Abstract
We present a study on segmentation of leaf images restricted to semi-controlled conditions, in which leaves are photographed against a solid light-colored background. Such images can be used in practice for plant species identification, by analyzing the distinctive shapes of the leaves. We restrict our attention to segmentation in this semi-controlled condition, providing us with a more well-defined problem, which at the same time presents several challenges. The most important of these are: the variety of leaf shapes, inevitable presence of shadows and specularities, and the time constraints required by interactive species identification applications. We evaluate several popular segmentation algorithms on this task. Different datasets of leaf images are used, with manually segmented images serving as ground truth for quantitative comparisons. We observe that many of the methods are not immediately applicable: they are either too slow or would require that important modifications be introduced. We thus present extensions to our previously published segmentation method, which are able to improve its performance. The previous approach was based on pixel clustering in color space. Our extensions introduce a graph cut step and the use of a training set of manual segmentations in order to adjust important parameters of the method. The new method is fast enough for an interactive application, while producing state-of-the-art results.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Agarwal et al. [1] and Belhumeur et al. [7, 26] proposed mobile systems for plant species identification via leaf recognition. These systems used the shape of a leaf to identify its tree species, and required that the leaves be photographed against a plain light-colored background. Large datasets of these leaf images were collected, giving us the opportunity to study a useful semi-controlled segmentation problem, which at the same time allows for objective assessments of different methods.
On the other hand, much research in image segmentation has been dedicated to the problem of general segmentation in uncontrolled settings. For example, Arbelaez et al. present a comparison of methods on images of several different types [4], which constitute the Berkeley Segmentation Dataset and Benchmark [3]. Similarly, Alpert et al. [2] collected a database with a variety of different image types. Thus, much previous work has been dedicated to designing general-purpose methods that have high agreement with users’ subjectively defined hand-drawn regions. The leaf segmentation problem we address here is different in that the environment is much more constrained. Furthermore, there is very little subjectivity involved in defining the ground truth for these images. Finally, we are also concerned in precisely locating segmentation boundaries. Precise boundary detection is important for plant identification since the leaf shape will be used as the main recognition cue. Again, this is in contrast to previous work, as precise boundary detection is usually not a prerequisite for general-purpose segmentation methods.
Though at first sight segmenting a leaf against a plain light-colored background seems easy due to the somewhat controlled conditions, in fact it poses significant challenges. The task requires that segmentations be produced in time that is suitable for an interactive application, and whose boundaries are faithful to the true leaf boundaries, sufficient enough to enable correct recognition.
Figure 1 presents examples of leaf images from the three datasets we experiment within this study, to illustrate the variety of leaves and acquisition conditions that must be dealt with. In Fig. 2, we demonstrate the difficulty of traditional methods in this problem. A series of undesirable results are shown, resulting from the GrabCut method (see Sect. 4.3) when applied in difficult scenarios. The GrabCut method was chosen here for illustration purposes since it is very well known and was one of the best performing on our datasets (see Sect. 5). Below we more generally identify a series of significant practical challenges, several of which were previously noted [26].
-
Speed is a major challenge for our application, since the segmentation method should work as part of an interactive system. Coupled with this, high-resolution images are required in order to capture fine-scale details, such as leaf serrations.
-
The datasets present a large variety of leaf shapes, due to the diversity among the different species. In particular, compound leaves are especially difficult to segment for traditional methods, due to their complex segmentation boundaries, some of which include several concavities and holes. Examples are presented in Fig. 2c, d.
-
Pine leaves were identified to present a special difficulty, since most of them occupy only a small fraction of the image. Many methods fail on these because they are biased towards producing larger segments. Figure 9 in Sect. 5 illustrates this situation well, by showing results of applying the GrabCut method to several photos of pines leaves.
-
The leaf images present natural variations in lighting. One of the most difficult problems on these datasets has been correctly segmenting out the shadows that the leaves cast on the background. Refer to Fig. 2a–c.
-
The color of a leaf can vary, producing difficulties as presented in Fig. 2h. In addition, some tree species have shiny leaves, occasionally producing specular highlights which can confuse segmentation methods, as in Fig. 2e, f.
-
The venation patterns on leaves can be very light-colored, presenting a strong contrast and creating a strong edge with the rest of the leaf, as in Fig. 2f, g.
Images illustrating the three datasets used in this study. Refer to Sect. 4.1 for descriptions of each dataset. For the Lab and Field datasets, leaf species were randomly selected to have their images shown. For the User dataset, images were randomly sampled without regard to species, since this dataset was not labeled
Some of the undesirable results obtained when applying GrabCut to a challenging set of images. See the text for a discussion, and Sect. 4.3 for a review of the GrabCut method and the setup used
In this work, we experiment with several available implementations of popular segmentation methods. In order to provide an objective evaluation, we manually segment images from two leaf datasets. We also present qualitative observations on these two datasets, as well as an additional third dataset, composed of images uploaded by users of the mobile leaf identification system. We observe that the segmentation methods do not immediately work well for leaf segmentation. First of all, several methods were not developed with speed as a requirement and, for reasonably sized images, cannot produce results in sufficient time. Other methods have their own specific inherent biases, and would require the introduction of important modifications in order to work well throughout the different leaf species. We thus present extensions to our previously proposed segmentation method, which was described in the work of Kumar et al. [26]. We add a graph cut step as well as learning of several parameters of the method, via a training a set of manually segmented images. The new approach is still fast, while producing improved segmentation results.
2 Related work
Several plant species identification approaches were presented for the ImageCLEF plant identification task [22]. The task’s datasets are composed of leaf images, along with their respective metadata. Some of the identification approaches proposed have a leaf segmentation step, while others do not. The scan and scan-like datasets are photographed against a uniform background and usually segmented using some variant of Otsu’s thresholding method [34]. For literature reviews of plant identification approaches using images of leaves, we refer the reader to [7, 22].
A more sophisticated leaf segmentation approach was presented by Cerutti et al. [13], in which polygonal shape models were used to constrain an active contour. Due to the variety of leaf shapes, modeling becomes a complex problem. The authors presented a model for leaves with multiple numbers of lobes, while a separate model was used for compound leaves. The approach seems to be especially beneficial when dealing with unconstrained environments (see below). Though it should work well on leaves with known shape, it could be prone to failure on leaves with shapes that were not modeled. This lead the authors to only consider Angiosperm species for segmentation in ImageCLEF [13]. A similar approach (though somewhat more simple) was presented by Manh et al. [29], in which a single shape model was used with the goal of segmenting leaves of a particular species of weed.
In the present work, we do not adopt any kind of shape prior, due to the difficulty of the modeling problem. Leaves can be simple, compound, or found grouped into clusters. More direct strategies such as that of Cerutti et al. [13], Nilsback and Zisserman [31] for flowers or by Kumar et al. for other object classes [25], do not appear to be sufficient for our problem. These kinds of shape priors involve a somewhat limited amount of flexibility, which makes it difficult to model the variety and complexity of leaf shapes.
Leaf segmentation is much more difficult on the photo ImageCLEF dataset, in which photographs are taken in unconstrained conditions. Some authors have attempted to use completely automatic approaches, but with limited success. Casanova et al. [12] experimented with k-means to cluster the image pixels into leaf and background. Yanikoglu et al. [39] assume the central region of the image contains the leaf, so that the largest cluster found from this central region (when performing histogram clustering) defines the leaf’s color. With this representative color in hand, an over-segmentation is found using watershed, whose segments are assigned to leaf or background according to the distance of their color to the reference leaf cluster color. Camargo Neto et al. [11] also approached the unconstrained problem, in the context of image analysis for weed control. Their approach begins by finding leaf pixel candidates based on a color index. The final segmentation is found by creating a fragmentation of the set of candidate leaf pixels, whose fragments are then merged in a procedure that favors the formation of convex shapes.
Most works, however, applied traditional interactive segmentation techniques in order to obtain reasonable results on ImageCLEF’s photo images. Casanova et al. [12] used mean shift [16] to produce an over-segmentation of the image. A user would then indicate some background and foreground segments, which were used in a merging phase as to label the remaining segments. Cerutti et al. [13] used an interactive version of their polygonal model method, in which a user initially marks a region inside the leaf, providing a color model for leaf pixels. Yanikoglu et al. [39] use the marker-based version of the watershed transform, while Arora et al. [5] adopted the interactive GrabCut system [35]. Though user interaction is interesting in certain situations, it becomes impractical for large datasets with thousands of images or more [22]. In this study, we focused only on fully automatic approaches.
A system capable of working with photos of leaves on uncontrolled backgrounds would be very appealing from the perspective of a user. However, it is not clear to what extent such a system should rely on image segmentation. As noted by Bakic et al. [6], shape boundary features of segmented leaves become unreliable in this scenario, leading several authors to work with interactive segmentation techniques or avoid the problem altogether [22]. It is important to note that dealing with uncontrolled conditions requires approaches that are different in nature to the ones we study here, which are more appropriate in our semi-controlled scenario.
We should note that for the task of plant identification, leaf shape is an extremely important cue [7]. In order to use the leaf’s shape for recognition, it is not sufficient to provide only a coarse description of where the leaf is, but it is required that segmentation boundaries faithfully represent the true leaf shape. Due to this challenge, the authors of [7] collected datasets in which leaves were photographed against a plain light-colored background. We approach the leaf segmentation problem under this semi-controlled condition. The current problem is thus substantially different from previous work on segmentation in uncontrolled environments [22, 32, 38], in which a precise segmentation boundary is not as crucial.
In our experiments, we use images taken in semi-controlled conditions, either in a laboratory setting, or taken with mobile devices against plain light-colored backgrounds (see Sect. 4.1). Our datasets were collected from a real-world functioning system for plant species identification via mobile devices [26]. Images from our Lab and Field datasets are used in practice as labeled training data within the system’s recognition engine, while the User images were taken by users of the mobile system, presenting us with a series of real challenges. We would like to point out that the leaf datasets from the ImageCLEF plant identification task [22] are also relevant for the segmentation task, but we did not experiment with them. However, the datasets we have used are comprehensive and allow us to arrive at relevant conclusions, since they include a wide variety of species and acquisition conditions, and were taken from a real-world functioning system. It is important to note that images from our Lab dataset, which contain pressed leaves taken under controlled conditions, are representative of those in ImageCLEF’s scan photos. At the same time, our Field and User images, which were taken with mobile devices with leaves placed against plain light-colored backgrounds, represent to a good extent those in the scan-like photos from ImageCLEF.
The segmentation method we present here extends upon our previous work [26], in which color space clustering was used to define foreground and background regions. Here, we introduce two major extensions. First, we add a graph cut [9] step, applied on the pixel probabilities provided by the clustering method. The graph cut technique is widely used and well established in image segmentation. Its attractiveness comes from its global optimization formulation along with the fast methods available to find its solution. The second extension we introduce consists of estimating model parameters via a learning phase, using manual segmentations. This removes the need for manual parameter tuning, though it requires the availability of a dataset of manually segmented images.
GrabCut was initially developed by Rother et al. for interactive image segmentation [35]. However, given an appropriate initialization, the method is shown to be very useful for image segmentation in general, side-stepping the need for user interaction. The GrabCut approach has some similarity to the approach we adopt here, in that we perform color space clustering, followed by a graph cut step. However, there are two important differences. First, GrabCut makes use of multiple iterations, which alternate between model estimation in pixel color space and graph cut in image space. Our approach is faster, defining the model in a single step, after which graph cut is applied. Second, GrabCut uses a Gaussian mixture to model each class, instead of single Gaussians as we do here, thus we require less computation. In Sect. 5, we present a comparative evaluation of GrabCut and our method, demonstrating these differences.
In this work, we learn certain graph cut parameters by selecting a range of parameter values and choosing the one that produces the lowest error on a training set of manually segmented images. This training strategy is able to work in a reasonable amount of time, since we end up only having two parameters to estimate. It has been noted—among others by Kumar and Hebert [27]—that training of random fields using pseudo-likelihood inference is known to produce unreliable parameter estimates. Using more precise inference techniques becomes very time-consuming, so that more recent work takes an alternative, two-phase approach, similar to what we adopt here. In this two-phase approach, first unary potential parameters are learned in isolation. Then, the parameters describing binary or higher order interactions are learned by cross-validation (or hold out validation). An illustrative example of this type of approach was presented by Kohli et al. [24].
A problem somewhat related to leaf segmentation is that of flower segmentation for species recognition. Nilsback and Zisserman [32] developed a flower recognition approach which relied on segmentation prior to feature extraction. The flower segmentation problem is particularly challenging because image backgrounds are uncontrolled, while at the same time the appearance of different classes of flowers varies widely. The problem was dealt with by learning foreground and background color distributions, and by defining a flexible shape prior to capture the structure of petals.
Chai et al. [14] later proposed a co-segmentation method for flower segmentation, which did not require manual segmentations or modeling of a shape prior, yet showed increased performance. In the current work, we make use of manually segmented leaf images, following a more traditional supervised line of work.
3 Proposed segmentation method
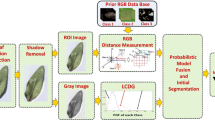
The segmentation method we propose here is an extension of our previous work [26]. Our method consists of the following steps, illustrated in Fig. 3 and described in more detail in the following sections. First, the image pixels are clustered into two groups in saturation-value color space, so that one group corresponds to leaf pixels, and the other, to background pixels. The clusters are found using the Expectation-Maximization (EM) algorithm to fit two normal distributions to the pixel data. This provides us with a probability indicating how likely each pixel is to belong to each cluster, according to the model from the normal distributions. The probabilities define the energies used in the graph cut step that follows. The graph cut formulation also allows us to incorporate edge information, encouraging the segmentation boundaries to follow strong image edges. Finally, false-positive regions are removed from the segmentation via a post-processing procedure.
Segmentation pipeline. The first step consists of EM clustering in saturation-value space. This provides us with probabilities that are used for defining energies in the graph cut formulation. After the graph cut step, a post-processing procedure is applied in order to eliminate false-positive regions
3.1 Expectation-Maximization in saturation-value space
The first step of our segmentation method uses the modified Expectation-Maximization (EM) clustering procedure in saturation-value color space that was described in our previous work [26]. For completeness, we summarize the approach here, then explain the improvement that we introduced for pixel weighting.
A mixture model composed of two normal distributions is fit to the pixel data in saturation-value space. If we denote a pixel’s saturation and value as \({\varvec{x}}\), the mixture model is
where \(p({\varvec{x}} | \varvec{\mu }_{f}, \varvec{\varSigma })\) and \(p({\varvec{x}} | \varvec{\mu }_b, \varvec{\varSigma })\) are normal distributions. \(\varvec{\mu }_{f}\) represents the mean of the foreground (leaf) distribution, while \(\varvec{\mu }_b\) is the mean of the background distribution. A common shared covariance matrix \(\varvec{\varSigma }\) is used. The set of model parameters is \(\varvec{\varTheta } = \{\varvec{\mu }_f, \varvec{\mu }_b, \varvec{\varSigma }\}\). Note that each normal distribution is assigned an equal weight of \(1/2\).
The model is fit using the EM algorithm. From an initial estimate of the model parameters \(\varvec{\varTheta }\), EM proceeds to find a local maximum of the data’s likelihood (see e.g., [8]). This can lead to undesirable solutions if the initial parameters are not set carefully. The means for the normal distributions are thus initialized near their expected values, so that they converge to the correct clusters when provided with pixel data from a new image. The covariance matrix is simply initialized as a multiple of the identity matrix.
In [26], a pixel weighting approach was used. Pixel weighting was introduced in order to correctly segment pine leaves in which only a small fraction of the total image pixels were leaf. A region in saturation-value space that was likely to contain leaf pixels was manually defined. Given a new leaf image, the pixels inside and outside the region would be weighted so that each set of pixels had equal total weight. These pixel weights were then used during the EM procedure. Here, we will adopt basically the same weighting procedure, except that we define the region that is likely to contain leaf pixels using a training set of manually labeled leaf pixels.
For the manually segmented images, we are provided a ground truth label for each pixel, which can either be leaf or background. For each of the leaf and background classes of pixels, this allows us to estimate a probability density function in saturation-value space, using kernel density estimation [8]. Given a new leaf image, we divide its pixels into two sets: pixels that are more likely to be leaf according to the kernel density estimate, and pixels that are more likely to be background. As in the previous approach, each set of pixels is then assigned a weight, so that each set has the same total weight during EM.
3.2 Graph cut
After running the EM procedure, we are able to compute an estimated probability that each pixel belongs to either leaf or background. We then include a graph cut step that uses these probabilities to determine the image segmentation, following the work of Boykov and Jolly [9]. For quickly finding the optimal graph cut, we employ the optimized algorithm by Boykov and Kolmogorov [10], whose implementation is provided by the authors.
Let \(\mathbf{y} = (y_1, y_2, \ldots , y_N)\) denote the binary segmentation we wish to solve for, with all \(y_i \in \{0, 1\}\), and \(N\) the total number of image pixels. The graph cut minimizes the energy function (or cost function)
\(R\) is the regional term defined by the unary potentials of the underlying Markov random field (MRF) formulation and \(B\) denotes the boundary term, which penalizes discontinuities in the cut, and corresponds to binary or higher-order potentials in the MRF formulation. First, for the regional term, we will use the posterior probabilities provided by the EM procedure. Let \(Pr(y_i = 0|{\varvec{x}}_i)\) and \(Pr(y_i = 1|{\varvec{x}}_i)\) denote, respectively, the probability that pixel \(i\) is background and leaf, given its features \({\varvec{x}}_i\). These probabilities are assigned according to the model estimated by EM. Then the regional term is written
Following [9], the boundary term is given by
and \(\mathcal N \) denotes the set of neighboring pairs of pixels in the image, which in our case we take to be the pairs of four-neighbors. From a grayscale version \(G\) of the image, we adopt the commonly used boundary coefficients
where \(G_i\) and \(G_j\) are, respectively, the gray values for pixels \(i\) and \(j\).
We work with a set of manually segmented training images, which allow us to estimate the graph cut parameters \(\lambda \) and \(\sigma \) (defined, respectively, in Eqs. 2 and 8). This is done by selecting a grid of \((\lambda , \sigma )\) pairs and choosing the one that produces the highest average accuracy when applied to the images in the training set.
3.3 Post-processing
After the graph cut step is applied to an image, we follow a straightforward post-processing procedure [26] to remove undesired false-positive regions. The procedure removes two type of false positives. The first type, which is very common to these images, consists of regions along the image boundaries which fall outside the light-colored sheet of paper or background used to create a contrast against the leaf. The second type consists of isolated regions present due to shadows, uneven backgrounds, or extraneous objects.
Post-processing consists of first performing a morphological dilation, then computing the connected components of the result. A connected component that has a large intersection with the image border relative to its area is then excluded as a false positive. Of the remaining connected components, the largest one is chosen to be the leaf.
4 Experimental evaluation
The datasets used in our experiments are presented in Sect. 4.1 below. In Sect. 4.2, we explain how methods that produce multiple segments (over-segmentations) are evaluated and compared to those that produce binary segmentations. All of the compared methods are listed along with their relevant settings in Sect. 4.3. The performance metrics used in the quantitative evaluation are discussed in Sect. 4.4, which are computed against ground-truth manually segmented images. To compute statistically significant differences between the performances of methods, we use the sign test, as described in Sect. 4.5. Later on, we present the experimental results in Sect. 5.
4.1 Datasets
We experiment on three datasets of leaf images, which are illustrated in Fig. 1.
-
1.
The first dataset, which we refer to as Lab, consists of leaves collected from trees in the Northeastern US by field botanists. These images have important particularities: their leaves were flattened by pressing prior to being photographed, and they were photographed under controlled lighting with a high-quality camera. The complete dataset has 4,221 images. The original images were manually cropped close to the leaves and then resized so that the size of their maximum dimension (width or height) would be 512 pixels. Of the 4,221 images, 30 were randomly selected to be manually segmented.
-
2.
The second dataset, which we call Field, consists of 1,042 images collected by researchers in the field using different mobile devices. These present varying acquisition poses, illumination conditions and amounts of blur, though an effort was made by the researchers for the images to be reasonably uniform. Of the 1,042 images, 786 present a size of 1,600 \(\times \) 1,200 pixels, while the remaining 256 have a size of 2,048 \(\times \) 1,536 pixels. 56 of the images were randomly selected to be manually segmented.
-
3.
Finally, the third dataset is from images uploaded by users of the mobile leaf identification system, which we will call User. 1,000 uploaded images were randomly selected in the period from July to October 2011, from users near New York City or Washington D.C. Only images that were appropriately taken according to instructions provided to users were accepted, leaving only 497 images. These images present more challenges than the ones from the Field dataset, since they contain an even greater variety of conditions. Users could choose to upload images in three different sizes: 640 \(\times \) 480 pixels (small), 960 \(\times \) 720 pixels (medium) and 1,024 \(\times \) 768 pixels (large). Of the 497 images analyzed, 317 were large, 147 were medium, and 33 were small. Since we did not manually segment these images, this dataset is only used for qualitative observations.
4.2 Evaluation of over-segmentations
Initially, we expected all methods we experimented with to be able to produce two segments, such that one would correspond to leaf and the other to background. In practice, however, we found that some methods, when set to produce just two segments, do not produce meaningful results on our images. Since this was so common, instead of abandoning these methods altogether, we resorted to experimenting with them by producing over-segmentations.
The over-segmentations are evaluated in two different manners. In the first evaluation, to obtain an upper bound on a method’s performance, we assign each segment to leaf or background as to yield the highest agreement with the ground truth as measured by pixel accuracy. This provides an optimistic evaluation, or upper-bound, on the method’s performance. Secondly, for a more realistic evaluation, we use a simple heuristic strategy to assign each of the segments to leaf or background, as follows. First we compute the median pixel saturation value of each segment. Then, we take the mid-point between the minimum and maximum of these median values. Segments that have median saturation that are larger than the mid-point are assigned leaf, while those with median saturation below the mid-point are assigned background. The heuristic provides a lower bound for a method’s performance, though in practice the heuristic works well when the number of segments is small, resulting in performance that is very similar to that obtained from the best possible assignment strategy.
For a fair overall comparison between methods, we would like to somehow assess how well they are solving the original leaf segmentation problem. The most relevant situation for us is thus when the number of segments produced by the over-segmentations is small. Conversely, when the number of segments produced is large, the segmentation method itself is not solving the original problem, but only part of it, so we do not find these results relevant for comparison purposes. As we will see in Sect. 5, when methods that produce over-segmentations are set to produce a small number of segments, the heuristic assignment strategy gives results that are very similar to the results from the best assignment. Thus, when performing comparisons to methods that produce binary segmentations, we will always use the results obtained from the heuristic strategy.
Though the upper bounds obtained by the best possible assignment strategy can be very high when the number of segments produced is large, for small and intermediate numbers of segments, they provide us with relevant information. First, when the number of segments is small, the best assignment strategy provides similar performance to the heuristic assignment strategy. This lets us know that the heuristic strategy is performing as well as possible. Second, if we believe that for some particular intermediate numbers of segments, it is feasible to design a perfect assignment strategy, then for that number of segments, the best assignment’s performance should be interpreted as realistic.
4.3 Methods and settings
This section reviews the methods that were compared on our datasets. Any relevant adjustments and settings of each particular method are also presented. The methods to be tested were chosen due to their popularity and availability of implementation. The review of each of the evaluated methods follows below.
Otsu
As a baseline method, we convert the images to gray scale and apply a threshold, resulting in an image segmentation. The threshold is found using the method of Otsu [34]. We use the implementation provided in Matlab’s Image Processing Toolbox.
In order to understand Otsu’s threshold selection method, let us consider that the pixels in the image form an empirical probability distribution. The distribution can be computed from the image’s graylevel histogram by simply dividing all of the histogram frequency counts by the total number of pixels in the image. Otsu’s method considers classical criteria used in discriminant analysis [20] on the distribution in order to define a good threshold. A threshold on the gray level divides the image pixels into two classes. Intuitively, the criteria favor maximizing the resulting between-class variance while at the same time minimizing the within-class variance.
Suppose we have fixed a threshold, as to divide the image pixels into two classes. Let us denote the total density of the pixels in each class by \(\omega _0\) and \(\omega _1\) (so that \(\omega _0 + \omega _1 = 1\)), their means by \(\mu _0\) and \(\mu _1\), and their variances by \(\sigma _0^2\) and \(\sigma _1^2\). Additionally, let us denote the mean and variance of the complete data distribution by \(\mu _T\) and \(\sigma _T^2\). Then we can define the between-class variance as \(\sigma _B^2 = \omega _0(\mu _0 - \mu _T)^2 + \omega _1(\mu _1 - \mu _T)^2\). The criterion function that Otsu’s method maximizes is \(\eta = \sigma _B^2 / \sigma _T^2\). In practice, the optimal threshold is found by simply computing the criterion function \(\eta \) for each possible gray level and choosing the one that maximizes it.
Graph-based image segmentation (GBIS)
Felzenszwalb and Huttenlocher proposed an efficient graph-based image segmentation method [19], and have provided their own implementation, which we experiment with here.
The method begins by defining a graph where each pixel is a vertex and edges connect nearby pixels together. Each edge is assigned a weight that indicates the dissimilarity between its pixels. The graph implicitly defines an initial segmentation, such that each image pixel forms its own segment. The segmentation algorithm proceeds to analyze the edges in order of increasing weight, so that the most similar pixel pairs are analyzed first. For each edge being analyzed, if its pixels currently belong to different segments, we consider whether or not we should merge these segments. Segments are merged whenever the edge weight measuring pixel dissimilarity is small relative to the minimum internal difference (defined below) between the segments. Intuitively, we merge segments when the pair of pixels analyzed does not indicate that there is an image edge between them, by presenting low dissimilarity relative to their minimum internal differences, which measure the natural within-segment variations.
Here, we review the criterion used by the method for deciding when to merge segments. Denote \(V\) the set of graph nodes, which is initially the set of all image pixels. Denote the set of \(m\) edges linking pixels together as \(E\), and the weight of an edge \(e \in E\) by \(w(e)\). The internal difference of a segment \(C \subseteq V\) is defined as the largest weight in the minimum spanning tree MST\((C,E)\) of the segment:
Given a pair of segments \((C_1, C_2)\) being considered, their minimum internal difference is defined as
where the threshold function \(\tau \) is defined as \(\tau (C) = k / |C|\), with \(|C|\) denoting the number of pixels in segment \(C\). The original internal difference Int\((C)\) can occasionally (by chance) be very small, especially when dealing with smaller segments. Without the \(\tau (C)\) term within MInt, this would impede certain small segments from ever merging. The \(\tau (C)\) term corrects for this effect.
The most relevant parameter of the method is \(k\) above which defines the threshold function \(\tau \), roughly controlling the minimum size of a segment. We experimented with the range \(k \in \) {1, 10, 100, 1,000, 4,000}.
In the implementation we use, there are edges between four-neighboring pixels and their dissimilarity is defined as the \(L_2\) (Euclidean) distance between their feature vectors. A feature vector for a pixel is defined as \((x, y, r, g, b)\), where \((x, y)\) is the location of the pixel in the image, and \((r, g, b)\) is the color of the pixel.
Throughout, we set the remaining relevant settings as follows. These settings have shown to work well on our different sets of leaf images. The GBIS method applies a Gaussian filter to smooth the image slightly, in order to compensate for digitization artifacts without significantly affecting the content. We set the standard deviation of the Gaussian used to smooth the image to \(\sigma = 0.5\) pixels. The final step of the method is to apply a post-processing, where very small segments are merged to their most similar neighboring segments. We have set the minimum size a segment is allowed to have (so that it is not merged to a neighbor) to \(0.5\,\%\) of the image size.
Mean shift Comaniciu and Meer proposed the use of mean shift [16] as a general clustering procedure, having applied it to image segmentation. We used the speed optimized implementation of mean shift provided in the EDISON system [15]. Clustering is performed jointly in spatial coordinates and in the L*u*v* color space.
For each pixel in an image, the mean shift procedure finds a local maximum of a density function defined from the image data. Each pixel is then associated with its respective maximum. The density function is defined as a kernel density estimate, computed from the image data, and thus requires a kernel bandwidth to be defined. When mean shift is used for image segmentation, two bandwidths are used: \(h_s\), which is defined in geometric image space, and \(h_r\), defined in the feature space, which in this case is the L*u*v* color space. The main advantage of the mean shift procedure is that the local maximum of the density function associated with each pixel can be found relatively quickly.
More specifically, a given image pixel \(\mathbf{x}\) can be described by its spatial coordinates \(\mathbf{x}^s\) and L*u*v* features \(\mathbf{x}^r\), such that \(\mathbf{x} = (\mathbf{x}^s, \mathbf{x}^r)\). An image with \(N\) pixels \(\mathbf{x}_1,\ldots , \mathbf{x}_N\) defines the density function of interest
where \(K\) is the kernel function. The kernel function we adopt is
where \(k\) is a function of a single variable called profile, which defines the shape of the kernel, and \(C\) is a normalizing constant.
Using large image space bandwidths, \(h_s\), is beneficial, resulting in more correct and complete segmentations. In practice, however, the use of a large spatial bandwidth is very time consuming, so that we have fixed this bandwidth at \(h_s = 30\) pixels. After having fixed this parameter, varying the feature space bandwidth \(h_r\) will determine the number of segments in the result. We have experimented with the range \(h_r \in \{5, 10, 20\}\).
Figure 4 presents mean shift execution times as a function of the bandwidth parameters \(h_s\) and \(h_r\). The times shown are averages taken over the manually labeled images from the Lab dataset, after having resized them so that their largest dimension was 700 pixels. See Sect. 5 for specifications of the machine used.
GrabCut
Rother et al. presented the GrabCut [35] system for the purpose of interactive image segmentation. An open source implementation of the method is available within the OpenCV library [33], which we use in this study.
Though our application is not interactive, we take advantage of GrabCut’s sophisticated segmentation algorithm. Their segmentation approach can be seen as an extension to the graph cut algorithm, which was reviewed in Sect. 3.2. The important difference is that GrabCut assumes that the color distributions of the foreground and background classes are unknown. The distributions are modeled in color space each by a Gaussian Mixture Model (GMM). The segmentation problem then consists of jointly finding the best set of pixel labels (which define the segmentation), along with the best set of GMM parameters, as to minimize a Gibbs energy function. The energy defined is analogous to the cost from Eqs. 2–8, with the main difference being the addition of the GMM terms.
More specifically, the energy to be minimized is
where \(U\) denotes the data term, and will be defined as the log-likelihood of the pixels’ GMM probabilities, and \(V\) denotes a smoothness term, based on neighboring pixels, as described further below. \(\mathbf{y} = (y_1, y_2, \ldots , y_N)\) denotes the binary segmentation, with all \(y_i \in \{0, 1\}\), and \(N\) the total number of image pixels. Similarly, \(\mathbf{z} = (z_1, z_2, \ldots , z_N)\) with each \(z_i\) containing the RGB values for pixel \(i\), and \(\mathbf{k} = (k_1, k_2, \ldots , k_N)\) with each \(k_i\) indicating the unique GMM component to which pixel \(i\) is assigned (GrabCut uses hard component assignments for speed purposes.) Finally, \(\varvec{\theta }\) contains the set of GMM parameters of both classes.
The smoothness term \(V(\mathbf{y}, \mathbf{z})\) is analogous to the boundary term in Eq. 8, but defined in color space:
Each of the terms above is described in turn. \(\gamma \) is a model parameter indicating the relative importance of the data and smoothness terms \(U\) and \(V\). \(\mathcal N \) denotes the set of neighboring pairs of pixels in the image, which is taken to be the pairs of four-neighbors. \(\delta \) is defined as in Eq. 7. \(z_i\) and \(z_j\) are, respectively, the RGB values for pixels \(i\) and \(j\). \(\beta \) is another method parameter, which is computed on a per-image basis from an estimate of the image noise. On the other hand, \(\gamma \) above assumes a fixed value, which must be set manually. We set \(\gamma = 2\) so that the method works well across the different varieties of leaf images.
In order to minimize the energy function, an iterative scheme was devised [35]. Each iteration begins by updating the GMM models according to the running set of labels, and then using the updated models to compute new class likelihoods for each pixel. The class likelihoods are then used to find a new set of labels via the graph cut method, and the whole process is restarted and iterated until convergence.
The method requires an initial estimate of the GMM parameters to be supplied. This estimate is computed in practice by setting an initial image map, which provides one of four possible labels for each pixel: “certainly foreground”, “probably foreground”, “certainly background”, and “probably background”. We carefully initialize the image map in the following manner: pixels on the image border are labeled as “certainly background”; pixels with high saturation and low value are labeled as “probably foreground”; and the remaining pixels are labeled as “probably background”.
Multiscale normalized cut (MSNcut)
The normalized cut (Ncut) criterion for image segmentation was proposed by Shi and Malik [37], along with a method to approximate its solution. The normalized cut approach treats the segmentation problem as a node partitioning problem, where image pixels are considered as nodes in a graph. Thus, the partition also defines a cut of the graph edges. The novelty of the normalized cut lies in allowing such a graph-based approach to not only consider the dissimilarity between different groups of pixels, but to also compensate for the within group similarities.
We experimented with the normalized cut implementation provided by its authors. The method seems to work better as the graph connection radius increases, which adds more edges to the graph being cut. However, increasing the connection radius is prohibitively slow, rendering this method impractical for our application. Thus, we did not include experiments with normalized cut, and instead worked with the multiscale version, described below.
Cour et al. [17] presented a multiscale algorithm to solve a constrained version of the normalized cut problem (MSNcut). Their method allowed the use of graphs with longer-range dependencies, in effect allowing for the use of larger images with improved results. The method decomposes the weight matrix, which represents all graph edge weights, as a sum over multiple scales. At smaller scales, more pixels are considered for forming edges, but the connection radius is limited, while at larger scales, pixels are sub-sampled more sparingly, allowing the connection radius to increase. The problem formulation considers the graphs at different scales simultaneously, with an added explicit constraint that the segmentation obtained be consistent across scales. The problem of finding the optimal partition is then approximated by a constrained optimization problem, whose solution can be found in time linear in the number of pixels.
The authors provide an implementation on their website, which we use here. The implementation allows graph edge weights to be defined by a combination of two terms: pixel color similarity, and an intervening contours [28] similarity. Using the intervening contour produced worse results on our images, so we only use pixel color similarities to define the edge weights.
Increasing the graph connection radii improved results and we have set them as large as possible, up to the memory limitation of our machine. As an example of the memory consumption of the algorithm, if we are working with an image of size 700 \(\times \) 525, we can choose to use four different scales to work with, as done in the original authors’ implementation. Let us denote the set of connection radii associated to each scale as \(R = \{r_1, r_2, r_3, r_4\}\), where \(r_i\) is measured in pixels. Under this setting, if we set \(R_1 = \{6, 9, 12, 18\}\), the method consumes a peak of 2.1 GB, whereas for a larger \(R_2 = \{10, 15, 20, 30\}\), the memory peak is of 4.9 GB. In practice, when dealing with images of this size, we use the set of radii \(R_2\). We varied the number \(k\) of segments output by the method within the range \(k \in \{2, 10, 20, 30, 40\}\).
Segmentation by weighted aggregation (SWA)
Sharon et al. [36] introduced segmentation by weighted aggregation (SWA). SWA is a multiscale graph-based approach based on forming aggregates, which are represented as nodes in graphs. Edges initially link neighboring pixels, with weights that represent pixel similarity. The overall objective of the method is to segment an image into multiple salient regions. A saliency criterion is defined, which measures the weight of edges that leave the group of graph nodes relative to the weight of edges within the group of nodes. This is in the same spirit of the normalized cut criterion (see Sect. 4.3), though the exact formulated criterion is different.
The main part of the method proceeds in a bottom-up fashion. It begins by assigning every pixel to one of a large group of small aggregates, each with a representative node. These assignments are done based on pixel affinities, which are stored in the graph edge weights. Aggregates are then recursively aggregated together, leading to a series of graphs, each at a different level of coarseness. The result is a pyramid of graphs, from which the segmentations will be later obtained. At any given level, aggregation is done by selecting a set of representative nodes, such that each of the non-representative nodes are strongly connected to at least one representative. The relationship between the aggregates at successive levels is stored in an interpolation matrix, computed from the edge weights. The main observation here is that the saliency computed at a given level can be approximately represented by the saliency at its corresponding successive coarser level. The aggregation process is such that at any given level of the pyramid, aggregates are allowed to have pixels that overlap.
After building the pyramid of aggregates, a segmentation can finally be obtained through a top-down procedure. First, from the pyramid, a set of the most salient aggregates is selected. Note that in principle the selected salient aggregates can be from different levels of the pyramid. Each aggregate in this set is repeatedly projected down onto finer levels via the interpolation matrices that were computed when building the pyramid as to compute weights that link the finer level aggregates with their higher level representatives. At the finest level, each image pixel will be assigned to the aggregate with which it has the highest linking weight.
Compared to other methods, an important advantage of this type of approach is that the larger aggregates allow for an appropriate extraction of texture features, which would not be possible at finer levels [21]. At the same time, descending to lower levels allows the fine details of the segments to be preserved. Also of importance is that the multiscale strategy allows the method to be linear time in the number of pixels.
The authors have provided an implementation of the method on their website. It has a series of adjustable parameters, though we have found reasonable results by using the default parameter settings. The algorithm is capable of producing a collection of segmentations, each with a different level of coarseness, corresponding to a different level of the pyramid. We denote the pyramid level here by \(c\), whose range starts at \(1\) (which produces the coarsest segmentation, usually containing only two segments). We experimented with \(c \in \{1, 2, 4, 6, 8\}\) and left the other parameters at their default values.
Global Pb with Oriented Watershed Transform and Ultrametric Contour Map (gPb+OWT+UCM)
Arbelaez et al. [4] presented an approach for segmentation with a series of steps. It starts out by computing a multiscale version of the probabilistic boundary detector due to Martin et al. [30]. From this initial boundary map, a global boundary map is computed using a spectral clustering formulation. This global boundary map is used to produce a super-segmentation of the image via the Oriented Watershed Transform. Finally, a hierarchical collection of segmentations, represented using the Ultrametric Contour Map, is computed from the Oriented Watershed Transform. We have experimented with the implementation provided by the authors of the method. Due to its memory requirements and our required image resolution, we were not able not perform comprehensive experiments with this method. See Sect. 5 for a discussion.
Expectation-Maximization (EM)
We experiment with a Matlab implementation of our previous leaf segmentation method as described by Kumar et al. [26]. We follow the segmentation steps as proposed in the paper, including the pixel weighting procedure during EM. For consistency between the methods, we do not include their stem removal step. It should be noted that we do not include the speed optimizations from [26], so that the times reported here are significantly larger.
Expectation-Maximization with trained pixel weighting (EM+TW)
This version of the method includes the trained pixel weighting used during EM, as opposed to using a hand-drawn delineation for the weighting scheme (see Sect. 3). Since this method requires a training set of manually segmented images, it is evaluated via twofold cross-validation.
Expectation-Maximization with trained pixel weighting and graph cut (EM+TW+GC)
The method we propose here, based on Expectation-Maximization, and followed by a graph cut step, is described in Sect. 3. As with the previous method, it is evaluated via twofold cross-validation.
4.4 Performance metrics
We evaluate the following performance measures for each of the different methods compared. For the quantitative analysis, we experimented on the images from the Lab and Field datasets which have manual segmentations. This allows us to compute measures of the following three important segmentation characteristics: how well the pixels from a segmentation agree with the ones from its respective manual segmentation; how well its boundary matches the boundary from the manual segmentation; and how similar the features computed from a given segmentation are to the ones computed from the manual segmentation. We will describe the metrics used to assess these characteristics further below. We also time the methods, since we are concerned with using them in an interactive application.
For quantifying the degree of agreement between pixels from a method’s segmentation and a manual segmentation, accuracy is a very intuitive measure. However, when only a fraction of the pixels belong to the leaf class, accuracy is very insensitive. We also measure pixel precision, recall and F-measure (F\(_{1}\) score) in this case, with the F-measure acting as a reasonable overall summary of performance [2].
The pixel agreement measures defined above are not sensitive to important image features that have only a small number of pixels, such as leaf serrations, or thin leaf tips. Thus, we also include measures of boundary agreement. Again, we use precision, recall and the F-measure, but computed over the boundary pixels. For deciding whether a point on the boundary is considered a true positive, false positive, true negative or false negative, we find a correspondence between the points of the boundaries produced by the method and the ground truth. The correspondence is done using the assignment procedure described by Martin et al. in the appendix of [30], whose code is provided along with the Berkeley Segmentation and Boundary Detection Benchmark and Dataset [3].
Finally, to give us an idea of the effect of the different methods on system performance, we use shape features computed from the segmentations. The shape features we use are the histograms of curvature over scale (HoCS) [26], with an implementation provided by the authors. After normalizing the histograms, we compute their similarity. Denote \(a = (a_1, \ldots , a_n)\) the features obtained from a given segmentation method and \(b = (b_1, \ldots , b_n)\) the corresponding features from the manual segmentation. The similarity is defined by their histogram intersection as \(s(a, b) = \sum _{i=1}^n \min (a_i, b_i)\).
4.5 Testing for statistically significant differences
We use hypothesis tests in order to compare a given pair of methods according to a performance metric. In general, a reasonable idea about which method performs better can be obtained by simply comparing the means or medians of the methods on a given dataset. However, especially on smaller datasets, there is some variance associated with these mean and median values. Hypothesis testing allows us to attach a confidence to a comparison, by computing the probability of the observed outcome under the (null) hypothesis that the two methods in fact have equivalent performance.
In our quantitative experiments, we adopt the sign test. The discussion and notation below follow Dixon and Mood [18]. Given a metric and a pair of methods to be compared, we treat the value of the metric obtained by each method when applied to leaf images as a random variable. It is difficult to make any strong assumptions about the probability distributions of our metrics, due to their complex nature. Thus, we resort to the sign test, which makes very few assumptions about its underlying distributions. A downside of the sign test is that it has reduced statistical power relative to others such as the paired \(t\) test (i.e., it is more conservative).
Suppose an observed leaf dataset has \(n\) images. For each image \(i \in \{1,\ldots , n\}\), we compute a pair of metrics produced by the two segmentation methods, which we denote \((x_i, y_i)\). The sign test takes into consideration only the signs of the differences \(x_i - y_i\). The main assumptions of the test are the following. First, it assumes that there is a fixed (and unknown) probability \(p = Pr(x_i > y_i)\), with \(0 < p < 1\). In other words, \(p\) is the probability that, for any pair of observations \((x_i, y_i)\), we will have \(x_i > y_i\). Second, it is assumed that the different observation pairs \((x_i, y_i), i = 1, \ldots , n\) are independent of each other. These weak assumptions contrast with, for example, those of the paired \(t\) test, which requires that the differences between paired observations be normally distributed.
Our null hypothesis is that \(p = 1/2\), which is equivalent to assuming that the median difference in the metrics resulting from the two methods is zero. Given the nature of our metrics, it is safe to assume that \(Pr(x_i = y_i) = 0\), so that \(Pr(x_i < y_i) = 1 - p\). Thus, if we denote by \(w\) the number of pairs for which \(x_i > y_i\), then under the null hypothesis \(w\) will follow a binomial distribution of probability \(1/2\).
All tests we perform are two-tailed, since we cannot make any prior assumptions about which of the two methods being compared is better. To perform the test, we count the number of pairs \(w\) for which \(x_i > y_i\) and \(n - w\) for which \(x_i < y_i\). Let \(r\) denote the smaller of the two counts, i.e., \(r \!=\! \min \{w, n \!-\! w\}\). Given an observed value of \(r\), the corresponding \(p\) value is \(Pr(R \le r)\), where \(R\) denotes a random variable from the same distribution that generated \(r\). Computation of the \(p\) value \(Pr(R \le r)\) is done by adding up the values of the binomial distribution that correspond to \(R \le r\), which will span both of its tails [18]. We set the significance value to the commonly adopted \(\alpha = 0.05\). That is, if we obtain a \(p\) value smaller than \(0.05\), we reject the null hypothesis and we call a given difference between methods significant.
In our quantitative experiments, we study two different leaf populations in turn: the first is that of images taken in laboratory settings (for which we experiment with the Lab dataset), and the second is that of images taken by researchers in the field (for which we use the Field dataset). These datasets are described in Sect. 4.1.
5 Results
We first present a comparison of execution times. All experiments were performed on a machine with 2 quad-core Intel Xeon CPUs, at 2.13 GHz clock speed and 4 MB cache. The machine had 12 GB RAM. All manually labeled images from the Lab dataset were used for measuring average times. Each image was resized so that its maximum dimension (either height or width) was set to a predetermined value, while preserving its aspect ratio. Figure 5a presents the average execution time per image as a function of image size. In order to better visualize the times for the faster methods, these are again plotted in greater detail in Fig. 5b. Observe in particular that, when the largest image dimension is set to 700 pixels, GrabCut takes around 7 s per image. Though GrabCut is among the fastest tested methods, this speed will not be satisfactory for many interactive applications. For mean shift, MSNcut, SWA, and gPB + OWT + UCM, the average computation times for images with their largest dimension set to 700 are above 50 s, which restricts their applicability to our problem.
It is important to point out some particularities of the previous experiment when the largest image dimension was set to 700 pixels. Note first that, at 700 pixels, the execution times are not available for gPB + OWT + UCM. This was due to the memory requirements of the method, which were not met by the machine. Note also that in general the time for MSNcut increases with image size. However, for the case when the largest image size is 700 pixels, due to our limited memory, we decreased the graph connection radii across scales. This had the side effect of not increasing the method’s execution times, though the results with smaller radii tend to be worse.
For the remaining experiments that follow, we resized all images so that their maximum dimension was 700 pixels. This resolution preserves most of the leaf image details, allowing us to capture thin stems and small-scale leaf serrations. We have excluded the gPB + OWT + UCM method from the remaining analysis, since it would require introducing some major modifications in order to run at the desired resolution without running out of memory.
As a principal metric to summarize performance, we measure the boundary agreement between the segmentation produced by a given method and the corresponding ground-truth manual segmentation. This agreement can be quantified by the F-measure, computed as described in Sect. 4.4. A more complete set of results is presented in the supplementary material, though usually all of the measures follow the same trends. The boundary agreement F-measure has the advantage of being sensitive to differences in the shapes of the segmentations, which will finally be used for leaf identification. A brief discussion on the merits of the different metrics was presented in Sect. 4.4.
Figures 6 and 7 show the distribution of F-measures on the Lab and Field datasets. These were computed on the 30 Lab images and 56 Field images for which manual segmentations were available. In Fig. 6, when a method’s name is marked with an asterisk, it indicates that the method was used to produce over-segmentations of the images, which were then evaluated according to the best possible assignment of the segments to leaf and background. The best possible assignment was determined using the ground truth manual segmentations, and provides an upper-bound on the method’s performance (see Sect. 4.2). On the other hand, the absence of an asterisk on a method that produces over-segmentations indicates that the segments were assigned according to the heuristic strategy described in Sect. 4.3. In order to obtain a fair comparison with methods that produce binary segmentations, in Fig. 7, methods that produce over-segmentations always had their segments assigned according to the heuristic strategy (denoted without an asterisk). All methods were run with and without the post-processing procedure described in Sect. 3.3. We report only the result that produced the best mean boundary F-measure: in the figures, when a method is marked with a superscript \(P\), it indicates post-processing improved the result and is therefore reported, whereas the absence of the \(P\) indicates post-processing did not improve the results, so that the result without post-processing is reported. This gives us a more meaningful comparison between methods. In any case, it is important to point out that the post-processing procedure can be very beneficial for certain methods, as shown later in Fig. 12.
Performance of methods which produce over-segmentations and require a parameter to be chosen. (For a summary of the results of all methods, including those without parameters to be chosen, refer to Fig. 7.) Boundary agreement F-measures are shown for images from the Lab and Field datasets. Higher values indicate better performance. There were a total of 30 Lab images and 56 Field images. Methods marked with an asterisk (whose over-segmentations were evaluated according to the best possible assignment of segments to leaf and background) have been shaded in gray. The boxes in the plots contain the second and third quartiles, while the vertical red line indicates the median value
Figure 6 shows only the methods which produce over-segmentations, having a parameter that is varied throughout a range. Table 1 shows the average number of segments produced by these methods for each of their parameter settings. In Fig. 6, note first that the methods marked with an asterisk improve with the number of segments that they produce, up to the point of achieving very high F-measures.Footnote 1 For very fine segmentations, though, the methods are not really solving the original leaf segmentation problem, but only part of it. Thus, we are not interested in comparing its results in this case to those of methods that directly produce binary segmentations. On the other hand, when the number of segments produced is small, note from Fig. 6 that the performance of the heuristic assignment is very close to the upper bound performance given by the best assignment. This lets us know that in this case the heuristic is working as well as possible. This is the most interesting case for us for comparison purposes: a fairer comparison to methods that produce binary segmentations is obtained when the over-segmentation methods are set to produce few segments as to try, as much as possible, to solve the original segmentation problem. Finally, note that, for the methods evaluated with heuristic assignments, the performance has a peak at a certain parameter setting, at which point using either a finer or coarser segmentation will result in a performance decrease.
In Fig. 7, all methods are present, with only the best performing parameter settings reported, chosen according to the highest mean boundary F-measure. In the figure, the methods that produce over-segmentations were evaluated using the heuristic assignment strategy, as to allow for a fair comparison. Figure 7 shows that EM + TW, and EM + TW + GC are consistently the highest scoring on both datasets. They are followed closely by EM, GrabCut, MSNcut and SWA. As was previously shown in Fig. 5, MSNcut and SWA are significantly slower than other methods, which puts them at a practical disadvantage. On the other hand, EM and EM + TW are the fastest of the best performing methods, followed closely by EM + TW + GC, then GrabCut. Otsu, GBIS, and mean shift in general produce worse results.
We performed several sign tests to compare different pairs of methods, as described in Sect. 4.4. The p values for the tests comparing boundary F-measures are shown in Tables 2 and 3, respectively, for the Lab and Field datasets. Again, here the methods that produce over-segmentations were evaluated using the heuristic assignment strategy. The performance differences follow the same trends on both datasets, but note that statistical significance appears much more frequently on the Field dataset. The corresponding tables for the other performance measures are presented in the supplementary material.
Figure 8 presents the results from Table 3 in the form of a graph. When there is a statistically significant difference between a pair of methods, as measured by boundary agreement F-measures on the Field dataset, there is an arrow going from the better method to the worst. The absence of an arrow indicates that the difference between methods was not significant. Methods are grouped together into the same node when they present the exact same set of differences between other methods and no difference amongst themselves. Note in Fig. 8 that SWA was better than many of the other methods. Upon further investigation, it was noted that, most of the time, SWA produces results that are only slightly superior. However, there are some few images for which SWA produces large mistakes that would interfere with recognition, whereas other methods do not. This is reflected in the lower average performance of SWA, which is shown in Fig. 7. The sign test ignores the severity of these mistakes, as it avoids making any assumptions about the distributions of the performance metrics.
Graph showing statistically significant differences between boundary agreement F-measures on the Field dataset. An arrow indicates that there was a statistically significant difference between a pair of methods. Methods are grouped together into the same node when they present the same set of differences between other methods and no difference amongst themselves
We would also like to point out the following regarding the hypothesis tests. On the Field dataset, among all four metrics (pixel accuracy, pixel F-measure, boundary F-measure, and HoCS feature similarity), at significance \(\alpha = 0.05\), there is a fairly consistent separation of the methods into two groups. The first group contains the better performing methods, and is composed of EM, EM + TW, EM + TW + GC, GrabCut, mean shift and SWA. Their performances are not consistently different amongst themselves and are fairly consistently better than the methods in the second group. The second group is composed of Otsu, GBIS, and MSNcut, which can also be seen grouped into the bottom node of Fig. 8.
Finally, we experimented on the larger datasets: all of the Lab images (excluding the ones that have manual segmentations, which were used for parameter adjustment); all of the Field images (again excluding those with manual segmentations); and all of the User images, whose segmentation parameters were set using images with manual segmentations from the Field dataset. Due to the large number of images in these datasets, it was only practical to experiment with the fastest methods, namely Otsu, GBIS, EM, EM + TW, EM + TW + GC, and GrabCut. The other methods showed themselves to be too slow for our application (see Fig. 5).
In order to make differences between methods evident on these large datasets, we adopt the following procedure. Given a pair of methods that we would like to compare, we order all the images in the dataset by how similar the segmentations produced by both methods are. For example, given methods \(A\) and \(B\), the image for which the segmentations produced by \(A\) and \(B\) are most dissimilar should appear first, while the image which produces the most similar segmentations should appear last. Here, we measure similarity between segmentations using their overlap ratio (see e.g., [23]), defined simply as the number of pixels in the intersection of the segmentations, divided by the number of pixels in their union.
The above procedure for comparing pairs of methods is motivated by the following observations. It would be prohibitively time-consuming to provide manual segmentations for very large sets of images, given that manually segmenting out the complex shapes of leaves is a labor-intensive process. At the same time, given a pair of methods, by viewing the images for which the resulting segmentations are most dissimilar, we are able to quickly understand some of their major differences. Examples of this behavior will be shown in the figures that follow. In particular, in the majority of cases, simply by looking at the most dissimilar results on a given set of images, it is easy to see which method is performing better.
A major mode of failure for all methods is small pine leaves, which only occupy a small fraction of the image. EM, EM + TW, and EM + TW + GC perform much better on this type of image due to the pixel weighting procedure, though there is still some room for improvement. Figure 9 compares EM + TW + GC\(^P\) with GrabCut\(^P\) on the complete Lab dataset, making evident the difficulty of traditional methods on small pine leaves. Next, we would like to note that on the Lab dataset, trained pixel weighting brings an important improvement over weighting using a manually delineated region in saturation-value space. Figure 10 illustrates this by comparing EM and EM + TW on the Lab images. We would also like to note the effect of adding a graph cut step to EM. Graph cut improves the results by requiring more compact segmentations and by being able to position the segmentation boundaries over image edges. This tends to fix errors such as those due to specularities, cast shadows, or leaves with uneven colors. Figure 11 illustrates this by showing a comparison between EM + TW and EM + TW + GC on the User dataset, where the difference between the two methods is more pronounced. Finally, another common issue is the presence of false positives in the outer regions of the images. These are caused either due to poor illumination or an unexpected absence of the light-colored background. The post-processing step we add to the various methods is able to fix this problem in most cases, as exemplified in Fig. 12.
Segmentation results on the Lab dataset illustrating the difficulty of traditional methods with pine leaves. From top to bottom: original image, result of GrabCut\(^P\) (GrabCut, plus post-processing), and result of EM + TW + GC\(^P\) (EM with trained pixel weighting and graph cut, followed by post-processing). The images are ordered by overlap ratio between the two segmentations, so that the left-most image has the most dissimilar segmentations, with the similarity increasing as we move right. In order to better illustrate the range of differences, every eighth image is shown. Most images have been cropped closely to the leaves after segmentation, for better visualization. The original image sizes range from about two to five times larger in each dimension
Segmentation results on the Lab dataset illustrating the effects of training for EM pixel weighting. From top to bottom original image, result of EM (with pixel weighting using a manually delineated region), and result of EM + TW (EM with trained pixel weighting). The images are ordered by overlap ratio between the two segmentations, so that the left-most image has the most dissimilar segmentations, with the similarity increasing as we move right. In order to better illustrate the range of differences, every eighth image is shown. The first image and the third to last image show zoomed-in details of the originals for better visualization
Segmentation results on the User dataset illustrating the effects of adding the graph cut step. From top to bottom original image, result of EM + TW, and result of EM + TW + GC. The images are ordered by the overlap ratio between the two segmentations, so that the left-most image has the most dissimilar segmentations, with the similarity increasing as we move right
Segmentation results on the User dataset illustrating the effects of post-processing. From top to bottom original image, result of EM, and result of EM\(^P\) (EM with post-processing). The images are ordered by the overlap ratio between the two segmentations, so that the left-most has the most dissimilar segmentations, with the similarity increasing as we move right. In order to better illustrate the range of differences, every eighth image is shown. Note the post-processing procedure was in error on the left-most image, though in all the other images it improved the segmentations. The post-processing error on the leftmost image occurred due to the following. In post-processing, we begin by dilating the segmentation, so that close by segments get merged together. This caused the leaf part of the segmentation to merge to the large segment that surrounds it. This merger was not expected by our algorithm and resulted in the error. The surrounding segment with which the leaf segment merged falls outside the sheet of paper in the original image, and ideally should be eliminated. However, in this case the complete merged segment was later judged to be lying along the image boundary, and was eliminated as a whole
After visually assessing all of the results, we have the following qualitative observations. As noted in Sect. 1, the following general difficulties were noted on these datasets: images with small pine leaves; complex compound leaves; uneven illuminations; cast shadows; specularities; natural variations in color; and venations. Overall, even for EM + TW + GC, which performs very well, there appears to be a good amount of room for improvement due to these difficulties. The User images proved to be much more challenging than the Field and Lab, due to the large variety of imaging conditions.
6 Conclusion
We have presented a study on efficient segmentation of leaves in semi-controlled conditions. The recent development of interactive applications for plant species identification based on computer vision created a situation in which this kind of segmentation method would be useful. The results showed that several general-purpose segmentation algorithms do not work satisfactorily on this problem when they are tested over large datasets containing a variety of species. Some of the methods experimented with are too slow, or have steep memory requirements. Other methods were able to work reasonably well, but would require important modifications in order to produce competitive results across the different leaf species.
In order to address the segmentation problem, we have extended a previous segmentation method, based on color space clustering [26]. First, we use the graph cut formulation on top of the EM clustering results to help overcome problems due to shadows and specularities. This allows us to incorporate image edges as an important cue. Second, we adjust the method’s parameters using training. By introducing training with manual segmentations, the method was able to work well on different leaf datasets with minimal manual parameter adjustment. In practice, the resulting method is fast and presents state-of-the-art results. In our quantitative experiments, it consistently showed to be among the top performing methods, while the qualitative results clearly showed the benefits of the newly proposed extensions.
Considering that segmentation will be applied within an interactive application, one line of improvement is to allow a user to guide the segmentation process, by interactively indicating leaf and background regions such as in the work of Boykov and Jolly [9]. Another line of improvement could consider more complex models, by adding pixel classes corresponding to cast shadows, specularities, or venations, and restrictions on the relative colors and positions between different classes, or on their shapes.
Notes
GBIS is an exception, since even at the smallest observation scale (\(k = 1\)), it still does not produce a fine enough segmentation.
References
Agarwal, G., Belhumeur, P., Feiner, S., Jacobs, D., Kress, W.J., Ramamoorthi, R., Bourg, N.A., Dixit, N., Ling, H., Mahajan, D., Russell, R., Shirdhonkar, S., Sunkavalli, K., White, S.: First steps toward an electronic field guide for plants. Taxon 55(3), 597–610 (2006)
Alpert, S., Galun, M., Basri, R., Brandt, A.: Image segmentation by probabilistic bottom-up aggregation and cue integration. In: Computer Vision and Pattern Recognition (CVPR), pp. 1–8 (2007)
Arbelaez, P., Fowlkes, C., Martin, D., Malik, J.: Berkeley segmentation and boundary detection benchmark and dataset (Accessed 2011). http://www.eecs.berkeley.edu/Research/Projects/CS/vision/grouping/resources.html
Arbelaez, P., Maire, M., Fowlkes, C., Malik, J.: Contour detection and hierarchical image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 33(5), 898–916 (2011)
Arora, A., Gupta, A., Bagma, N., Mishra, S., Bhattacharya, A.: A plant identification system using shape and morphological features on segmented leaflets. In: Working Notes of CLEF 2012 Conference (2012)
Bakic, V., Yahiaoui, I., Mouine, S., Litayem, S., Ouertani, W., Verroust-Blondet, A., Goëau, H., Joly, A.: Inria IMEDIA2’s participation at ImageCLEF 2012 plant identification task. In: Working Notes of CLEF 2012 Conference (2012)
Belhumeur, P., Chen, D., Feiner, S., Jacobs, D., Kress, W., Ling, H., Lopez, I., Ramamoorthi, R., Sheorey, S., White, S., Zhang, L.: Searching the world’s herbaria: A system for visual identification of plant species. In: European Conference on Computer Vision, pp. 116–129 (2008)
Bishop, C.M.: In: Pattern recognition and machine learning. Information Science and Statistics. Springer, New York (2006)
Boykov, Y., Jolly, M.P.: Interactive graph cuts for optimal boundary & region segmentation of objects in N-D images. In: International Conference on Computer Vision, vol. 1, pp. 105–112 (2001)
Boykov, Y., Kolmogorov, V.: An experimental comparison of min-cut/max-flow algorithms for energy minimization in vision. IEEE Trans. Pattern Anal. Mach. Intell. 26(9), 1124–1137 (2004)
Camargo Neto, J., Meyer, G.E., Jones, D.D.: Individual leaf extractions from young canopy images using Gustafson-Kessel clustering and a genetic algorithm. Comput. Electron. Agric. 51(1), 66–85 (2006)
Casanova, D., Florindo, J.B., Gonçalves, W.N., Bruno, O.M.: IFSC/USP at ImageCLEF 2012: Plant identification task. In: Working notes of CLEF 2012 Conference (2012)
Cerutti, G., Antoine, V., Tougne, L., Mille, J., Coquin, D., Vacavant, A.: ReVeS Participation—Tree species classification using random forests and botanical features. In: Working Notes of CLEF 2012 Conference (2012)
Chai, Y., Lempitsky, V., Zisserman, A.: BiCoS: A bi-level co-segmentation method for image classification. In: International Conference on Computer Vision (2011)
Christoudias, C., Georgescu, B., Meer, P.: Synergism in low-level vision. In: International Conference on Pattern Recognition, vol. 4, pp. 150–155 (2002)
Comaniciu, D., Meer, P.: Mean shift: a robust approach toward feature space analysis. IEEE Trans. Pattern Anal. Mach. Intell. 24(5), 603–619 (2002)
Cour, T., Benezit, F., Shi, J.: Spectral segmentation with multiscale graph decomposition. In: Computer Vision and Pattern Recognition, pp. 1124–1131 (2005)
Dixon, W.J., Mood, A.M.: The statistical sign test. J. Am. Stat. Assoc. 41(236), 557–566 (1946)
Felzenszwalb, P., Huttenlocher, D.: Efficient graph-based image segmentation. Int. J. Comput. Vis. 59, 167–181 (2004)
Fukunaga, K.: Introduction to Statistical Pattern Recognition, 2nd edn. Academic press, San Diego (1990)
Galun, M., Sharon, E., Basri, R., Brandt, A.: Texture segmentation by multiscale aggregation of filter responses and shape elements. In: Computer Vision and Pattern Recognition (CVPR), pp. 716–723 (2003)
Goëau, H., Bonnet, P., Joly, A., Yahiaoui, I., Barthélémy, D., Boujemaa, N., Molino, J.: The ImageCLEF 2012 plant identification task. In: Working Notes of CLEF 2012 Conference (2012)
Heckemann, R.A., Hajnal, J.V., Aljabar, P., Rueckert, D., Hammers, A.: Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. NeuroImage 33(1), 115–126 (2006)
Kohli, P., Ladick, L., Torr, P.H.S.: Robust higher order potentials for enforcing label consistency. Int. J. Comput. Vis. 82(3), 302–324 (2009)
Kumar, M., Torr, P., Zisserman, A.: Obj Cut. In: Computer Vision and Pattern Recognition, vol. 1, pp. 18–25 (2005)
Kumar, N., Belhumeur, P.N., Biswas, A., Jacobs, D.W., Kress, W.J., Lopez, I.C., Soares, J.V.B.: Leafsnap: a computer vision system for automatic plant species identification. In: European Conference on Computer Vision, pp. 502–516 (2012)
Kumar, S., Hebert, M.: Discriminative random fields. Int. J. Comput. Vis. 68(2), 179–201 (2006)
Leung, T., Malik, J.: Contour continuity in region based image segmentation. In: European Conference on Computer Vision, pp. 544–559. Springer, Berlin (1998)
Manh, A.G., Rabatel, G., Assemat, L., Aldon, M.J.: Weed leaf image segmentation by deformable templates. J. Agric. Eng. Res. 80(2), 139–146 (2001)
Martin, D., Fowlkes, C., Malik, J.: Learning to detect natural image boundaries using local brightness, color, and texture cues. IEEE Trans. Pattern Anal. Mach. Intell. 26(5), 530–549 (2004)
Nilsback, M.E., Zisserman, A.: Delving into the whorl of flower segmentation. In: BMVC (2007)
Nilsback, M.E., Zisserman, A.: Automated flower classification over a large number of classes. In: Indian Conference on Computer Vision, Graphics and Image Processing (2008)
Open source computer vision library (OpenCV). http://opencv.org/
Otsu, N.: A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9(1), 62–66 (1979)
Rother, C., Kolmogorov, V., Blake, A.: “GrabCut”: Interactive foreground extraction using iterated graph cuts. In: SIGGRAPH, pp. 309–314 (2004)
Sharon, E., Galun, M., Sharon, D., Basri, R., Brandt, A.: Hierarchy and adaptivity in segmenting visual scenes. Nature 442(7104), 719–846 (2006)
Shi, J., Malik, J.: Normalized cuts and image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 22(8), 888–905 (2000)
Welinder, P., Branson, S., Mita, T., Wah, C., Schroff, F., Belongie, S., Perona, P.: Caltech-UCSD birds 200. Tech. Rep. CNS-TR-2010-001, California Institute of Technology (2010)
Yanikoglu, B., Aptoula, E., Tirkaz, C.: Sabanci-Okan System at ImageClef 2012: Combining features and classifiers for plant identification. In: Working Notes of CLEF 2012 Conference (2012)
Acknowledgments
The authors would like to gratefully acknowledge Peter N. Belhumeur, Neeraj Kumar, and Arijit Biswas for helping organize the collections of images used in this work. W. John Kress, Ida C. Lopez, and collaborators at the Smithsonian Institution’s Department of Botany collected and curated the Lab and Field datasets. The authors are grateful to Aditya Malik for manually segmenting several of the leaf images and for helpful discussions. We would also like to acknowledge the authors of the several segmentation methods whose publically available implementations we have used. This work was supported by National Science Foundation grants #0968546, #0325867, and #1116631.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Soares, J.V.B., Jacobs, D.W. Efficient segmentation of leaves in semi-controlled conditions. Machine Vision and Applications 24, 1623–1643 (2013). https://doi.org/10.1007/s00138-013-0530-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00138-013-0530-0