Abstract
Maize (Zea mays L.) is a highly versatile crop with huge demand of nitrogen (N) for its growth and development. N is the most essential macronutrient for crop production. Despite being the highest abundant element in the atmosphere (~ 78%), it is scarcely available for plant growth. To fulfil the N demand, commercial agriculture is largely dependent on synthetic fertilizers. Excessive dependence on inorganic fertilizers has created extensive ecological as well as economic problems worldwide. Hence, for a sustainable solution to nitrogenous fertilizer use, development of biological nitrogen fixation (BNF) in cereals will be the best alternative. BNF is a well-known mechanism in legumes where diazotrophs convert atmospheric nitrogen (N≡N) to plant-available form, ammonium (NH4+). From many decades, researchers have dreamt to develop a similar symbiotic partnership as in legumes to the cereal crops. A large number of endophytic diazotrophs have been found associated with maize. Elucidation of the genetic and molecular aspects of their interaction will open up new avenues to introgress BNF in maize breeding. With the advanced understanding of N-fixation process, researchers are at a juncture of breeding and engineering this symbiotic relationships in cereals. Different breeding, genetic engineering, omics, gene editing, and synthetic biology approaches will be discussed in this review to make BNF a reality in cereals. It will help to provide a road map to develop/improve the BNF in maize to an advance step for the sustainable production system to achieve the food and nutritional security.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The world population is expected to grow to ~ 10 billion by 2050. Hence, to fulfil the food requirements agricultural production needs to be increased by 56% (Bloch et al. 2020). Cereals represent biggest share among crops in food basket. However, for sustainable agricultural production, the supply of sufficient nitrogen (N) is the biggest challenge to the cereal crops with 25 to 30% of nitrogen use efficiency (NUE). Worldwide, it has been estimated that plants require approximately 150 to 200 million tonnes of mineral N, out of which only about 100 million tonnes of N is produced through the industrial Haber–Bosch process (Unkovich et al. 2008). Industrial N production creates a number of environmental issues by emission of 3.6 kg CO2-eq. per kg of N manufactured (Fossum 2014). On the contrary biological nitrogen fixation (BNF) provides a win–win situation under changing climate scenario. In BNF, without any cost the atmospheric N is converted into reduced forms by a group of prokaryotes called diazotrophs, such as cyanobacteria, free-living soil bacteria, associative bacteria (Azospirillum) and symbiotic bacteria (Rhizobium and Bradyrhizobium) (Postgate 1982). It has been evaluated that BNF generates approximately 80–90% of N available to plants diversity (Rascio and Rocca 2013). Through BNF, a total of 139 to 175 million tonnes of N is added to the earth’s ecosystem and out of which 35–44 million tonnes N is contributed by symbiotic associations (Chafi and Bensoltane 2009; Bano and Iqbal 2016). Hence, it is one of the major contributors to the total biospheric N accounting for 30–50% of the overall N in crop fields (Ormeno-Orrillo et al. 2013). BNF is an integral process in leguminous crops, while cereal crops lack this ability. Considering the economic and environmental concerns related to N fertilizers, it will be a viable and sustainable approach to harness this mechanism in maize, which is produced in maximum quantity in the world, to boost its production sustainably.
Maize is the highest yielding cereal crop being cultivating in about 170 countries globally and covering ~ 194 million hectares area with 1147.6 million thousand tonnes production (FAOSTAT 2020). Maize is principally utilized for feed and industrial purposes, besides directly consumed as food or processed food in various parts of the world (Fig. 1). Its proportion of consumption varies considerably from country to country like in the USA 35.0% of corn is used for feed, 34.7% for ethanol production,13.5% for exports, 8% for dried distiller grains (DDGs), 1.5% for starch, 7.5% for human consumption and 1% for seed (NCGA 2020), whereas in India, ~ 60% of the total maize grain is consumed as feed, 14% for industrial purposes, 17% as food, 7% as processed foods, and 4% for other purposes including seed (Rakshit and Chikkappa 2018). Although maize has wider adaptability, several factors (biotic and abiotic) constrain its production and productivity. Among the abiotic stresses, low nutrient and water availability directly hamper the physiological processes of maize and affect yield in turn (Gonzalez-Dugo et al. 2010).

Source: OECD/FAO 2020)
Global consumption of maize (
N is the principal macronutrient playing a major role in plant physiology. Being the prime component of amino acids, chlorophyll, adenosine triphosphate (ATP) and nucleic acids, it is the most vital nutrient required in the largest quantity for maize production (Wang et al. 2017). For the production of one ton of crop biomass, almost 9–11 kg of N is required (Anuar et al. 1995). In maize, the increased application of N is directly proportional to biomass, grain yield, leaf number and shelling percentage (Nunes et al. 1996; Bakht et al. 2006). Its deficiency constrains proper maize development and growth. Although N is the most abundant element in the atmosphere (78.1%) and soil reserves (2–20 t ha−1) (Bockman et al.1990), but its availability for crops is limited as plants utilize it in reduced forms only, viz. nitrate (NO3−) or ammonium (NH4+) (Huang et al. 2000). The concentration of NO3− and NH4+ is very dynamic and largely depends on the soil edaphic environment.
Crop plants access N directly either from chemical fertilizers, manures or atmosphere through processes like BNF and lightning (Vance 2001). From 1960s, the application of synthetic nitrogenous fertilizers for crop production has increased nine times, and over the next 40 years, it is expected to increase further by 40 to 60% (Sutton et al. 2013). In 2017, for total agricultural production, approximately 109 million tonnes of the nitrogenous fertilizers were consumed worldwide (FAO 2017). It is projected that by 2050, there will be a need for additional 443 million tonnes of maize production that will require approximately 27–63 million tonnes of N (Alexandratos and Jelle 2012). Maize is a heavy feeder crop and highly responsive to N application; thus, the demand for nitrogenous fertilizers is also expected to increase many folds. Organic fertilizers are insufficient to meet the total N requirement. Hence, excessive use of chemical fertilizers costs us large sums of money in addition to human and environmental health (Glendining 2009). Globally, out of total N applied to crops through fertilizers, only 47% is converted into grains and the remaining is either turned in organic matter or lost through erosion, leaching, runoff and emitted in gaseous forms, viz. ammonia (NH3), nitrous oxide (N2O), dinitrogen (N2) or nitrogen dioxide(NO2) (Lassaletta et al. 2014). Hence, all these issues necessitate the search for a sustainable alternative approach to supply N for cereal crops production, including maize. In this review suitability of different breeding and biotechnological approaches to turn BNF a reality in maize with more N availability to increase yield and reduced environmental concerns have been discussed. It will give a direction for breeding-based new approaches to reduce the nitrogenous fertilizers dependency and promote the eco-friendly productive system for heavy N executive maize cropping system.
Need for biological nitrogen fixation in maize
For a sustainable solution to nitrogenous fertilizers use, BNF in cereals has been a hope since 1970s. Manufacturing nitrogenous fertilizers is a highly energy-consuming process requiring six times more energy than phosphorus (P) and potassium (K) fertilizers production (Da Silva et al. 1978). Reducing dependence on these fertilizers by developing BNF an integral mechanism in crop development will be a profitable venture. Studies depict that globally, BNF produces nearly 200 million tonnes of N annually (Graham 1992; Peoples et al. 2009) and out of which 50% of crop field N is contributed by diazotrophic bacteria using nitrogenase enzyme (Ramírez-Puebla et al. 2019). Thus, BNF lures the attention of agricultural researchers to incorporate this trait in staple and economically important food crops (Table 1). However, due to the complex nature of the BNF process, there are various constraints at the technological level to successfully address this trait. Hence, N-fixation in cereals remains an unresolved issue for researchers.
Several research groups diligently worked to achieve this target likewise a project on “Assessing Opportunities for Nitrogen Fixation in Rice (Oryza sativa)" was conducted under the aegis of the International Rice Research Institute (IRRI) with many other countries during 1994–2001 (Ladha and Reddy 2000). Under the project, the rice nodulin genes (ENOD40) were characterized in addition studied the role of chitin in nodulation, explored genetic programs for root endosymbiosis, and proposed modification of lateral root as a more achievable goal in the short period with BNF ability. Similarly, many other projects have also been initiated to introgress BNF in cereals funded by the Bill and Melinda Gates Foundation (BMGF, USA), the National Science Foundation (NSF, USA), the Biotechnology and Biological Sciences Research Council (BBSRC, UK) and the Indian Council of Agricultural Research (ICAR, India) (Sharma et al. 2016). In 2016, the India-UK N-fixation centre has been launched at the ICAR-Indian Institute of Soil Science, Bhopal (India), with the objectives of genetic engineering of rhizobia and rice endophytes, and improving BNF. They have engineered the rhizobia and characterized endophytes in rice with studying their mechanism of infection (ICAR 2016). Engineering Nitrogen Symbiosis for Africa (ENSA) project funded by BMGF has purposed synthetic biology as a viable approach to engineer N-fixation in cereals (Rogers and Oldroyd 2014). They have discovered a similar pathway for lateral roots and nodule development indicating that a substantial part of nodule formation machinery is already present in cereals which can be further engineered for N-fixation (Katharina et al. 2019). A group of scientist at Lethbridge Research and Development Centre, Canada, have successfully developed nif genes cluster with 16 essential genes and directly introgress it in wheat mitochondria. Now the team is working to regenerate the nif genes-enriched wheat plantlets (King 2019). Currently, the maize research is mainly focused on multiple traits like high yield, disease-pest resistance, and fertilizer use efficiency, but study regarding the interaction between maize and diazotrophs is a still ignorant area to tap it on.
Various N-fixing bacteria such as Azospirillum, Azoarcus and Herbaspirillum inhabit in endophytic associations with maize in intercellular plant tissues without causing any disease (Rosenblueth and Martínez-Romero 2006). It provides a scope to explore this trait in maize breeding to fix N naturally. Plant-associated diazotrophs also benefit the crop growth and development by promoting the mechanisms like the synthesis of vitamins and phytohormones, stimulation of nutrient uptake (solubilize and mineralize inorganic and organic phosphate), reduction in ethylene synthesis and improving resistance against pathogenic microbes, etc. (Berge et al. 1990; Triplett 1996; Hallmann et al. 1997). Therefore, under suitable conditions plant growth or health can also be benefitted from a diazotrophic association (Boddey et al. 2000; Chelius and Triplett 2000). Considering all these, the integration of the BNF mechanism with maize breeding strategies will be helpful to achieve sustainable agricultural production (Riggs et al. 2001; Kennedy et al. 2004). Maize is not a plant with N-fixation ability. Hence, no experimental data is available to estimate its effect on maize yields. However, an analysis of energy cost on legumes like soybean (Glycine max) may give a rough idea of energy cost levied in maize by N-fixation. The assessment has shown that soybean with nodules consume 19% of photosynthate, but this will compensate with N-fixation, nitrate assimilation and other indirect benefits like C sink maintenance that will increase photosynthesis, H2 released in the process will enhance plant growth and other positive environmental consequences (Ladha and Reddy 2000).
Different approaches available for nitrogen fixation
N-fixation is a dynamic and highly energy demanding process (Fig. 2a). All N-fixing prokaryotes i.e. diazotrophic bacteria fix N by owning nitrogenase enzyme system, which hydrolyses 16 ATPs for one molecule of N-fixation, making it a most expensive metabolic process (Simpson and Burris 1984) (Fig. 2b). Diazotrophic bacteria are highly diverse in phylogeny and ecological niche and have a synchronized interaction with the host plant for N-fixation. These diazotrophic bacteria can be free-living or in symbiotic associations with crop plants. The BNF is an extremely sensitive process influenced by nutrient and environmental conditions. It enables to supply all or part of crop N requirements through endosymbiotic, associative and endophytic interactions (King and Purcell2005; Liu et al. 2011). Free-living diazotrophs constitute a small fraction of plant rhizosphere ecosystem including cyanobacteria like Anabaena and Nostoc and other genera such as Azotobacter, Beijerinckia and Clostridium. Many prokaryotes fix N symbiotically (Rhizobium, Bradyrhizobium, Mesorhizobium and Sinorhizobium) with the host where plants provide sugars to the microorganisms and in exchange microbes provide fixed N for host plant growth (Graham and Vance 2003). Symbiotic N-fixation is observed in water fern Azolla with Cyanobacteria Anabaena azollae, actinorhizal trees and shrubs with actinomycete Frankia (non-legume N fixers), and in legumes with Rhizobium and Bradyrhizobium bacteria. Several microorganisms also show the associative or endophytic symbioses such as Azospirillum lipoferum, Azospirillum brasilense, Azoarcus evansii and Herbaspirillum seropedicae in a wide variety of crop plants including cereals.
Schematic representation of biological nitrogen fixation (BNF). a For effective symbiotic association development mainly two classes of genes are required: nodulation and nitrogen fixation genes. Nodulation genes, e.g. nodABC, interact with host-specific nodulin genes and plant flavonoids. Nitrogen fixation genes possess the structural genes (nif and fix) to encode nitrogenase (nifHDK) enzyme that is the lead actor for nitrogen fixation; b metabolic reaction of BNF is highly expensive as it consumes 16 ATP to fix one molecule of N; c the key actor nitrogenase genes assembly—the genes are coloured by their function as blue (nitrogenase), green (cofactor biosynthesis, shading corresponds to operons), and yellow (e− transport) and a horizontal bar indicates overlapping genes; d mature nitrogenase enzyme with electron-transport chain catalyses nitrogen fixation process (Temme et al. 2012; Laranjo et al. 2014)
The BNF is a unique natural system under the partnership of plants and diazotrophs for capturing atmospheric N and processing it into a usable form of N through enzymatic reduction. The key enzyme that fixes atmospheric N, i.e. nitrogenase is highly conserved and complex present in free-living and symbiotic diazotrophs which enables them to participate in various types of associations/interactions with their host plants. The proteins and non-proteins involved in the constitution of the nitrogenase enzyme make it sensitive to the presence of oxygen (Burén and Rubio 2017). Thus, there are some obligate and facultative anaerobic bacteria also like Clostridium pasteurianum and Klebsiella oxytoca, respectively, which fix N in the complete absence of oxygen. Other microbes like Azotobacter vinelandii, being the obligate aerobes, carry out N-fixation activity by shielding nitrogenase and consuming oxygen through cytochrome oxidases (Mahmud et al. 2020). Endosymbiosis between legumes and rhizobia is the most conventional N-fixing system in agriculture. In legume nodule, rhizobia reside intracellularly and fix N symbiotically in cytoplasmic vesicles. Endosymbiosis is the most evolved, stable and efficient N-fixing mechanism as it creates reducing conditions for nitrogenase, a pathway to transfer N-fixation product to host and protect against oxygen and antagonistic bacteria (Quispel 1991). Other than nodular structures, there are other modes too for intracellular endosymbiotic N-fixation in plants likewise in non-legume Gunnera plants, cyanobacteria Nostoc fix N in mucilage-secreting glands of stem. It establishes similar symbiosis like legume-rhizobium by invading intracellularly in gland cells (Werner 1995). Rhizobium also fix N in many leguminous species (subfamilies Papilionoideae, Caesalpinioideae and Mimosoideae) without developing nodules (Bryan et al. 1996). In maize, an indigenous landrace Sierra Mixe grown in Oaxaca province of Mexico possess extensive aerial root system that secretes carbohydrate rich (mainly arabinose, fucose and galactose) mucilage and fix about 29–82% N (Van Deynze et al. 2018). Many other bacteria which colonize intracellularly for N-fixation have been detected in many plant species like banana (Musa sp.) (Thomas and Reddy 2013; Thomas and Sekhar 2014.), peach palm (Bactris gasipaes) (de Almeida et al. 2009), scotch pine (Pinus sylvestris) (Pirttilä et al. 2000), cactus and non-nodulating legumes (Sambukumar et al. 2015). This indicates that nodule formation is not a compulsory mode for intracellular N-fixation in crop plants. Several diazotrophic bacteria have been known for N-fixation in maize by establishing rhizospheric or endophytic associations like Azospirillum, Klebsiella, Pantoea, Herbaspirillum, Bacillus, Rhizobium etli and Burkholderia (Chelius and Triplett 2000; Dong et al. 2001; Gutiérrez-Zamora and Martinez 2001; Caballero-Mellado et al. 2004; Perin et al. 2006). These diazotrophs reside intercellularly in plant tissues without causing any disease and are part of so-called endophytic diazotrophs (Montañez et al. 2009). Various studies have been undertaken to estimate the effect of these diazotrophs on maize yield (Table 2). Estrada et al. (2002) isolated the most abundant N-fixing endophyte, Burkholderia associated with maize and further confirmed that these isolates densely colonize into maize tissues which subsequently enhances yield. Riggs et al. (2001) carried out a study to identify maize-endophyte associations and their effect on maize production under both laboratory and field conditions and a significant increase in yield was observed with the association of Klebsiella pneumoniae and Herbaspirillum seropedicae endophytes. Pandey et al. (1998) conducted an experiment using the strains of Azotobacter chroococcum and Azospirillum brasilense in local maize cultivars and found significant improvement of 1–1.5 fold in maize production in tropical conditions. Shaharoona et al. (2006) isolated several rhizo-bacterial strains from the maize field with a significant positive correlation between 1‐aminocyclopropane‐1‐carboxylic acid (ACC) deaminase activity of diazotrophs and maize root elongation. All these studies indicated that a number of endophytes are there to increase yield and biomass of maize, but the application of these inoculants is not reliable as they are highly environment dependent. Therefore, researchers are exploring to directly introgress nif genes in cereals through genetic engineering or breeding.
Potential genetic resources for assessing nitrogen fixation in maize
Screening or development of germplasm possessing N-fixation capability is the most crucial and prerequisite step to integrate BNF with the breeding process. Many important crops have been screened for BNF like wheat (Jain et al. 1986), maize (Von Bülow and Doebereiner 1975; Baldani et al. 1997; De-Polli et al. 1982), sugarcane (Reis et al. 2000), rice (Boddey et al. 1995) and other plants (Tapia-Hernandez et al. 2000). Several non-pathogenic diazotrophic bacteria have been detected in maize and there is a need to discover maize germplasm that would be benefitted in association with these endophytic bacteria (Triplett 1996). Estrada et al. (2002) screened maize varieties to isolate N fixer endophyte from the root tissues. The N-fixing genus Burkholderia isolates were identified in field-grown maize tissues and its wild relative, teosinte (Z. mays ssp. mexicana). The study speculated that the strains of Burkholderia species have a kind of primitive symbiosis with teosinte from the domestication period. The mucilage in the aerial root is also reported to be an important harbinger of such symbiotic associations (Dennis et al. 2010; Li et al. 2011). Teosinte, the wild relative of maize possesses an extensive aerial roots system with mucilage secretion (in mild quantity) and nitrogenase enzyme activity. The acetylene reduction assay (ARA) indicates that mucilage production with N-fixation ability is an ancient trait of maize.
Sierra Mixe, an indigenous maize landrace possessing similar mechanism with Burkholderia strains in its mucilage has been reported. This hypothesized that this trait has been potentially introgressed into the landrace from teosinte during the post domestication period (Van Deynze et al. 2018). Earlier also, Raju et al. (1972) observed N-fixing capacity in a hybrid Pioneer 3773 grown in Oregon, USA. They isolated a strain Enterobacter cloacae from plant rhizosphere and roots with the ability to fix N and producing gel, but the N-fixation rate was very low. It is speculated that the production of gel helps to protect against oxygen and facilitate N-fixation in roots. The similar feature of producing mucilage exudate as in aerial roots of Sierra Mixe have been detected in other cereal crops also like wheat (Sinha et al. 2002), sorghum (Werker and Kislev 1978; Li et al. 2014) and barley (Carter et al. 2019) indicating that it is an ancient common feature prevalent in cereal crops. Hence, screening of maize germplasm to identify new genotypes possessing different and unique mechanisms of N-fixation should be focused on to further utilize them in breeding programmes.
Genetic Basis of Nitrogen Fixation
The most evolved legume-rhizobia interaction is very complex at the genetic level. To develop such symbiosis in cereals deeper insight about the gene families involved in N-fixation process is needed. There are a number of gene sets from both host and diazotrophs responsible for BNF (Fig. 2c).
Host-specific genes
Nodulin genes: The host plant genes involved in a symbiotic association are nodulin genes. Nodulin genes are specifically expressed to develop nodule structure, morphogenesis, size, and number of nodules which together influence N-fixation rate. Nodulin genes are of two types: C-Nodulins, which are present in all legumes, and S-Nodulins, which are species specific (Lodha and Nainawatee 1993). Several genes for nodulin specific proteins (S-Nodulins) have been identified in many leguminous crops such as nodulin-35 of Glycine max, glutamine synthetase in Phaseolus vulgaris, apyrase in Dolichos biflors and Gs-50 in Glycine soja (Etzler et al. 1999; Stacey et al. 2000). The genetic dissection of nodulin genes is prerequisite for genetic manipulation of BNF by analysing their sequence, regulatory mechanism and function. Some early nodulins (like ENOD2, ENOD40, ENOD12) are activated by Rhizobium signalling molecules, while late nodulins like leghaemoglobin, sucrose synthase, glutamine synthetase and nodulin-26 are involved in the physiological adaptation of nodule development (Brewin 2001).
Bacterial Sym genes
In N-fixing bacteria, number of gene sets are responsible to develop the symbiotic association. These are nod, nif and fix genes, which are collectively called as Sym genes. In fast-growing rhizobial strains Sym genes are found in large plasmid, while in slow-growing strains these are present on bacterial chromosome only.
Nod genes
The bacterial nod gene set is responsible for nodule formation in interaction with host nodulin genes. Based on function, there are several nod genes present in bacterial strains (Table 3) involved to induce and control nodule formation. This major structural modification in infected host plant cells enclose the bacteroid inside providing the suitable conditions for nitrogenase expression (Verma et al. 1986).
Nif genes
The nif gene family is highly conserved among diazotrophs and encodes for MoFe-dependant nitrogenase complex enzyme, which is the key component to catalyse atmospheric N into reduced forms (NH4+) (Raymond et al. 2004). Various nif genes have been identified and characterized structurally and functionally (Galton and Smith 1993; Parvez and Wani 2007) (Table 3). Besides encoding nitrogenase, nif genes also play a role to encode several regulatory proteins needed in N-fixation. The expression of nif genes is sensitive to oxygen or fixed N concentrations. Nif genes vary in their regulatory mechanism as in R. melilotus and R. leguminoserum they expresses under single operon, while in B. japonicum and R. phaseoli reside on different operons (Ow et al. 1985). It has been observed that by direct cell contact only nif genes can be transferred in between bacteria so this seems a significant aspect to work on to transfer these genes in plants.
Fix genes
It is the other gene family which plays a critical role in regulation and metabolism of oxygen concentration for nif genes expression. Nif genes perform another important function of regulating these fix genes. Many fix genes have been identified and characterized. The fix genes are regulated under three core operon structures such as fixABCX, fixGHIS and fixNOPQ. The fixABCX operon regulates gene transcription under low oxygen concentration, while fixNOPQ operon encodes cytochrome cbb3 oxidase complex that metabolizes oxygen by the bacteroid (Black et al. 2012).
Unlike conventional BNF process through nodulation, Van Deynze et al. (2018) identified sequence of six core nif genes (nifH, nifD, nifE, nifK, nifN, and nifB) from diazotroph associated with aerial roots of Sierra Mixe maize landrace. However, the genetic basis of N-fixation in aerial root associated with mucilage of landrace Sierra Mixe and its microbial inoculum is still unexplored. It can be hypothesized that this association is either environmental or seed-borne or other factors. It indicates that nif genes are key actors in N-fixation whether through forming nodules or in root mucilage. But as per current understanding, this novel way of BNF in maize has not yet been harnessed because Sierra Mixe has grown only in restricted areas. Further, the mucilage production largely depends on environmental conditions. Hence, currently this mechanism has been proposed as a model to study the association between cereal crops and diazotrophs to support N-fixation in cereals like maize, rice, sorghum and wheat. It will help to create a scaffold to understand diazotrophic activities and their functions in cereals which can be further utilized to increase their N-fixation potential by enhancing the mucilage production by practicing genetic selection or modifying the structural and regulatory pathways of the microbes associated with mucilage production. Further, there is need to understand at the molecular level that how these nif genes are able to express without creating anaerobic conditions in root mucilage or any other components are involved for their regulation. Thus, for future, it is important to study the genetic basis of BNF in Sierra Mixe, detection of the diazotrophs involved in its association, and the mechanism of their microbial recruitment which will open up new avenues for research into a potentially novel model of the BNF in maize.
Other genes
Various important accessory genes are also involved to fix N successfully, viz. exo gene that produces exopolysaccharide, hup gene that helps in hydrogen uptake, gln gene for glutamine synthase, dct genes to transport dicarboxylate, nfe genes to efficiently develop nodules and competitiveness, ndv-1,2 gene for glucans synthesis and lps genes for lipopolysaccharide production (Laranjo et al. 2014).
Bacterial secretion systems
After induction of nodule formation the next step is an infection of bacteria in plants tissues and their maturation to N-fixing bacteroids. Many gene families are involved, but protein secretion systems are a crucial group of genes to infect plant with diazotrophs. There are seven groups of bacterial secretion systems, viz. Types I to VII in addition to the fimbrial chaperone–usher pathway (Black et al. 2012). These systems have diversified functions like assembling surface structures to contact target cells, deliver DNA or protein effectors, affect host range, produce nodulation outer proteins (Nops), virulence and conjugal transfer or communication between microsymbionts and host.
Exopolysaccharide production
In symbiotic association, production of exopolysaccharide (EPS) is critical for the initial bacterial invasion to form indeterminate types of nodules on legumes (Skorupska et al. 2006). EPS plays a role in nodules induction by facilitating both infection thread initiation and bacterial release in symbiosis with the host (Kelly et al. 2013). In Sinorhizobium–Medicago (alfalfa) model, Sinorhizobium meliloti invades and differentiates inside its host plant alfalfa by producing exopolysaccharides succinoglycan (EPSI) and galactoglucan (EPSII) which induce the formation of infection thread (Jones et al. 2007). In absence of EPS, the competition to nodulate host plant increases leading to a diminished capacity to infect host (López-Baena et al. 2016). The comparison among genome of various diazotrophs shows that there is genetic complexity in EPS regulation that is still needed to understand. During evolution, different species have adopted diverse sets of genes which indicates that for universal symbiome production of EPS is not a necessary feature (Black et al. 2012).
Different breeding approaches for introducing nitrogen fixation ability in maize
Breeding for symbiotic N-fixation is a promising area to research. The N-fixation is a very dynamic trait controlled by multiple genes in both diazotrophic bacteria and plant species in addition to affected by edaphic and abiotic factors. From last many years, huge efforts have been made to isolate, identify and test diverse diazotrophs from cereals with an inherent ability to fix N, but practically still it has a long way to go to turn it a reality. There are several factors/constraints that hinder it to incorporate in maize or any other cereal crop; for example (1) it requires the association and coordination of totally two different organisms (plants and microbes) to mutually function and fix N in a new environment (Rice et al. 2000), (2) N-fixation requires nearly 30 essential genes to function in a coordinated manner which is tightly regulated in their host under a very sensitive pathway (Rogers and Oldroyd 2014), (3) a large number of variables like plant genotype, diazotroph’s strain, environmental factors (water, soil, O2 permeability, salinity and nutrient cycle), etc., highly affect BNF, making it a very sophisticated and tricky process to manipulate (Rice et al. 2000; Serraj et al. 2001; Chaudhary et al. 2008; Hartmann et al. 2009; Chaparro et al. 2014; Pfeiffer et al. 2017), (4) several plant defence alkaloids (like salicylic acid, phytoalexins) show an inhibitory effect on host–microbes interaction (Lebeis et al. 2015), (5) from plant to plant, the quorum-sensing signal for diazotrophs varies (Venturi and Keel 2016), which also alters the bacterial genes expression, (6) nitrogenase expression is highly sensitive to oxygen and needs anaerobic conditions to function that makes it more challenging to implement this strategy, and (7) as N-fixation is a high energy demanding process so, it is also unclear whether cereal hosts can provide this much energy with reducing conditions to endure nitrogenase catalysis (Van Velzen et al. 2018).
To resolve all the issues, one methodology is to study a model system of any naturally existing non-rhizobial N-fixing bacterium like in sugarcane diazotroph that fix N intracellularly by colonizing in root systems (Cocking et al. 2006) or Sierra Mixe landrace producing mucilage with N-fixation ability. Following that different breeding approaches should be practiced to develop/identify maize germplasm with BNF traits. To identify a maize genotype with the ability to fix N and compatible with BNF fixers is a challenging and crucial task to accomplish. The traits that stimulate N-fixation in non-leguminous plants are called as N-fixation supportive (NFS) traits (like nodulation traits, fixed nitrogen, shoot dry weight, root traits) so genetic variability and heritability of such traits should be traced to use in breeding programs. The genetics of mucilage production in aerial roots needs to dissect and the ways to exploit it for field level product development should be explored. In legume-rhizobium the defence alkaloids show an inhibitory effect on their interaction (Lebeis et al. 2015). Likewise in cereals also their effects on diazotrophs should be accessed. Correlation of N-fixation ability with the plant attributes like acid-tolerant plants (exude dicarboxylic acids) that shows higher N-fixation rate (Christiansen-Weniger et al. 1992) needs to be studied to develop BNF plants type. Mutagenesis can also be applied to develop novel diazotroph strains with improved N-fixation ability in cereal crops (Maier and Brill 1978). Ling et al. (2013) showed that by applying reversible mutation it is possible to manipulate both the nodulation and N-fixation genes without affecting host–microbes association in cereals.
Molecular breeding approach
Molecular breeding approaches provide opportunities for mapping and introgression of the essential genes for nodulin, nitrogenase, lectins and malate dehydrogenase into recipient plants. However, till now in any crop optimized expression of all N-fixation genes have not been worked out. One of the conventional approaches in maize will be the mapping of the genes/QTLs for NFS traits from the screened maize cultivars measuring total mucilage production, amount of aerial roots, total N-fixation and N use efficiency. Application of molecular markers is the primary approach to validate the expression of nitrogenase biosynthesis and N-fixation (Schmid and Hartmall 2007). Initially, the RFLP (Restriction fragment length polymorphism) markers with nif gene probe and PCR (Polymerase chain reaction) fingerprinting were used in cyanobacterium to identify the gene diversity (Plazinski et al. 1985). Ueda et al. (1995) also detected diazotrophic bacteria in rice using PCR amplified nifH sequences. Rai et al. (2014) isolated different restriction fragments using nifH RFLP markers analysis from the soil samples. Hence, the construction of a molecular markers library will be an efficient way to divulge the genetic diversity of uncharacterized diazotrophs in the maize rhizosphere.
There are many root architectural traits like root length, surface area, diameter, volume, root hairs and other nodulation traits involved in BNF. These traits are genetically controlled by polygenes or quantitative trait loci (QTLs). Therefore, identification of major QTLs for these traits will be a vital objective of maize breeding to enhance N-fixation. Akiyoshi et al. (2012) mapped 34 QTLs in Lotus japonica for controlling BNF traits like nodule number, nodule weight, acetylene reduction activity (ARA) per plant, ARA per nodule number and ARA per nodule weight etc. Similarly, Santos et al. (2013) identified two QTLs for shoot dry weight, three QTLs for nodule number, and one QTL for nodule dry weight with a phenotypic variance of 15.4, 13.8, and 6.5%, respectively, from a soybean RIL population. Therefore, such studies should be extended to track QTLs for BNF traits in maize and cereal crops to identify the contrasting genotypes.
The direct transfer of nif genes into maize should be attempted through simultaneously or one by one introgression of vital genes required for the biosynthesis of FeMo-Co unit and nitrogenase function. Evolutionary genomics of N-fixation suggests that introgression of BNF ability into non-legumes require only a few genetic elements by concatemerizing bacterial genetic constitution (Bailey-Serres et al. 2019). Thus, to introgress nitrogenase requires minimum three genes (nifH, nifD, and nifK) of bacterial genetic units (Yang et al. 2018) (Fig. 2d). A novel model for N-fixation as identified in Sierra Mixe landrace of maize (Van Deynze et al. 2018) also has its own limitations like it is long in duration with taller plants and is not directly suitable for cultivation purposes. Hence, the transfer of BNF traits from this landrace through marker-assisted breeding to adopted accessions of maize will be a fruitful venture (Fig. 3). Meanwhile, further understanding the mechanisms of symbiosis in plants and diazotrophs with high-throughput genotyping will also be beneficial to implement the innovative breeding approaches to introgress N-fixation traits into maize.
Omics Approaches
To elaborate the molecular basis of BNF traits, different functional genomics approaches like transcriptomics, proteomics and metabolomics can also be performed on rhizobial symbiosis. The integration of different omics data will help in better understanding of rhizobial cellular activities, their adaptation and homology with other functional diazotrophs. Metabolic databases like KEGG, MetaCyc and BioCyc (Caspi et al. 2008) provides huge data to study differentially regulated pathways. It will further be useful in selection and prioritization of candidate genes suitable for mutagenesis and validation experiments (Lardi and Pessi 2018). Recently omics approaches have been utilized as a viable tool to identify and isolate diazotrophs from cereal crops such as in rice (Bao et al. 2014), sugarcane (Fischer et al. 2012), sorghum (Kiwamu et al. 2019) and sweet potato (Ipomoea batatas) (Terakado-Tonooka et al. 2013). From obtained omics results microbes with nitrogenase proteins should be isolated and phylogenetic markers can be predicted. From the metagenome sequence, the nitrogenase gene sequences can be computed using nitrogenase databases. Kiwamu et al. (2019) carried out an omics study to identify functional N-fixing diazotrophs (Bradyrhizobium) associated with sorghum. Proteome analysis revealed that three NifHDK proteins of Bradyrhizobium species were consistently found across sample replicates. Thus, omics data will be used to identify and isolate different functional diazotrophic strains in maize with better understanding their regulatory mechanism. Metagenome and proteome data can be further used to study the genetic basis of N-fixation in mucilaginous aerial roots of landrace Sierra Mixe and its microbial inoculum.
Genome Editing
Genome editing, a novel approach facilitates modification in the genome of any living organism by inserting, deleting, modifying, or replacing any DNA sequence at site-specific locations. It allows remodelling of N-fixation regulation by diazotrophs without the inclusion of any trans-elements. After identification of efficient diazotrophs closely associated with the crop, gene editing will allow disrupting the regulatory pathways linked to N-fixation, sensing and assimilation. It can replace native promoters of nif genes from already characterized promoters of different strains permitting N-independent expression of nif genes. Such remodelling via promoters allows microbes to adapt to different niche and environmental conditions. Bloch et al. (2020) isolated a strain of Kosakonia sacchari from maize plants and through gene editing decoupled the nitrogenase enzyme biosynthesis from the regulatory networks that respond to exogenous cellular N. The edited non-transgenic strains were capable to express nitrogenase genes in the corn rhizosphere under various levels of applied N fertilizers in the greenhouse as well as field conditions. In comparison with initial field count, the overall colonization was significantly higher, but all the remodelled strains show a decrease in colonization which suggests that higher expression of nitrogenase activity led to a fitness cost so we have to optimize its expression. Hence, through gene editing by optimum modification of the regulatory pathways of diazotrophs can make this process accessible to cereal crops too to boost crop production sustainably.
Genetic engineering
Genetic engineering is one of the most viable approach to harness symbiotic N-fixation by modulating the mechanisms and signalling system (Oldroyd and Dixon 2014). Molecular and genetic dissection of diazotrophs with their interaction to non-leguminous crops opens a new door to engineer nodule development or other mode of symbiosis (Oldroyd et al. 2011; Oldroyd 2013). For genetic manipulation of multiple gene complexes (nodulin, nod and nif genes) there is need of a series of events to engineer BNF in maize. Hence, by thoroughly analysing all the feasible engineering approaches we should strategies our objectives by utilizing the different advanced biotechnological tools to develop BNF in maize (Table 4; Fig. 4).
-
1.
Introgression of nitrogenase genes into maize
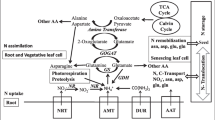
Engineering strategies for BNF: a introgression of nitrogenase gene into maize mitochondria or chloroplast, b development of maize root-nodule symbiosis (RNS) as in legume. AmtB—ammonia transporter; AMCs—arbuscular mycorrhizal components; cyt bd—cytochrome bd; Glu—glutamate; Gln—glutamine; GS—glutamine synthetase; LCOs—lipochitooligosaccharides; Mo—molybdenum; NH3—ammonia; N2ase—nitrogenase; Nod factors—nodulation factors; S—sulphur
To introduce BNF in maize one needs to construct the bacterial genes assembly encoding the nitrogenase enzyme with optimized N-fixation capability. As genetics of both mitochondria and chloroplast resembles to diazotroph plasmid constitution so these will be the perfect site for nif genes expression as well as will provide the sufficient energy (ATP) required for nitrogenase expression (Biswas and Gressho 2014; Curatti and Rubio 2014) (Fig. 4a). In chloroplast, the transcription and translation signals also match more closely with the microbes. But still, organelle transformation also has some challenges to encounter as in chloroplast for optimum transcription of nif genes, there is a need to replace nif gene promoter with suitable chloroplast promoter. Further, oxygen generated during photosynthesis will be detrimental for the nitrogenase enzyme. To address these challenges, we need to separate both the photosynthesis and N-fixation processes by regulating the expression of nif genes in the night when photosynthesis is absent or in non-photosynthetic parts such as roots (Peoples et al. 2009). López-Torrejón et al. (2016) engineered nifH, nifM, nifU and nifS from Azotobacter vinelandii into yeast cell and depicts that functional nitrogenase protein can be formed if only NifH and NifM polypeptide is targeted into the mitochondrial background jointly. In another study, Buren et al. (2017) targeted nine nif genes from A. vinelandii (nifH, nifD, nifU, nifK, nifM, nifS, nifE, nifB and nifN) into mitochondria and generate an active NifDK tetramer that is an essential component for nitrogenase expression in a eukaryotic cell. In tobacco (Nicotiana spp.) NifH and NifM genes have been transferred in chloroplast generating an active NifH protein with low expression (Ivleva et al. 2016). Therefore, for stable expression, efficient transformation methodology with the ideal nif genes set is needed, and their amino acid sequence need to be optimized to enhance stability in eukaryotic cells without negotiating with nitrogenase activity (Allen et al. 2017; Buren and Rubio 2017).
-
2.
Development of maize root-nodule symbiosis (RNS) as in legume
To develop root-nodule symbiosis, the arbuscular mycorrhizal (AM) associations in maize should be approached where the crop plant develops endosymbiotic associations with endomycorrhizal fungi (Fig. 4b). In legumes and cereals, symbiotic signalling (SYM) pathways with similar genetic constituents play a role to promote AM symbiosis (Gutjahr et al. 2008). Similar genetic components are involved in AM and RNS development in legumes making it a “common symbiosis pathway” (CSP) (Markmann and Parniske 2008). Recently, phylogenomic studies also indicate that any species with AM associations can be turned into symbiont N fixer by introgressing small sets of genes (Griesmann et al. 2018). Hence, this association should be focused to research on developing a functional signalling pathway to support RNS in maize (Reddy et al. 2013; Delaux et al. 2015; Mus et al. 2016).
-
3.
Maize rhizosphere engineering for enhanced diazotrophs growth and colonization
Mostly the diazotrophs population density is way too low in maize tissues to fix adequate N. Hence, it is important to develop a strategy to enhance diazotrophs colonization in maize roots to increase N-fixation. Nutritional resources also influence the diazotrophs population in the rhizosphere (Oger et al. 1997; Savka and Farrand 1997). A specialized carbon source will encourage microbe colonization by establishing appropriate signals between diazotrophs and maize (Mus et al. 2016). Host plant roots secrete polysaccharide mucilage to stimulate microbe colonization by providing significant carbon source in the rhizosphere. In pea (Pisum sativum), root mucilage promotes 3–25% growth of diazotrophic bacteria like Rhizobium leguminosarum, Burkholderia cepacia, and Pseudomonas fluorescens (Knee et al. 2001). Opines are the chemical compounds produced by rhizobial strains in legume nodules and are found to be suitable for chemical signalling between plants and rhizosphere bacteria (Murphy et al. 1995; Gordon et al. 1996). In this direction, rhizosphere engineering could be tried in maize to produce a metabolite that favour diazotrophs growth (Rossbach et al. 1994). It has been attempted in barley where rhizopine biosynthesis genes were successfully transferred by expressing a synthetic pathway for the production of rhizopine scyllo-inosamine (Geddes et al. 2019). It helps to produce synthetic signalling networks in between plant rhizosphere and microbes to regulate the targeted bacterial genes expression. In analogous to natural signalling in legume-rhizobium symbioses, rhizopine engineered transkingdom signalling controls the synthetic symbioses in cereal crops for N delivery and fixation. So overall engineering the rhizosphere regions, different genes and signalling system at the perfect site will enable the BNF a success in maize.
Amidst all these promises, genetic engineering has many challenges to work upon. Most importantly efficient control of the multigene expression is the biggest challenge. Modification of the complex regulatory pathways, construction of large synthetic multigene cassettes, characterization of wide range promoters and their regulatory control in many organisms are some of the major challenges. Finally, even if all these challenges are overcame release of such genetically modified plant need to pass regulatory barriers, which may become a challenge.
Synthetic biology
Synthetic biology, a field of science, facilitates the redesigning of useful organisms/microbes with new and valuable abilities by engineering. Due to large nif genes machinery the transfer of nitrogenase in crop plants is complicated. There is need to simplify this complex process and develop robust genetic components that perform nitrogenase related functions in diverse cellular environments. Thus, reduction in nitrogenase genetic components is prerequisite to transfer in eukaryotic hosts. Synthetic biology will facilitate to overcome nitrogenase related challenges by regrouping genes into synthetic gene products by harnessing the knowledge of nitrogenase biosynthesis and catalysis steps. A few studies have been carried out in this direction where considering Klebsiella pneumoniae or K. oxytoca as nif genes sources, genes structures were reconstituted and reorganized by splicing nonessential genetic components to transfer in E. coli (Temme et al. 2012; Yang et al. 2014). The finding depicts that many genes are not required for artificial N-fixation (ANF) system in E. coli. This enables to engineer a minimal nitrogenase system in organelles comprising the structural and metallocluster biosynthesis genes. Wang et al. (2013) and Smanski et al. (2014) replaced the complex regulatory elements of nif gene clusters with a simple expression system which enables the expression of nitrogenase enzyme under different genetic and physiological conditions in eukaryotic organelles. Yang et al. (2017) utilized synthetic biology to reform electron-transport components (ETCs) of ferredoxin-NADPH oxidoreductases (FNRs) and ferredoxins molecules derived from plant organelles (chloroplast and mitochondria) which enable a reduction in microbial gene number required for nitrogenase engineering to generate a putative N-fixing plant. The results showed that the FNR–ferredoxin module from organelles is active in both natural and engineered nitrogenase enzyme depicting the potential for future engineering of diazotrophs in maize crop. Hence, with a detailed understanding of all the components required for BNF, nitrogenase can be engineered to simplify the complexity of the process by reducing targeted gene number into cereal crops.
Future perspectives
Cereal crops with N-fixation ability have been a dream of researchers since many decades. Maize is N exhaustive crop as it requires a millions tonne of N for high production, while heavy dose of N with low (20–30%) nitrogen use efficiency creates a number of side effects on the soil, water, and environmental pollution with millions dollar investment. Therefore, to transfer N-fixation traits in maize is the need of hour to solve the above issues for advance sustainability. Breeding or engineering for BNF in maize is a cumbersome task to develop the N-fixation ability. Hence, exploration of natural and phylogenetic diversity is needed to promote the BNF potential in maize. Technological advancements in next-generation sequencing, gene editing and synthetic biology will enable the dissection and manipulation of diazotrophs and plants at an unprecedented scale. For establishment of efficient symbiotic N-fixation, functional genomics will elucidate the molecular basis underlying this process for the maize. Large datasets can be generated through transcriptomics, proteomics and metabolomics by using faster and sensitive instruments to monitor the end products of gene expression in between host and diazotrophs and assess the physiological changes in symbiosis-specific pathways. The amalgamation of high-throughput and innovative technologies such as genome sequencing, transposon sequencing, and proteomics will enable to identify novel essential rhizobial genes, undetected coding genes, monitoring gene expression at different stages of symbiotic interaction and elucidate genetic elements required for an efficient symbiosis. Synthetic biology will open up the scope to shorten functional gene clusters so that the nif gene machinery can be successfully introgressed into heterologous hosts. There is need to study the non-leguminous model systems likewise in Sierra Mixe to harness it through breeding or engineering to enhance N-fixation rate. Development of large omics datasets for integrative analysis and mining of symbiotic genes will enable to identify the candidate genes to be used in breeding programs. Metagenomic studies are likely to identify microbiome at rhizosphere, non-rhizosphere, endosphere, phyllosphere and spermosphere, which may potentially be involved in BNF. However, this requires extensive computational facilities and precise sample handling, which is a limiting factor in low resource infrastructure and with untrained manpower. Directed investment in this direction is prerequisite to make BNF a reality in non-leguminous plants with the potential significant benefits. Integrating the prospects of plant and microbial diversity with genetic engineering will facilitate short- or long-term solutions to increase agricultural production for advance sustainability.
References
Akiyoshi T, Takahiro G, Ryo A, Shao-hui Z, Susumu A, Akihiro S (2012) Quantitative trait locus analysis of symbiotic nitrogen fixation activity in the model legume Lotus japonicus. J Plant Res 125(3):395–406
Alexandratos N, Jelle B (2012) World agriculture towards 2030/2050: The 2012 revision.
Allen RS, Tilbrook K, Warden AC, Campbell PC, Rolland V, Singh SP et al (2017) Expression of 16 nitrogenase proteins within the plant mitochondrial matrix. Front Plant Sci 8:287. https://doi.org/10.3389/fpls.2017.00287
Anuar AR, Shamsuddin ZH, Yaacob O (1995) Contribution of legume-N by nodulated groundnut for growth of maize on an acid soil. Soil Biol Biochem 27(4–5):595–601. https://doi.org/10.1016/0038-0717(95)98637-4
Bailey-Serres J, Parker JE, Ainsworth EA, Oldroyd GED, Schroeder JI (2019) Genetic strategies for improving crop yields. Nature 575:109–118. https://doi.org/10.1038/s41586-019-1679-0
Bakht J, Ahmad S, Tariq M, Akber H, Shafi M (2006) Response of maize to planting methods and fertilizer N. J Agric & Biol Sci 1:8–14
Baldani J, Caruso L, Baldani VL, Goi SR, Döbereiner J (1997) Recent advances in BNF with non-legume plants. Soil Biol Biochem 29(5–6):911–922
Baldani JI, Baldani VLD (2005) History on the biological nitrogen fixation research in graminaceous plants: Special emphasis on the Brazilian experience. Anais da Academia Brasileira de Ciências 77:549–579
Bano SA, Iqbal SM (2016) Biological nitrogen fixation to improve plant growth and productivity. Int J Agri Innov Res 4(4):596–599
Bao Z, Okubo T, Kubota K, Kasahara Y, Tsurumaru H, Anda M et al (2014) Metaproteomic identification of diazotrophic methanotrophs and their localization in root tissues of field-grown rice plants. Appl Environ Microbiol 80:5043–5052. https://doi.org/10.1128/AEM.00969-14
Belay N, Sparling R, Daniels L (1984) Dinitrogen fixation by a thermophilic methanogenic bacterium. Nature 312:286–288
Bennett AB, Pankievicz VC, Ané JM (2020) A model for nitrogen fixation in cereal crops. Trends Plant Sci 25(3):226–235
Berge O, Fages J, Mulard D, Balandreau J (1990) Effects of inoculation with Bacillus circulans and Azospirillum lipoferum on crop-yield in field grown maize. Symbiosis 9:259–266
Biswas B, Gressho PM (2014) The role of symbiotic nitrogen fixation in sustainable production of biofuels. Int J Mol Sci 15:7380–7397
Black M, Moolhuijzen P, Chapman B, Barrero R, Howieson J, Hungria M, Bellgard M (2012) The genetics of symbiotic nitrogen fixation: comparative genomics of 14 rhizobia strains by resolution of protein clusters. Genes 3(1):138–166
Bloch Sarah E, Ryu M, Ozaydin B, Broglie R (2020) Harnessing atmospheric nitrogen for cereal crop production. Curr Opin Biotech 62:181–188
Bochman OC, Kaarstad O, Lie OH, Richards I (1990) Agriculture and fertilizers: a report from Agriculture Group Norsk Hydro. Oslo, Norway 193:117–119
Boddey R, De Oliveira O, Urquiaga S, Reis V, De Olivares F, Baldani V, Döbereiner J (1995) Biological nitrogen fixation associated with sugar cane and rice: contributions and prospects for improvement. Plant Soil 174(1/2):195–209
Boddey RM, Da Silva LG, Reis V, Alves BJR, Urquiaga S (2000) Assessment of bacterial nitrogen fixation in grass species. In: Triplett EW (ed) Prokaryotic nitrogen fixation: a model system for the analysis of a biological process. Horizon Scientific Press, Poole, pp 705–726
Brewin NJ (2001) Nodulins. In: Brenner S, Miller JH (eds) Encyclopedia of genetics. Academic Press, pp 1334–1335. https://doi.org/10.1006/rwgn.2001.1641
Bryan JA, Berlyn GP, Gordon JC (1996) Towards a new concept of the evolution of symbiotic nitrogen fixation in the Legumnosae. Plant Soil 186:151–159
Burén S, Rubio LM (2017) State of the art in eukaryotic nitrogenase engineering. FEMS Microbiol Lett 365:fnx274
Burén S, Young EM, Sweeny EA, Lopez-Torrejón G, Veldhuizen M, Voigt CA, Rubio LM (2017) Formation of nitrogenase NifDK tetramers in the mitochondria of Saccharomyces cerevisiae. ACS Synth Biol 6:1043–1055
Caballero-Mellado J, Martínez-Aguilar L, Paredes-Valdez G, Los Santos PED (2004) Burkholderia unamae sp nov. an N2-fixing rhizospheric and endophytic species. Int J Syst Evol Microbiol 54(4):1165–1172
Carter AY, Ottman MJ, Curlango-Rivera G, Huskey DA, D’Agostini BA, Hawes MC (2019) Drought-tolerant barley: II. Root tip characteristics in emerging roots. Agronomy 9(5):220
Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, Latendresse M, Paley S, Rhee SY, Shearer AG, Tissier C et al (2008) The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 36:D623–D631
Chafi MH, Bensoltane A (2009) Vicia faba (L), a source of organic and biological manure for the Algerian arid regions. World J Agric Sci 5(6):698–706
Chaparro JM, Badri DV, Vivanco JM (2014) Rhizosphere microbiome assemblage is affected by plant development. ISME J 8:790–803. https://doi.org/10.1038/ismej.2013.196
Chaudhary M, Adu-Gyamfi J, Saneoka H, Nguyen N, Suwa R, Kanai S, El Shemy H, Lightfoot D, Fujita K (2008) The effect of phosphorus deficiency on nutrient uptake, nitrogen fixation and photosynthetic rate in mashbean, mungbean and soybean. Acta Physiol Plant 30:537–544
Chelius MK, Triplett EW (2000) Diazotrophic endophytes associated with maize. In: Triplett EW (ed) Prokaryotic nitrogen fixation: A model system for the analysis of a biological process. Horizon Scientific Press, Poole, pp 779–791
Christiansen-Weniger C, Groneman AF, van Veen JA (1992) Associative N2 fixation and root exudation of organic acids from wheat cultivars of different aluminium tolerance. Plant Soil 139:167–174. https://doi.org/10.1007/BF00009307
Cocking EC, Stone PJ, Davey MR (2006) Intracellular colonisation of roots of Arabidopsis and crop plants by Gluconacetobacter diazotrophicus. Vitro Cell Dev Biol Plant 42(1):74–82
Curatti L, Rubio LM (2014) Challenges to develop nitrogen-fixing cereals by direct nif -gene transfer. Plant Sci 225:130–137
Da Silva JG, Serra GE, Moreira JR, Conçalves JC, Goldemberg J (1978) Energy balance for ethyl alcohol production from crops. Science 201:903–906
de Almeida CV, Andreote FD, Yara R, Tanaka FAO, Azevedo JL, de Almeida M (2009) Bacteriosomes in axenic plants: endophytes as stable endosymbionts. J Microbiol Biotechnol 25:1757–1764
Delaux PM, Radhakrishnan G, Oldroyd G (2015) Tracing the evolutionary path to nitrogen-fixing crops. Curr Opin Plant Biol 26:95–99. https://doi.org/10.1016/j.pbi.2015.06.003
Dennis PG, Miller AJ, Hirsch PR (2010) Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol Ecol 72(3):313–327
De-Polli H, Boyer CD, Neyra CA (1982) Nitrogenase activity associated with roots and stems of field-grown corn (Zea mays L.) plants. Plant Physiol 70(6):1609–1613
Dobbelaere S, Croonenborghs A, Thys A, Ptacek D, Vanderleyden J, Dutto P, Labandera-Gonzalez C et al (2001) Responses of agronomically important crops to inoculation with Azospirillum. Funct Plant Biol 28(9):871–879
Döbereiner J (1966) Azotobacter paspali sp. nov. uma bactéria fixadora de nitrogênio na rizosfera de Paspalum. Pesq Agropec Bras 1:357–365
Dong Y, Glasner JD, Blattner FR, Triplett EW (2001) Genomic interspecies microarray hybridization: rapid discovery of three thousand genes in the maize endophyte, Klebsiella pneumoniae 342, by microarray hybridization with Escherichia coli K-12 open reading frames. Appl Environ Microbiol 67(4):1911–1921
Estrada P, Mavingui P, Cournoyer B, Fontaine F, Balandreau J, Caballero-Mellado J (2002) A N2-fixing endophytic Burkholderia sp. associated with maize plants cultivated in Mexico. Can J Microbiol 48(4):285–294
Etzler ME, Kalsi G, Ewing NN, Roberts NJ, Day RB, Murphy JB (1999) A nod factor binding lectin with apyrase activity from legume roots. PNAS 96(10):5856–5861
FAO (Food and Agricultural Organization) (2017). http://www.fao.org/faostat/en/#data/RFN. Accessed 25 April 2020
FAOSTAT (2020). http://www.fao.org/faostat/en/#data/QC. Accessed 25 April 2020
Fischer D, Pfitzner B, Schmid M, Simões-Araújo JL, Reis VM, Pereira W et al (2012) Molecular characterisation of the diazotrophic bacterial community in uninoculated and inoculated field-grown sugarcane (Saccharum sp.). Plant Soil 356:83–99. https://doi.org/10.1007/s11104-011-0812-0
Fossum JP (2014) Calculation of Carbon Footprint of Fertilizer Production, Open information note. Oslo: Yara International ASA. Available at: http://yara.com/doc/122597_2013_Carbon_footprint-of_AN_Method_of_calculation.pdf.
Gaby JB, Buckley DH (2011) A global census of nitrogenase diversity. Environ Microbiol 13:1790–1799
Galton JR, Smith RJ (1993) Nitrogen fixation. In: Lea PJ, Leagood RC (eds) Plant biochemistry and molecular biology. Willey, UK, p 420
Geddes BA, Paramasivan P, Jorin A, Thompson AL, Christensen K, Jorrin B, Brett P, Oldroyd CSJ, GED, Poole PS, (2019) Engineering transkingdom signalling in plants to control gene expression in rhizosphere bacteria. Nat Commun 10:1–11
Glendining MJ, Dailey AG, Williams AG, Van Evert FK, Goulding KWT, Whitmore AP (2009) Is it possible to increase the sustainability of arable and ruminant agriculture by reducing inputs? Agric Syst 99(2–3):117–125
Gonzalez-Dugo V, Durand JL, Gastal F (2010) Water deficit and nitrogen nutrition of crops: A review. Agron Sustain Dev 30:529–544
Gordon DM, Ryder MH, Heinrich K, Murphy PJ (1996) An experimental test of the rhizopine concept in Rhizobium meliloti. Appl Environ Microbiol 62:3991–3996
Graham PH (1992) Stress tolerance in rhizobium and Bradyrhizobium, and nodulation under adverse soil conditions. Can J Microbiol 38:475–484
Graham PH, Vance CP (2003) Legumes: Importance and constraints to greater use. Plant Physiol 131:872–877
Griesmann M, Chang Y, Liu X, Song Y, Haberer G, Crook MB, Billault-Penneteau B, Lauressergues M, Keller J, Imanishi L et al (2018) Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science 361:6398
Gutiérrez-Zamora ML, Martınez-Romero E (2001) Natural endophytic association between Rhizobium etli and maize (Zea mays L.). J Biotechnol 91(2–3):117–126
Gutjahr C, Banba M, Croset V, An K, Miyao A, An G et al (2008) Arbuscular mycorrhiza–specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell 20:2989–3005. https://doi.org/10.1105/tpc.108.062414
Hallmann J, Quadt-Hallmann A, Mahaffee WF, Kloepper JW (1997) Endophytic bacteria in agricultural crops. Can J Microbiol 43(10):895–914
Han Y, Lu N, Chen Q, Zhan Y, Liu Wei L, Zhu B, Lin M, Yang Z, Yan Y (2015) Interspecies transfer and regulation of Pseudomonas stutzeri A1501 Nitrogen Fixation Island in Escherichia coli. J Microbiol Biotechnol 25:1339–1348
Hartmann A, Schmid M, van Tuinen D, Berg G (2009) Plant-driven selection of microbes. Plant Soil 321:235–257. https://doi.org/10.1007/s11104-008-9814-y
Huang YZ, Feng Z, Fuzhu Z (2000) Study on loss of nitrogen fertilizer from agricultural fields and countermeasure. Journal of Graduate School, Academia Sinica 17(2):49–58
ICAR (2016) India-UK Nitrogen Fixation centre inaugurated at ICAR-IISS, Bhopal. https://icar.org.in/node/4523. Accessed 1 January 2021
Ivleva NB, Groat J, Staub JM, Stephens M (2016) Expression of active subunit of nitrogenase via integration into plant organelle genome. PLoS ONE 11:e0160951
Jain DK, Rennie RJ (1986) Use of spermosphere model for the screening of wheat cultivars and N2-fixing bacteria for N2 fixation. Can J Microbiol 32(4):285–288
Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC (2007) How rhizobial symbionts invade plants: the Sinorhizobium–Medicago model. Nat Rev Microbiol 5:619–633
Katharina S, Lilley JLS, Lee T, Tamvakis I, Kohlen W, Bailey PC, Thomas A, Luptak J, Ramakrishnan K, Carpenter MD, Mysore KS, Wen J, Ahnert S, Grieneisen VA, Oldroyd GED (2019) Nodule inception recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Curr Biol 29(21):3657-3668.e5. https://doi.org/10.1016/j.cub.2019.09.005
Kelly SJ, Muszyński A, Kawaharada Y, Hubber AM, Sullivan JT, Sandal N, Carlson RW, Stougaard J, Ronson CW (2013) Conditional requirement for exopolysaccharide in the Mesorhizobium-Lotus symbiosis. Mol Plant Microbe Interact 26:319–329. https://doi.org/10.1094/MPMI-09-12-0227-R
Kennedy Ivan R, Choudhury ATMA, Kecskés ML (2004) Non-symbiotic bacterial diazotrophs in crop-farming systems: can their potential for plant growth promotion be better exploited? Soil Biol Biochem 36(8):1229–1244
King CA, Purcell LC (2005) Inhibition of N2 fixation in soybean is associated with elevated ureides and amino acids. Plant Physiol 137:1389–1396
King C (2019) On the way to Nitrogen-fixing cereals. https://www.seed.ab.ca/on-the-way-to-nitrogen-fixing-cereals/. Accessed 1 January 2021
Kiwamu M, Hara S, Morikawa T, Wasai S, Kasahara Y, Koshiba T, Yamazaki K, Fujiwara T, Tokunaga T (2019) Identification of nitrogen-fixing Bradyrhizobium associated with roots of field-grown sorghum by metagenome and proteome analyses. Front Microbiol 10:407
Knee EM, Gong FC, Gao M, Teplitski M, Jones AR, Foxworthy A, Mort AJ, Bauer WD (2001) Root mucilage from pea and its utilization by rhizosphere bacteria as a sole carbon source. Mol Plant-Microbe Interact 14:775–784
Ladha JK, Reddy PM (eds) (2000) The quest for nitrogen fixation in rice. In: Proceedings of the 3rd Working Group Meeting on Assessing Opportunities for Nitrogen Fixation in Rice, International Rice Research Institute, Makati, p 354.
Laranjo M, Ana A, Solange O (2014) Legume growth-promoting rhizobia: An overview on the Mesorhizobium genus. Microbiol Res 169(1):2–17
Lardi M, Pessi G (2018) Functional genomics approaches to studying symbioses between legumes and nitrogen-fixing rhizobia. High-Throughput 7(2):15
Lassaletta L, Billen G, Grizzetti B, Anglade J, Garnier J (2014) 50 year trends in nitrogen use efficiency of world cropping systems: the relationship between yield and nitrogen input to cropland. Environ Res Lett 9(10): Article ID 105011
Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M et al (2015) Plant microbiome. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349:860–864. https://doi.org/10.1126/science.aaa8764
Li R, Han Y, Lv P, Du R, Liu G (2014) Molecular mapping of the brace root traits in sorghum (Sorghum bicolor L. Moench). Breed Sci 64(2):193–198
Li YJ, Fu YR, Huang JG, Wu CA, Zheng CC (2011) Transcript profiling during the early development of the maize brace root via Solexa sequencing. FEBS J 278(1):156–166
Ling J, Zheng H, Katzianer DS, Wang H, Zhong Z, Zhu J (2013) Applying reversible mutations of nodulation and nitrogen-fixation genes to study social cheating in Rhizobium etli-legume interaction. PLoS ONE 8(7):e70138
Liu Y, Wu L, Baddeley JA, Watson CA (2011) Models of biological nitrogen fixation of legumes. In: Lichtfouse E et al (eds) Sustainable Agriculture, vol 2. Springer, Berlin, pp 883–905
Lodha ML, Nainawatee HS (1993) Biological nitrogen fixation. In: Mehta SL, Lodha ML, Saini PV (eds) Recent avances in plant biochemistry. IARI, Phillipines, pp 173–224
López-Baena FJ, Ruiz-Sainz JE, Rodríguez-Carvajal MA, Vinardell JM (2016) Bacterial molecular signals in the Sinorhizobium fredii-soybean symbiosis. Int J Mol Sci 17:755. https://doi.org/10.3390/ijms17050755
López-Torrejón G, Jiménez-Vicente E, Buesa JM, Hernandez JA, Verma HK, Rubio LM (2016) Expression of a functional oxygen-labile nitrogenase component in the mitochondrial matrix of aerobically grown yeast. Nat Commun 7:11426. https://doi.org/10.1038/ncomms11426
Mahmud K, Makaju S, Ibrahim R, Missaoui A (2020) Current progress in nitrogen fixing plants and microbiome research. Plants 9(1):9
Maier RJ, Brill WJ (1978) Mutant strains of Rhizobium japonicum with increased ability to fix nitrogen for soybean. Science 201(4354):448–450
Markmann K, Parniske M (2008) Evolution of root endosymbiosis with bacteria: How novel are nodules? Trends Plant Sci 14:77–86. https://doi.org/10.1016/j.tplants.2008.11.009
Montañez A, Abreu C, Gill PR, Hardarson G, Sicardi M (2009) Biological nitrogen fixation in maize (Zea mays L.) by 15 N isotope-dilution and identification of associated culturable diazotrophs. Biol Fert Soils 45(3):253–263
Murphy P, Wexler W, Grzemski W, Rao J, Gordon D (1995) Rhizopines—Their role in symbiosis and competition. Soil Boil Biochem 27:525–529
Mus F, Crook MB, Garcia K, Costas AG, Geddes BA, Kouri ED, Paramasivan P, Ryu MH, Oldroyd GED, Poole PS et al (2016) Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl Environ Microbiol 82:3698–3710
NCGA (2020) World of corn. National corn growers association. http://www.worldofcorn.com#/. Assessed 15 August 2020
Nunes GHS, Silva PSL, Nunes SGH (1996) Response of maize to nitrogen levels and weeds control. Ciencia-e-Agrotecnologia 20:205–211
O’Hara GW (1998) The role of nitrogen fixation in crop production. J Crop Prod 1:115–138
OECD/FAO (2020) OECD/FAO Agricultural Outlook. https://doi.org/10.1787/agr-outl-data-en. Accessed 20 August 2020
Oger P, Petit A, Dessaux YJ (1997) Genetically engineered plants producing opines alter their biological environment. Nat Biotechnol 15:369
Oldroyd GE (2013) Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11:252
Oldroyd GE, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45:119–144
Oldroyd GED, Dixon R (2014) Biotech solutions to the nitrogen problem. Curr Opin Biotechnol 26:19–24
Ormeno-Orrillo E, Hungria M, Martínez-Romero E (2013) Dinitrogen-fixing prokaryotes. In: Rosenberg EFDL (ed) Prokaryotes: Physiology and Biochemistry. Springer, Berlin, pp 427–451
Ow David W, Gu Q, Xiong Y, Zhu J-B, Shen S-C (1985) Regulation of Klebsiella pneumoniae nitrogen fixation gene promoters by regulatory proteins ntrC, nifA, and nifL. In: Evans HJ, Bottomley PJ, Newton WE (eds) Nitrogen fixation research progress. Springer, Dordrecht, pp 461–467
Pandey A, Sharma E, Palni LMS (1998) Influence of bacterial inoculation on maize in upland farming systems of the Sikkim Himalaya. Soil Biol Biochem 30(3):379–384
Parvez S, Wani S (2007) Prospects of nitrogen fixation in rice. Asian J Plant Sci 6:203–213
Peoples MB, Brockwell J, Herridge DF, Rochester IJ, Alves BJR, Urquiaga S, Boddey RM, Dakora FD, Bhattarai S, Maskey SL et al (2009) The contributions of nitrogen-fixing crop legumes to the productivity of agricultural systems. Symbiosis 48:1–17
Perin L, Martínez-Aguilar L, Castro-González R, Santos PEDL, Cabellos-Avelar T, Guedes HV, Reis VM, Caballero-Mellado J (2006) Diazotrophic Burkholderia species associated with field-grown maize and sugarcane. Appl Environ Microbiol 72(5):3103–3110
Pfeiffer S, Mitter B, Oswald A, Schloter-Hai B, Schloter M, Declerck S et al (2017) Rhizosphere microbiomes of potato cultivated in the High Andes show stable and dynamic core microbiomes with different responses to plant development. FEMS Microbiol Ecol 93:fiw242. https://doi.org/10.1093/femsec/fiw242
Pirttilä AM, Laukkanen H, Pospiech H, Myllyla R, Hohtola A (2000) Detection of intracellular bacteria in the buds of Scotch pine (Pinus sylvestris) by in situ hybridization. Appl Environ Microbiol 66:3073–3077
Plazinski J, Innes RW, Rolfe BG (1985) Expression of Rhizobium trifolii early nodulation genes on maize and rice plants. J Bacteriol 163:812–815
Postgate JR (1982) The Fundamentals of Nitrogen Fixation. Cambridge University Press, New York, NY
Quispel A (1991) Critical evaluation of prospects for nitrogen fixation with non-legumes. Plant Soil 137:1–11
Rai S, Singh DK, Annapurna K (2014) Dynamics of soil diazotrophic community structure, diversity, and functioning during the cropping period of cotton (Gossypiumhirsutum). J Basic Microbiol 55(1):62–73
Raju PN, Harold JE, Ramon JS (1972) An asymbiotic nitrogen-fixing bacterium from the root environment of corn. Proc Natl Acad Sci 69(11):3474–3478
Rakshit S, Chikkappa GK (2018) Perspective of maize scenario in India: way forward. Maize J7(2):49–55
Ramírez-Puebla ST, Ormeño-Orrillo E, Rogel MA, López-Guerrero MG, López-López A, Martínez-Romero J, Negrete-Yankelevich S, Martínez-Romero E (2019) La diversidad de rizobios nativos de México a la luz de la genómica. Rev Mex Biodivers 90:902681
Rascio N, Rocca La N (2013) Biological nitrogen fixation. Reference module in earth systems and environmental sciences. Elsevier, ISBN: 9780124095489
Raymond J, Siefert JL, Staples CR, Blankenship RE (2004) The natural history of nitrogen fixation. Mol Biol Evol 21:541–554
Reddy PM, Altúzar-Molina AR, Ortiz-Berrocal M, Medina-Andrés R, López-Sámano M, Martínez-Aguilar L et al (2013) Predisposition and redesigning of genetic networks of rice for accommodating nitrogen-fixing rhizobial symbiosis. In: Muralidharan K, Siddiq EA (eds) International Dialogue on Perception and Prospects of Designer Rice. Society for Advancement of Rice Research, Hyderabad, pp 245–257
Reis Junior FB, Reis VM, Urquiaga S, Döbereiner J (2000) Influence of nitrogen fertilisation on the population of diazotrophic bacteria Herbaspirillum spp. and Acetobacterdiazotrophicus in sugar cane (Saccharum spp.). Plant Soil 219(1–2):153–159
Rice WA, Clayton GW, Olsen PE, Lupwayi NZ (2000) Rhizobial inoculant formulations and soil pH influence field pea nodulation and nitrogen fixation. Can J Soil Sci 80:395–400
Riggs PJ, Chelius MK, Iniguez AL, Kaeppler SM, Triplett EW (2001) Enhanced maize productivity by inoculation with diazotrophic bacteria. Funct Plant Biol 28(9):829–836
Rogers C, Oldroyd GE (2014) Synthetic biology approaches to engineering the nitrogen symbiosis in cereals. J Exp Bot 65(8):1939–1946
Rosenblueth M, Martínez-Romero E (2006) Bacterial endophytes and their interactions with hosts. Mol Plant-Microbe Interact 19(8):827–837
Rosenblueth M, Martínez-Romero E (2004) Rhizobium etli maize populations and their competitiveness for root colonization. Arch Microbiol 181:337–344. https://doi.org/10.1007/s00203-004-0661-9
Rossbach S, McSpadden B, Kulpa D, Rasul G, Ganoof M, de Bruijn FJ (1994) Use of rhizopine synthesis and catabolism genes to monitor soil bacteria and to create biased rhizospheres. Mol Ecol 3:610–611
Rubio LM, Ludden PW (2008) Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu Rev Microbiol 62:93–111
Sambukumar S, Affeldt K, Friedrich D, Rausch S, Reddy P, James E et al (2015) Exploring the Rhizobial interactions with non-nodulating legume Gleditsia triacanthos. In: 23rd North American conference on symbiotic nitrogen fixation, Ixtapa, Mexico pp 6–10.
Santi C, Bogusz D, Franche C (2013) Biological nitrogen fixation in non-legume plants. Ann Bot 111(5):743–767
Santos MA, Geraldi IO, Garcia AA, Bortolatto N, Schiavon A, Hungria M (2013) Mapping of QTLs associated with biological nitrogen fixation traits in soybean. Hereditas 150(2–3):17–25
Savka MA, Farrand SK (1997) Modification of rhizobacterial populations by engineering bacterium utilization of a novel plant-produced resource. Nat Biotechnol 15:363
Schmid M, Hartmann A (2007) Molecular phylogeny and ecology of root associated diazotrophic and Proteo-bacteria. In: Elmerich C, Newton WE (eds) Associative and endophytic nitrogen-fixing bacteria and cyanobacterial associations. Springer, Dordrecht, pp 41–71
Serraj R, Bona S, Purcell LC, Sinclair TR (2001) Role of soil-oxygen enrichment in the response of nitrogen fixation to drought stress in field-grown soybean. Soil Crop Sci Soc Fla Proc 60:88–93
Shaharoona B, Arshad M, Zahir ZA (2006) Effect of plant growth promoting rhizobacteria containing ACC-deaminase on maize (Zea mays L.) growth under axenic conditions and on nodulation in mung bean (Vigna radiata L.). Lett Appl Microbiol 42(2):155–159
Sharma P, Devi G, Sharma M, Mamrutha HM, Venkatesh K, Tiwari V, Singh GP, Sharma P (2016) Biological nitrogen fixation in cereals: an overview. J Wheat Res 8(2):1–1
Simpson F, Burris R (1984) A nitrogen pressure of 50 atmospheres does not prevent evolution of hydrogen by nitrogenase. Science 224:1095–1097
Sinha Roy S, Mittra B, Sharma S, Das TK, Babu CR (2002) Detection of root mucilage using an anti-fucose antibody. Ann Bot 89(3):293–299
Skorupska A, Janczarek M, Marczak M, Mazur A, Król J (2006) Rhizobial exopolysaccharides: Genetic control and symbiotic functions. Microb Cell Fact 5(7):1–19
Smanski MJ, Bhatia S, Zhao D, Park Y, Woodruff LBA, Giannoukos G, Ciulla C et al (2014) Functional optimization of gene clusters by combinatorial design and assembly. Nat Biotechnol 32(12):1241
Smil V (2001) Enriching the Earth; Massachusetts Institute of Technology: Cambridge. MA, USA, p 2001
Soumare A, Diedhiou AG, Thuita M, Hafidi M, Ouhdouch Y, Gopalakrishnan S, Kouisni L (2020) Exploiting biological nitrogen fixation: A route towards a sustainable agriculture. Plants 9(8):1011
Stacey G, Day RB, Reddy PM, Cohn J, Koh S, Okada M, Ito Y, Shibuya N, Ladha JK (2000) Chitin perception in legumes and rice: What distinguishes a nodulating plant. In: Ladha JK, Reddy PM (eds) The quest for nitrogen fixation in rice. Proceedings of the third working group meeting on assessing opportunities for nitrogen fixation in rice, 9–12 Aug 1999. International Rice Research Institute, Los Baños, Laguna, Philippines. Makati City (Philippines), pp 273–289
Stevenson FJ, Cole MA (1999) Cycles of soils: Carbon, nitrogen, phosphorus, sulfur, micronutrients. Wiley, Hoboken, NJ
Sutton MA, Bleeker A, Howard CM, Bekunda M, Grizzetti B, de Vries W, van Grinsven HJM, Abrol YP, Adhya TK, Billen G, Davidson EA, Datta A, Diaz R, Erisman JW, Liu XJ, Oenema O, Palm C, Raghuram N, Reis S, Scholz RW, Sims T, Westhoek H, Zhang FS, with contributions from Ayyappan S, Bouwman AF, Bustamante M, Fowler D, Galloway JN, Gavito ME, Garnier J, Greenwood S, Hellums DT, Holland M, Hoysall C, Jaramillo VJ, Klimont Z, Ometto JP, Pathak H, Plocq Fichelet V, Powlson D, Ramakrishna K, Roy A, Sanders K, Sharma C, Singh B, Singh U, Yan XY, Zhang Y (2013) Our nutrient world: The challenge to produce more food and energy with less pollution. In: Global overview of nutrient management. Centre for Ecology and Hydrology, Edinburgh on behalf of the Global Partnership on Nutrient Management and the International Nitrogen Initiative
Tapia-Hernandez A, Bustillos-Cristales MR, Jimenez-Salgado T, Caballero-Mellado J, Fuentes-Ramirez LE (2000) Natural endophytic occurrence of Acetobacter diazotrophicus in pineapple plants. Microb Ecol 39(1):49–55
Temme K, Zhao D, Voigt CA (2012) Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca. Proc Natl Acad Sci 109(18):7085–7090
Terakado-Tonooka J, Fujihara S, Ohwaki Y (2013) Possible contribution of Bradyrhizobium on nitrogen fixation in sweet potatoes. Plant Soil 367:639–650
Thomas P, Reddy MK (2013) Microscopic elucidation of abundant endophytic bacteria colonizing the cell wall—plasma membrane perispace in the shoot-tip tissue of banana. AoB PLANTS 5:plt011
Thomas P, Sekhar AC (2014) Live cell imaging reveals extensive intracellular cytoplasmic colonisation of banana by normally non-cultivable endophytic bacteria. AoB PLANTS 6:plu002
Triplett EW (1996) Diazotrophic endophytes: Progress and prospects for nitrogen fixation in monocots. Plant Soil 186(1):29–38
Ueda T, Suga Y, Yahiro N, Matsuguchi T (1995) Remarkable N2 -fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J Bacteriol 177:1414–1417
Unkovich M, Herridge D, Peoples M, Cadisch G, Boddey B, Giller K, Alves B, Chalk P (2008) Measuring plant-associated nitrogen fixation in agricultural systems, ACIAR Monograph No. 136
Van Deynze A, Zamora P, Delaux PM, Heitmann C, Jayaraman DK, Rajasekar S, Graham D et al (2018) Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLoS Bio 16(8):e2006352
Van Velzen R, Holmer R, Bu F, Rutten L, van Zeijl A, Liu W, Santuari L, Cao Q, Sharma T, Shen D et al (2018) Comparative genomics of the nonlegume Parasponiareveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proc Natl Acad Sci USA 115:E4700–E4709
Vance C (2001) Symbiotic nitrogen fixation and phosphorus acquisition. Plant nutrition in a world of declining renewable resources. Plant Physiol 127:391–397
Venturi V, Keel C (2016) Signaling in the rhizosphere. Trends Plant Sci 21:187–198. https://doi.org/10.1016/j.tplants.2016.01.005
Verma DPS, Fortin MG, Stanley J, Mauro VP, Purohit S, Morrison N (1986) Nodulins and nodulin genes of Glycine max. Plant Mol Biol 7(1):51–61
Von Bülow JF, Döbereiner J (1975) Potential for nitrogen fixation in maize genotypes in Brazil. Proc Natl Acad Sci 72(6):2389–2393
Wang X, Yang JG, Chen L, Wang JL, Cheng Q, Dixon R, Wang YP (2013) Using synthetic biology to distinguish and overcome regulatory and functional barriers related to nitrogen fixation. PLoS ONE 8(7):e68677
Wang Y, Janz B, Engedal T, de Neergaard A (2017) Effect of irrigation regimes and nitrogen rates on water use efficiency and nitrogen uptake in maize. Agric Water Manag 179:271–276
Werker E, Kislev M (1978) Mucilage on the root surface and root hairs of Sorghum: heterogeneity in structure, manner of production and site of accumulation. Ann Bot 42(4):809–816
Werner D (1995) Symbiosis of plants and microbes. In: Tikhonovich IA, Provorov NA, Romanov VI, Newton WE (eds) Nitrogen fixation: Fundamentals and applications: proceedings of the 10th international congress on nitrogen fixation. St Petersburg: Springer BV:241
Winogradsky S (1895) Recherche sur assimilation de l’azote libre de l’atmosphere par ls microbes. Arch Sci Biol 3:297–352
Yang J, Xie X, Xiang N, Tian ZX, Dixon R, Wang YP (2018) Polyprotein strategy for stoichiometric assembly of nitrogen fixation components for synthetic biology. Proc Natl Acad Sci 115(36):E8509–E8517
Yang J, Xie X, Yang M, Dixon R, Wang YP (2014) Reconstruction and minimal gene requirements for the alternative iron-only nitrogenase in Escherichia coli. PNAS 111(35):E3718–E3725
Yang J, Xie X, Yang M, Dixon R, Wang YP (2017) Modular electron-transport chains from eukaryotic organelles function to support nitrogenase activity. PNAS 114(12):E2460–E2465
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Rajeev K. Varshney.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Associative
-
Such bacteria are present in close association of plant roots and rarely observed in plant tissues. There is high competition with other soil microbes, so these are present in low density, for example Azospirillum and Azotobacter.
- Biological nitrogen fixation
-
It is a process to convert atmospheric nitrogen (N2) into ammonia by nitrogenase enzyme through a specific group of microorganisms. In this process atmospheric nitrogen (N2) is incorporated into the tissue of certain plants.
- Diazotrophs
-
These are prokaryotic bacteria that are able to fix atmospheric nitrogen (N2) into more usable form for plants such as in ammonia.
- Endophytics
-
These bacteria colonizes intercellular plant roots without inducing any extra structures, but these bacteria do not survive well in soil. Their interactions with plants appear to be less complex and provide more appropriate conditions for effective nitrogen fixation, for example Azoarcus spp., Herbaspirillum seropedicae and Gluconobacter.
- Endosymbionts
-
These are bacteria which reside intracellularly of roots inducing differentiated structure like nodules. These display complex interaction between microbes and host with facilitating the best appropriate conditions for effective nitrogen fixation, for example Rhizobium, Frankia, and Anabaena azollae.
Rights and permissions
About this article
Cite this article
Sheoran, S., Kumar, S., Kumar, P. et al. Nitrogen fixation in maize: breeding opportunities. Theor Appl Genet 134, 1263–1280 (2021). https://doi.org/10.1007/s00122-021-03791-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-03791-5