.
Meanwhile, we assess the accuracy rates of identification results in the proposed scheme via statistical measures, including accuracy, sensitivity and specificity. Equations (7)–(9) are shown as follows:
where accuracy is the degree of closeness of measurements of a quantity to that quantity’s true value. Sensitivity measures the proportion of actual positives which is correctly identified and specificity measures the proportion of negatives which is correctly identified.
The result listed in Table 2 shows that the position of abnormal erythrocytes can be figured out completely; moreover, erythrocytes can be effectively separated from the overlapping regions. The proposed method can filter the abnormal erythrocytes better and more effectively, providing information with reduced FP rates for medical workers.
3.2 Analysis of the proposed features and results of various classifiers
In the classifier, we input 14 features into three classifiers: the Bayes classifier, logistic model trees and rules classifier, and observe the classification performance. The test mode uses tenfold cross-validation and the 87 test data as shown in Table 3. Moreover, the classification results are shown in Tables 3, 4, 5 and 6. True positive describes the type of hemolytic anemia correctly categorized. The FP describes the types of hemolytic anemia incorrectly categorized. As seen in Table 3, the different classifiers applied in the proposed method all achieve accurate and effective classification of the four types of hemolytic anemia. We observe that the results for the thalassemia have lower recognition rate than other types of hemolytic anemia can be correctly classified. Moreover, thalassemia is an inherited blood disorder characterized by less hemoglobin than normal. Hemoglobin is the substance in erythrocytes that allows them to carry oxygen. The low hemoglobin may make the image of erythrocytes appear pale in some cases. Based on collected erythrocytes images, it may result in shape deformation after further image processing and lower recognition rate than the other types. Furthermore, the FP rate is very low representing very small number of incorrectly classified instances of abnormal erythrocytes. After checked, those instances are those without prominent characteristics on 14 features for classification.
4 Conclusions
In this paper, we present an automatic method for segmentation and classification of abnormal erythrocytes in blood smears. The proposed techniques offer three major contributions for the proposed techniques: First, overlapping erythrocytes in blood smears are split by using a procedure based on 8-connection chain codes. Second, normal and abnormal erythrocytes are recognized using directional information from chain codes. Finally, hemolytic anemia is classified into its four subtypes by incorporating three novel types of features including differential value of chain codes, irregularity of erythrocytes and variation of eight directions. To sum up, we can recognize whether erythrocytes are abnormal or not, segment overlapping erythrocytes effectively and meanwhile classify the types of hemolytic anemia accurately using the aforementioned classifiers. In conclusion, we reduce the time required for doctors to make judgments and improve the efficiency of treatment.
References
Doi, K.: Computer-aided diagnosis in medical imaging: historical review, current status and future potential. Comput. Med. Imaging Gr. Off. J. Comput. Med. Imaging Soc. 31(4–5), 198 (2007)
Shiraishi, J., Li, Q., Appelbaum, D., Doi, K.: Computer-aided diagnosis and artificial intelligence in clinical imaging. Semin. Nucl. Med. 41(6), 449–462 (2011)
Sharma, N., Aggarwal, L.M.: Automated medical image segmentation techniques. J Med. Phys./Assoc. Med. Phys. India 35(1), 3 (2010)
Clas, P., Groeschel, S., Wilke, M.: A semi-automatic algorithm for determining the demyelination load in metachromatic leukodystrophy. Acad. Radiol. 19(1), 26–34 (2012)
Adollah, R., Mashor, M.Y., Mohd Nasir, N.F., Rosline, H., Mahsin, H., Adilah, H.: Blood cell image segmentation: a review. In: 4th Kuala Lumpur International Conference on Biomedical Engineering (2008)
Ballarò, B., Florena, A.M., Franco, V., Tegolo, D., Tripodo, C., Valenti, C.: An automated image analysis methodology for classifying megakaryocytes in chronic myeloproliferative disorders. Med. Image Anal. 12(6), 703–712 (2008)
Shapiro, Linda G., Stockman, George C.: Computer Vision. Prentice-Hall, New Jersey (2001)
Pham, D.L., Xu, C., Prince, J.L.: Current methods in medical image segmentation. Annu. Rev. Biomed. Eng. 2(1), 315–337 (2000)
Sapna Varshney, S., Rajpal, N., Purwar, R.: Comparative study of image segmentation techniques and object matching using segmentation. In: Methods and Models in Computer Science. ICM2CS (2009)
Levin, A., Zomet, A., Peleg, S., Weiss, Y.: Seamless Image Stitching in the Gradient Domain. Computer Vision-ECCV (2004)
Dorini, L.B., Minetto, R., Leite, N.J.: White blood cell segmentation using morphological operators and scale-space analysis. In: Computer Graphics and Image Processing. SIBGRAPI (2007)
Vizireanu, D.N., Pirnog, C., Lãzãrescu, V., Vizireanu, A.: The skeleton structure—an improved compression algorithm with perfect reconstruction. J Digit. Imaging 14(2), 241–242 (2001)
Vizireanu, D.N., Halunga, S., Fratu, O.: A grayscale image interpolation method using new morphological skeleton. In: Telecommunications in Modern Satellite, Cable and Broadcasting Service. TELSIKS (2003)
Mohamed, M., Far, B.: An enhanced threshold based technique for white blood cells nuclei automatic segmentation. In: IEEE 14th International Conference on e-Health Networking, Applications and Services (Healthcom) (2012)
Hiremath, P.S., Bannigidad, P., Geeta, S.: Automated identification and classification of white blood cells (leukocytes) in digital microscopic images. IJCA Special Issue on “Recent Trends in Image Processing and Pattern Recognition” RTIPPR (2010)
Fatichah, C., Tangel, M.L., Widyanto, M.R., Dong, F., Hirota, K.: Parameter optimization of local fuzzy patterns based on fuzzy contrast measure for white blood cell texture feature extraction. J. Ref. J. Adv. Comput. Intell. Intell. Inform. 16(3), 412–419 (2012)
Sharif, J.M., Miswan, M.F., Ngadi, M.A., Salam, M.S.H., Mahadi bin Abdul Jamil, M.: Red blood cell segmentation using masking and watershed algorithm: a preliminary study. In: International Conference on Biomedical Engineering (ICoBE) (2012)
Khan, A.M., El-Daly, H., Rajpoot, N.M.: A Gamma–Gaussian mixture model for detection of mitotic cells in breast cancer histopathology images. In: 21st International Conference on Pattern Recognition (ICPR) (2012)
Hahn, K., Jung, S., Han, Y., Hahn, H.: A new algorithm for ellipse detection by curve segments. Pattern Recognit. Lett. 29(13), 1836–1841 (2008)
Jung, C., Kim, C., Chae, S.W., Oh, S.: Unsupervised segmentation of overlapped nuclei using Bayesian classification. IEEE Trans. Biomed. Eng 57(12), 2825–2832 (2010)
Freeman, H.: Computer processing of line-drawing images. ACM Comput. Surv. 6(1), 57–97 (1974)
Sánchez-Cruz, H., Bribiesca, E., Rodríguez-Dagnino, R.M.: Efficiency of chain codes to represent binary objects. Pattern Recognit. 40(6), 1660–1674 (2007)
Huang, D.Y., Wang, C.H.: Optimal multi-level thresholding using a two-stage Otsu optimization approach. Pattern Recognit. Lett. 30(3), 275–284 (2009)
Rodríguez, R.: A strategy for blood vessels segmentation based on the threshold which combines statistical and scale space filter: application to the study of angiogenesis. Comput. Methods Programs Biomed. 82(1), 1–9 (2006)
Yu, D., Pham, T.D., Zhou, X., Wong, S.T.: Recognition and analysis of cell nuclear phases for high-content screening based on morphological features. Pattern Recognit. 42(4), 498–508 (2009)
Theera-Umpon, N., Dhompongsa, S.: Morphological granulometric features of nucleus in automatic bone marrow white blood cell classification. IEEE Trans. Inf. Technol. Biomed. 11(3), 353–359 (2007)
Weka 3: Data Mining Software in Java. http://www.cs.waikato.ac.nz/ml/weka/ (2013)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, HM., Tsao, YT. & Tsai, SN. Automatic image segmentation and classification based on direction texton technique for hemolytic anemia in thin blood smears. Machine Vision and Applications 25, 501–510 (2014). https://doi.org/10.1007/s00138-013-0585-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00138-013-0585-y