Abstract
Key message
Hybrid durum has a promising yield potential coupled with good quality, but the efficiency of hybrid seed production must be improved.
Abstract
Hybrid breeding is a tremendous success story in many crops, but has not yet made a breakthrough in wheat, mainly due to inefficient hybrid seed production. In this study, we investigated the heterosis for grain yield and important quality traits in durum wheat of 33 hybrids built up from 24 parental lines, as well as the variation in anther extrusion and its genetic architecture in a vast collection of Central European elite durum lines. Average mid-parent heterosis for grain yield was 5.8%, and the best hybrids had a more than one ton per hectare higher grain yield than the best line cultivars. Furthermore, hybrids had a higher grain yield than lines at a given level of protein content or sedimentation value, underpinning their potential for a sustainable agriculture. However, seed set in our experimental hybrid seed production was low. We therefore evaluated 315 elite durum lines for visual anther extrusion, which revealed a large genetic variance and a heritability of 0.66. Results from association mapping suggest a mainly quantitative inheritance of visual anther extrusion with few putative QTL being identified, the largest one explaining less than 20% of the genotypic variance. Genome-wide prediction taking the four largest putative QTL into account yielded a mean cross-validated prediction ability of 0.55. Consequently, breeding for improved male floral characteristics is feasible in durum wheat, but should be mainly based on phenotypic selection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The introduction of hybrid breeding was a tremendous success story in many crops like maize, rye, or sugar beet (Coors and Pandey 1999; Geiger and Miedaner 1999). In bread wheat (Triticum aestivum ssp. aestivum), big efforts have recently been undertaken to evaluate the feasibility to shift from line to hybrid breeding (Miedaner et al. 2016; Thorwarth et al. 2018; Würschum et al. 2018). The promising heterosis for grain yield and disease resistance as well as a higher than expected grain yield at a given protein content underpin the potential of hybrid wheat to sustainably feed a growing world. For durum wheat (Triticum turgidum ssp. durum), a mid-parent heterosis for grain yield of 10% is reported in the literature, whereas heterosis for quality traits was close to zero or even negative (Gowda et al. 2010). However, to date only very few studies with low numbers of parental lines and hybrids are available, and thus, more research is required on the extent of heterosis in durum wheat.
Despite the promising heterosis in hybrid bread wheat, it has not yet reached a large market share compared with line varieties. This is mainly due to the inefficient hybrid seed production, which costs 2–3 times more than that required for line cultivars. A hybridization system has to be established that on the one hand warrants full sterility of the females in hybrid seed production and on the other hand fully fertile hybrids in farmers’ fields. All bread wheat hybrids in Central Europe are currently produced by a chemical castration method using Croisor®100. Alternatively, cytoplasmatic male sterility (CMS) could be used, but full restoration of fertility in hybrids is only achieved using several restorer genes, thus complicating efficient implementation of this method in wheat (Würschum et al. 2017a). Irrespective of the hybridization system, wheat lines must be identified that release plenty of viable pollen outside the floret to warrant cross-fertilization in hybrid seed production fields. The discovery of male sterility in wheat has offered prospects for the production of hybrid wheat seed on a commercial scale.
In bread wheat, numerous studies have been performed to investigate pollen dispersal outside the floret. It appears to depend on anther extrusion, size of anthers, the opening of the glumes, the awnless of the lemma, the size of the stigma, the duration of the stigma receptivity, the number of pollen grains per anther, the longevity of the pollen grain, and other factors (De Vries 1971; d’Souza 1970; Waines and Hegde 2003; Langer et al. 2014; Boeven et al. 2016; Muqaddasi et al. 2017). However, a recent publication using 31 male lines and two female testers in a crossing block experiment at two locations has shown that visual anther extrusion correlated with r = 0.76 (P < 0.001) with hybrid seed set on the sterilized female (Boeven et al. 2018). Thus, visual anther extrusion can serve as a rapid method to estimate the potential of a line as a male parent in hybrid wheat breeding. A large genetic variation and a high heritability for anther extrusion were shown for bread wheat, but genome-wide association mapping revealed a genetic architecture without large-effect QTL (Langer et al. 2014; Boeven et al. 2016; Muqaddasi et al. 2017). To date, no comparable studies are available for durum wheat.
In this study, we therefore evaluated 315 elite durum breeding lines from Central Europe for visual anther extrusion and genotyped them with genome-wide markers. Furthermore, we produced durum hybrids from six male and 19 female parental lines with the gametocide Croisor®100 and tested the hybrids in multi-location field trials. Our objectives were to (1) evaluate heterosis for grain yield and main quality traits in durum wheat, (2) investigate the genetic variance and heritability of anther extrusion in durum and determine its genetic architecture by association mapping, and (3) draw conclusions for hybrid durum breeding.
Materials and methods
This study is based on two different experiments with durum wheat (Triticum turgidum ssp. durum): The first experiment (Exp. 1) is based on 87 durum wheat breeding lines from the breeding program of the University of Hohenheim, 15 commercial varieties and 33 durum wheat hybrids. Hybrids were produced with the chemical hybridization agent Croisor®100 (Sintofen, Saaten Union Recherche) by using five of the breeding lines as male and nineteen as female parents. The hybrids and all lines were evaluated in yield trials at five locations in Germany and France in the growing season 2016/2017. The locations were Hohenheim (HOH, 48°43′07.3″N, 9°11′08.7″E, altitude 403 m, Germany), Eckartsweier (EWE,48°32′52.4″N, 7°52′32.5″E, altitude 140 m, Germany), Saint Jean d’Angély (SJDA, 45°55′34.5″N 0°14′45.0″W, France), Reuilly (REU, 47° 4′ 18″ N 1° 59′ 45″ E, France), and Semonville (SEM, 48°, 10′25″N, 1°53′56″E, France).
The yield trials were planted as augmented design, where five checks were replicated twice in each trial. The cultivation corresponded to a winter cropping system with sowing in autumn and harvest in the summer of the following year. The sowing density was 200 for hybrids and 380 seeds m−2 for lines, in which hybrids have to compensate by their tillering ability. This sowing density reduction is recommended by the breeding companies and used by bread wheat farmers to account for the considerably higher seed prices of hybrids. Net yield plot size ranged from 5 to 10 m2. All trials were treated with fertilizers (~ 200 kg nitrogen), several applications of fungicides against rusts, mildew, and Fusarium head blight, herbicides, and growth regulators according to farmers’ practice for intensive durum wheat production in the regions of the trial locations. Grain yield in tons per ha was recorded for all the genotypes at all locations, plant height in cm (PH) measured from the ground to the top of five representative ears, and heading time (HT) as the day in the year when 50% of the ears of a plot had fully emerged from the flag leaf were assessed at the two locations HOH and EWE. Six quality traits were assessed on the harvested samples from the field locations HOH, EWE, REU, and SEM. We assessed protein content (%; ICC standard method 159, ICC, Austria), sedimentation value (ml, ICC standard method 151, ICC, Austria), b-value of the semolina through chromameter of Konica Minolta CR-410 L*a*b, hectoliter weight (kg/hl, Marvin Grain Analyzer), and vitreosity (%, ICC standard method 129, ICC, Austria). However, quality was only assessed for the parental lines, hybrids, and a few checks, without evaluating the replicated checks at each individual location twice; thus, the genotype-by-location interaction could not be estimated for quality traits. The parental lines and hybrids as well as their BLUEs are shown in Suppl. Table S1.
In the second experiment (Exp. 2), we evaluated a set of 315 durum wheat lines consisting of 111 spring durum and 204 winter durum lines. These genotypes included elite varieties from Austria, France, Germany, Italy, and Spain, as well as numerous breeding lines from the breeding program of the University of Hohenheim (for details, see Rapp et al. 2018 and Suppl. Table S3). Field experiments were conducted at the location Hohenheim in the season 2014/2015 and 2016/2017 as well as at the location Oberer Lindenhof (OLI, 48°28′19.0″N, 9°18′29.3″E, altitude 700 m, Germany) in the season 2016/2017. All trials were grown in observation plots arranged as a partially replicated design with a replication factor of 1.18 (Williams et al. 2011). The cultivation again corresponded to a winter cropping system with sowing in autumn and harvest in the summer of the following year. All trials were treated with fertilizers (~ 200 kg nitrogen) and herbicides, but without growth regulators and fungicides as commonly performed in observation trials. During flowering stage, visual anther extrusion (VAEX) was assessed by visual scoring on a scale from 1 to 9 (1 = no anthers extruded, 9 = maximum anther extrusion) at all environments (Langer et al. 2014; Boeven et al. 2016). Furthermore, plant height and heading date were assessed as described for Exp. 1. All the lines are available upon request.
Phenotypic data analyses
Best linear unbiased estimators (BLUEs) for Experiment 1 and Experiment 2 were determined using the following mixed model:
where ykmn is the phenotypic observation of the kth genotype in the nth block of the mth location, gk the effect of the kth genotype, lm the effect of the mth location, bnm the effect of the nth block nested within the mth location, and ekmn the residual plot error associated with ykmn. For the calculation of genotype BLUEs, all effects except gk were modeled as random effects. For estimating variance components for Exp. 2, gk was additionally modeled as random effect.
For the estimation of variance components of Exp. 1, we wanted to dissect the genetic variance components into general combining ability (GCA) and specific combining ability (SCA) and we therefore analyzed the phenotypic data with the following model:
where ydijklmn is the phenotypic observation of the kth genotype, of the dth group in the nth block of the mth location, ad is a group effect for lines or hybrids, lm and bnm follow the same notation as above, aldm is the two-way interaction effect between the dth group and the mth location. We introduced dummy variables to estimate variance components for lines (female, males, checks, and additional lines) and hybrids. \( z_{k}^{L} \) is the dummy variable of the kth genotype in the group of lines, with \( z_{k}^{L} = 1 \) for checks and lines and \( z_{k}^{L} = 0 \) for hybrids. The coding of the dummy variable for hybrids is \( z_{k}^{H} = 1 \) for hybrids and \( z_{k}^{L} = 0 \) for checks and lines. With these dummy variables, the variance components can be estimated separately for parental lines and hybrids. pk is the effect of the kth line, \( g_{i}^{{\prime }} \) is the GCA effect of the ith female parental line, \( g_{j}^{{{\prime \prime }}} \) is the GCA effect of the jth male parental line, sij is the SCA effect of crosses between lines i and j, (pl)km is the two-way interaction effect between kth parental line and the mth location, \( \left( {g^{\prime } l} \right)_{im} \) and \( \left( {g^{\prime \prime } l} \right)_{jm} \) are the GCA-by-location interaction effects of female and male parental lines and εdijklmn the residual effect. As we had only 33 hybrids built up from 19 females and five males and as in durum no preliminary information existed for grouping lines into a male and female group, we calculated a joint GCA effect for parental lines. To obtain this combined GCA effect for parental lines as well as the GCA-by-location interaction effect, we used the and() function of asreml-r to overlay design matrices of \( g_{i}^{{\prime }} \) and \( g_{j}^{{{\prime \prime }}} \) as well as for \( \left( {g^{\prime } l} \right)_{im} \) and \( \left( {g^{\prime \prime } l} \right)_{jm} \). For more details, the reader is referred to following link: https://github.com/PaulSchmidtGit/Heritability/.
The group effect was modeled as fixed, all other effects were considered as random, and the error variance was modeled as heterogeneous for each location-by-group combination. Broad-sense heritability was calculated separately for lines and hybrids using group specific variance components following Piepho and Möhring (2007, formula 19):
where ϑ is the mean variance of a difference of two adjusted treatment means and \( \sigma_{\text{G}}^{2} \) the genetic variance.
For each combination of parental lines (Px), mid-parent performance (MP), mid-parent heterosis (MPH), and better-parent heterosis (BPH) were calculated. All analyses were performed with the statistical software R (R Core Team 2016) and the software ASReml 3.0 (Gilmour et al. 2009).
Genotypic data analyses
For Exp. 2, all lines were genotyped at Diversity Arrays Technology (Yarralumla, Australia) using genotyping by sequencing (DArTseq), which generates dominant silico-DArTs and codominant single-nucleotide polymorphism (SNP) markers. In the following, the notation for each marker is a combination of its Clone ID and the marker-type prefix “D” or “S” for silico-DArTs or SNP markers, respectively. We applied the following quality control criteria: Markers with more than 20% missing values as well as markers with a minor allele frequency (MAF) < 5% were removed from further analysis. After these quality checks, a total of 20,276 markers were available with 4.85% missing values. A low heterozygosity for SNP markers was observed, and thus, no SNP markers had to be removed due to heterozygosity. An imputation step was performed using the software package LD-kNNi (Money et al. 2015; Boeven et al. 2016) which yielded an imputation accuracy of 0.97. After imputation, markers with a minor allele frequency of < 5% were again removed and 12,591 markers remained that had passed the quality checks and had a known map position. These markers are shown in Suppl. Table S4.
Association mapping
We used a mixed model framework for genome-wide association mapping. Imputed marker data were tested for allelic association with genotypic trait values (BLUEs) using the “polygenic” function within the R package GenABEL (Aulchenko et al. 2007) incorporating a genome-wide kinship matrix (K matrix) to correct for familial relatedness (Yu et al. 2006). The K matrix was estimated by the “ibs” (option weight = “no”) function as implemented in GenABEL. Additionally, we performed the genome-wide association mapping but correcting for population structure by a PK model, i.e., by incorporating principal coordinates in addition to the kinship matrix. This yielded almost identical results (Suppl. Fig. S5) and was therefore disregarded from further analyses and discussion.
The marker effects were tested for significance by extracting the P values from the test for marker–trait association with one degree of freedom as implemented in the “mmscore” function in GenABEL. To account for multiple testing, we used a Bonferroni-corrected threshold of P < 0.05 (− log10(P) > 5.4).
In addition, we report putative QTL identified at an exploratory significance threshold for marker–trait associations of P < 0.001, which is comparable to a false discovery rate (FDR) threshold of 0.2.
The total proportion of explained genotypic variance (pG) was estimated by fitting all significant QTL in a linear model in the order of ascending P values to obtain \( R_{\text{adj}}^{2} \). The ratio \( pG = R_{\text{adj}}^{2} /h^{2} \) yielded the proportion of explained genotypic variance (Utz et al. 2000). The pG values of individual QTL were accordingly derived from the sums of squares of the QTL in this linear model (Würschum et al. 2015). We transferred the genetic map position of identified putative QTL to a physical map position by utilizing the clone sequences of the corresponding markers in a BLAST approach. We used the online tool BLASTn of the CNR InterOmics project with the reference genome of the wild emmer wheat accession “Zavitan” (Avni et al. 2017; CNR InterOmics 2017). The physical map position was assigned to a putative QTL, when the genetic and physical map positions were on the same chromosome and the identity of clone sequence and reference genome was ≥ 95%. The linkage disequilibrium (D′, R2, and P value) between markers of putative QTL on the same chromosomes was tested with TASSEL (Bradbury et al. 2007).
Genomic prediction
Genomic prediction was performed by ridge regression BLUP (RR-BLUP) (Meuwissen et al. 2001; Whittaker et al. 2000) with the R package “rrBLUP” (Endelman 2011; Endelman and Jannink 2012). In addition, we performed a weighted ridge regression best linear unbiased prediction (wRR-BLUP) by including the detected QTL from the genome-wide association mapping, which explained more than 5% of the genotypic variance, as fixed effects in the genomic prediction model (Zhao et al. 2014; Spindel et al. 2016; Boeven et al. 2016; Würschum et al. 2017b). For the two models, we employed fivefold cross-validation with random sampling and 1000 runs (Würschum et al. 2013, 2014). In each run, we randomly sampled 80% of the lines as training set to predict the remaining 20%. The prediction ability was then estimated as Pearson’s correlation coefficient between the observed and the predicted trait values. This procedure was repeated 1000 times.
Results
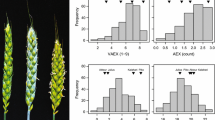
In a first experiment, we evaluated hybrids, their parental lines, and leading breeding lines of durum for agronomic and quality traits in multiple field tests across Germany and France. We observed a wide range of phenotypic values for all traits, yielding highly significant genetic variances and heritabilities in the expected range for the respective traits (Fig. 1a; Table 1, Suppl. Table S1). The hybrids had a considerably higher grain yield than their parental lines (Fig. 1a; Suppl. Table S1), resulting in on average positive values for mid-parent and better-parent heterosis. Furthermore, the best performing hybrid had a grain yield level of 9.6 tons per hectare as compared to the popular check variety “Miradoux” with only 8.5 tons per ha. For all quality traits, heterosis was close to zero or even negative. However, many of the hybrids with high grain yield had also a quality comparable to that of the high-quality commercial line variety “Miradoux” (Suppl. Table S1). Estimates for the variance due to GCA were almost all nonsignificant, and high amounts of variance due to SCA were identified for grain yield, protein content, and sedimentation volume. Genetic variances of parental lines were considerably larger for all traits than the variance due to GCA of the parents, and error variances of parental lines and hybrids were at a similar level. However, these estimates are based on only few parental lines and must therefore be taken with caution. We further investigated the genetic distance of the parental lines and its correlation with mid- and better-parent heterosis. For almost all traits, these correlations were low and partly with different signs for the different traits (Table 1).
In a second experiment, we investigated 315 durum lines on visual anther extrusion and observed a wide range of phenotypic values, following roughly a normal distribution and resulting in a highly significant genotypic variance (Fig. 1b; Table 2, Suppl. Table S3). A significant variance due to genotype-by-environment interaction and a high residual error, however, led to an only moderate heritability estimate of 0.66. The first two coordinates of a principal coordinate analysis of the 315 durum lines evaluated for visual anther extrusion explained only 14% and 4%, respectively (Fig. 2a). Nevertheless, the 111 spring durum lines seem to cluster slightly apart from the remaining winter durum lines as indicated also by the neighbor-joining tree (Fig. 2b). The durum lines with the highest visual anther extrusion were widely distributed across the neighbor-joining tree.
a Principal coordinate analysis of the 315 genotypes of Exp. 2 based on their molecular marker profiles. Percentages in parentheses refer to the proportion of variance explained by the first and second principal coordinate. b Neighbor-joining tree based on distance matrix. The winter durum lines are colored in red, and the spring durum lines are colored in green. The best 10 genotypes for visual anther extrusion (VAEX) are indicated in blue and the male lines of Exp. 1 in black (“Biensur” was a male line in Exp. 1 and belonged to the 10% best lines for VAEX in Exp. 2; for details, see Suppl. Table S3) (color figure online)
The genome-wide association mapping in Exp. 2 based on the exploratory significance threshold of P < 0.001 identified nine significant putative QTL for visual anther extrusion that jointly explained 61.9% of the genotypic variance (Figs. 3, 4; Table 3). The strongest putative QTL was identified on chromosome 7A, explaining almost 20% of the genotypic variance of visual anther extrusion. This putative QTL was also significant at the more stringent Bonferroni-corrected significance threshold. A second large putative QTL was identified on chromosome 3A, explaining 12% of the genotypic variance. As our durum lines were fixed for the dwarfing allele Rht-B1b, we could not investigate the influence of this dwarfing allele on visual anther extrusion (Boeven et al. 2016). We took the four putative QTL that explained more than 5% of the genotypic variance (7A_D1280335, 3A_D1093595, 2A_S2276567, 3A_S1250769) for a more detailed investigation. Interestingly, the majority of durum lines was already fixed for the positive allele at these putative QTL (Fig. 4a, b).
Manhattan plot of genome-wide association analysis for visual anther extrusion (VAEX). The lower dashed horizontal line corresponds to the exploratory significance threshold of P < 0.001, while the upper dashed line corresponds to the Bonferroni-corrected significance threshold of P < 0.05 (− log10(P) > 5.4)
a Boxplots showing the visual anther extrusion (VAEX) for genotypes carrying different alleles of the detected QTL explaining more than 5% of the genotypic variance (WD = winter durum, SD = spring durum). The numbers underneath the boxplots indicate the number of genotypes in the respective classes. b Boxplots showing genotypes carrying different allelic combinations of the three largest effect QTL
We further used these four putative QTL to simulate the potential of a marker-assisted prediction of visual anther extrusion. The mean prediction ability over 1000 runs was 0.52 by using only these four putative QTL (Fig. 5, MAS). In comparison, the standard ridge regression BLUP approach using all 12,591 markers yielded a lower cross-validated prediction ability with an average of 0.44 (Fig. 5, RR-BLUP). Combining the genome-wide markers with the four putative QTL in a weighted ridge regression BLUP approach yielded the highest prediction ability with an average of 0.55 (Fig. 5, wRR-BLUP).
Prediction ability for visual anther extrusion based on QTL explaining more than 5% of the genotypic variance (MAS), genome-wide prediction with ridge regression BLUP (RR-BLUP), or weighted ridged regression BLUP (wRR-BLUP) including the same QTL as for MAS as fixed effects in the model. Results are shown for fivefold cross-validation repeated 1.000 times
Discussion
Big efforts have recently been undertaken to investigate the potential of hybrid breeding in bread wheat (Triticum aestivum ssp. Aestivum). This revealed a substantial heterosis for grain yield (Longin et al. 2012, 2013; Corbellini et al. 2002; Perenzin et al. 1998; Borghi and Perenzin 1994), but hybrid seed production remains a major problem to be solved in this autogamous crop (De Vries 1971; d’Souza 1970; Waines and Hegde 2003; Langer et al. 2014; Boeven et al. 2016; Muqaddasi et al. 2017; Boeven et al. 2018). However, consequent selection on anther extrusion holds the potential to largely improve cross-pollination ability and therefore hybrid seed production in bread wheat (Muqaddasi et al. 2017; Boeven et al. 2016, 2018). This motivated us to investigate the potential and the feasibility of hybrid breeding in durum wheat. We therefore produced durum hybrids and evaluated them for their hybrid performance and their heterosis of important agronomic and quality traits (Exp. 1). In addition, we evaluated the available variation and the genetic architecture of anther extrusion in a vast collection of durum lines (Exp. 2).
Promising grain yield heterosis in durum wheat hybrids
The average mid-parent heterosis amounted to almost 6% for grain yield (Table 1), which was slightly less than the 10% reported previously for bread wheat or for durum wheat (Gowda et al. 2010; Longin et al. 2013). Nevertheless, the range of mid- and better-parent heterosis was large which might be explained by the use of parental lines, which have been selected due to floral traits and importance as line cultivar rather than on GCA. Thus, highest values of mid- and better-parent heterosis were similar to those reported for bread and durum wheat. Furthermore, the best hybrids had a more than one ton per hectare higher grain yield than important commercial checks like “Miradoux” and “Anvergur,” illustrating the high yield potential and the commercial competitiveness of durum hybrids (Suppl. Table S1). Interestingly, the hybrids with the highest yield were a combination of a winter durum line as male parent and a spring durum line as female parent. According to the neighbor-joining tree analysis in our durum wheat collection, spring and winter durum lines clustered quite separately (Fig. 2b). This might suggest that the combination of spring and winter type durum is a possible heterotic pattern, which, however, requires further validation with larger sets of hybrids and their evaluation across a range of target environments.
In accordance with results from bread wheat and a previous study on durum wheat hybrids (Gowda et al. 2010), negative values were found for mid- and better-parent heterosis of protein content and sedimentation value (Table 1). This is, however, not surprising when taking the negative correlation of grain yield and protein content into account. Thorwarth et al. (2018) and Rapp et al. (2018) therefore suggested to compare the yield potential of wheat lines and hybrids at the same level of protein content or sedimentation value. The authors clearly showed that wheat hybrids had a considerably higher grain yield at a given level of protein content or sedimentation value than wheat lines. In line with these findings, we found that durum hybrids also had a considerably higher yield at a given level of protein content or sedimentation value (Suppl. Table S1), underlining the potential of durum hybrids for a sustainable wheat production. For other important quality traits in durum wheat, like b-value, vitreosity, and hectoliter weight, we also observed heterosis values close to zero (Table 1). However, several hybrids with high grain yield had also a very good quality as compared to the high-quality reference commercial variety “Miradoux” and were considerably better than the commercial variety “Nobilis” (Suppl. Table S1). Consequently, durum hybrids combine a higher grain yield with good to very good quality and are therefore interesting for durum wheat production worldwide.
Hybrid seed production in durum
We used the chemical hybridization agent Croisor®100 to castrate the females in crossing blocks for experimental hybrid seed production. This gametocide is approved for commercial hybrid seed production in bread and durum wheat, and currently all bread wheat hybrids in Central Europe are produced with this gametocide. The castration with Croisor®100 was acceptable with almost 100% sterility in all 19 female parents used in our study. Small phytotoxic reactions were noted similar to those occurring in bread wheat, with slightly injured leaves and decoloration of leaves for some days after application. For 33 hybrid combinations, enough seeds for yield trials at five locations could be produced and we harvested up to 2 kg of good hybrid seed from a 6-m2 plot of a castrated female (Suppl. Table S2). This appears low, especially when compared to a normal seed yield of about 4 kg of a line in a plot of 6 m2. However, for hybrid seed production in bread wheat we harvested in a similar design up to 3 kg of hybrid seeds. We speculate that an optimization of the application protocol of Croisor®100 to durum wheat might improve hybrid seed yield in durum, but it will certainly remain considerably lower than that of pure lines.
Variation in cross-pollination ability in durum wheat
A major requirement for improved hybrid seed production is the identification of male lines with a high pollen shedding potential (De Vries 1971; d’Souza 1970; Waines and Hegde 2003; Langer et al. 2014; Boeven et al. 2016). In bread wheat, a recent publication using 31 male lines and two female testers in a crossing block experiment at two locations has shown that visual anther extrusion correlated with r = 0.76 (P < 0.001) with hybrid seed set on the sterilized female (Boeven et al. 2018). Thus, visual anther extrusion can serve as a rapid method to estimate the potential of a line as a male in hybrid wheat breeding. We therefore investigated visual anther extrusion in a large durum wheat collection comprising elite varieties and breeding lines from Austria, Canada, France, Germany, Hungary, Italy, and Spain (Suppl. Table S3). We observed a large and significant genetic variance for visual anther extrusion in this durum panel, thus enabling the selection of lines suited for the improvement in floral characteristics required for hybrid seed production (Table 2, Suppl. Table S3; Fig. 1b). However, the correlation between the BLUEs of the different locations was only around 0.5 resulting in a significant variance due to genotype-by-environment interaction and only a moderately high heritability of 0.66 (Table 2). This is considerably lower than recently reported for bread wheat (Langer et al. 2014; Boeven et al. 2016). Furthermore, the person who scored this experiment in durum also scored the trials for bread wheat, which were next to the durum trial. It was thus clearly visible that the highest anther extrusion in durum wheat was lower than the highest anther extrusion in bread wheat.
Nevertheless, across the 19 females we consistently achieved the highest hybrid seed set for the two winter durum lines “Sambadur” and “6.026/05/03” (data not shown). Both lines showed a good anther extrusion across the three locations with an average value close to 7 (Fig. 1b; Suppl. Table S3). Screening the large durum panel now identified other durum lines with an even higher visual anther extrusion than these two already acceptable male lines (Table 1, Suppl. Table S3; Fig. 1b). Additionally, these lines with highest visual anther extrusion were genetically quite different as visualized by their distribution across the neighbor-joining tree (Fig. 2b). Thus, we speculate that using durum lines with high anther extrusion as starting point for male lines and consequent selection for anther extrusion with optimization of the nicking of male and female lines and the gametozide could improve hybrid seed production of durum wheat in the future.
Genetic architecture of anther extrusion in durum wheat
Our genome-wide association mapping identified only four putative QTL, which explained more than 5% of the genotypic variance of visual anther extrusion (Fig. 3; Table 3). This is in agreement with studies from bread wheat, where despite high heritability estimates also only very few QTL with limited influence on the genetic variance were identified (Boeven et al. 2016; Muqaddasi et al. 2017).
Except for the putative QTL on chromosome 3A (S1250769), the majority of durum lines already carried the positive alleles at the other three major QTL identified in our study (Fig. 4). In particular, 167 of the 315 durum lines already carried three positive QTL alleles, with most of them belonging to the winter durum group. This is also reflected by the higher average of visual anther extrusion for winter than for spring durum (Fig. 4a) and underlines that the winter durum genotypes might be interesting as male heterotic group. In contrast, most spring durum varieties from the major durum production countries Italy, Spain, and France carried a maximum of two positive QTL alleles for visual anther extrusion.
Few of our putative QTL appeared to be at the same chromosomal regions as those QTL identified in bread wheat (Boeven et al. 2016). We therefore compared their physical positions in the bread wheat reference genome (IWGSC et al. 2018). For the three putative QTL in durum wheat (7A_D1280335, 3A_S06580, and 2B_D1133616) for which QTL were also reported on the same chromosomes in bread wheat, the physical distance between them was rather large, indicating that these markers identify different loci (Suppl. Table S6).
Combining the four putative major QTL identified in our study to predict anther extrusion via marker-assisted selection led to a cross-validated prediction ability of 0.52 (Fig. 5). In contrast, the genome-wide prediction with RR-BLUP delivered a slightly lower prediction ability, which is in line with findings from bread wheat (Boeven et al. 2016). Combining the major QTL with the genome-wide approach by a weighted RR-BLUP prediction led to the best prediction ability for visual anther extrusion of 0.55. However, this requires a genome-wide genotyping, while genotyping with only four markers tagging the QTL would cost just a fraction of that. Nevertheless, before they are utilized in breeding programs, these QTL for visual anther extrusion will need to be validated. Moreover, as many durum lines appear to be fixed already for the positive alleles at these QTL, further improvement in these lines can only be achieved by the genome-wide approach capturing additional small-effect QTL or by phenotypic selection.
Conclusions
Hybrid durum showed a promising yield potential coupled with good quality and a higher protein content at a given yield level than line varieties, thus making it appealing for a sustainable agriculture. As durum wheat is mainly produced in stressful environments (especially heat and drought), the improved yield stability of hybrids reported from other crops might also be of high interest, but warrants further research. However, the main bottleneck for successful hybrid durum breeding is an efficient hybrid seed production. We could show that the production of hybrid seed is feasible with the gametocide Croisor®100, which is commonly used in bread wheat, but improvements in its application protocol are required to reduce the seed yield gap to normal line varieties. In addition, durum lines with very good pollen shedding potential have to be identified as male lines. Visual anther extrusion appears as a rapid and robust method to identify good male lines, and we observed a large variation for this trait in elite durum that can be exploited for hybrid breeding. Our results on the genetic architecture of anther extrusion in durum point toward a quantitative nature of the trait with only few major- or moderate-effect QTL. Similar to bread wheat, we therefore recommend the following strategy to breed for good male lines: first, screening on visual anther extrusion in a large panel, then a more intensive evaluation of the best lines in multiple locations, and finally testing of the best males in crossing blocks to evaluate their seed set on female lines. The hybrid seed set then serves as the final selection criterion for the male lines and can additionally be used for a first hybrid yield test of the new male lines crossed with elite female testers to estimate their GCA. Collectively, our results show that hybrid durum wheat offers some advantages, but open questions remain on the economics of seed production and the future will show if hybrid durum can find its niche in the global cereal market.
Author contribution statement
CFHL designed the study and performed field trials; WA, PT, and MR performed all statistical analyses; WA, CFHL, and TW wrote the manuscript. All authors read and reviewed the manuscript.
References
Aulchenko YS, Ripke S, Isaacs A, van Duijn CM (2007) GenABEL: an R library for genome-wide association analysis. Bioinformatics 23:1294–1296
Avni R, Nave M, Barad O et al (2017) Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357:93–97
Boeven PHG, Longin CFH, Leiser WL, Kollers S, Ebmeyer E, Würschum T (2016) Genetic architecture of male floral traits required for hybrid wheat breeding. Theor Appl Genet 129:2343–2357
Boeven PHG, Würschum T, Julia R, Erhard E, Longin CFH (2018) Hybrid seed set in wheat is a complex trait but can be improved indirectly by selection for male floral traits. Euphytica 214:110
Borghi B, Perenzin M (1994) Diallel analysis to predict heterosis and combining ability for grain yield, yield components and bread-making quality in bread wheat (T. aestivum). Theor Appl Genet 89:975–981
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
CNR InterOmics (2017) CNR InterOmics Consortium. https://www.interomics.eu/wild-emmer-wheat-genome. Accessed 26 Oct 2017
Coors JG, Pandey S (1999) The genetics and exploitation of heterosis in crops. ASA, CSSA, and SSSA, Madison
Corbellini M, Perenzin M, Accerbi M, Vaccino P, Borghi B (2002) Genetic diversity in bread wheat, as revealed by coefficient of parentage and molecular markers, and its relationship to hybrid performance. Euphytica 123:273–285
d’Souza L (1970) Untersuchungen über die Eignung des Weizens als Pollenspender bei der Fremdbefruchtung, verglichen mit Roggen, Triticale und Secalutricum. Z Pflanzenzucht 63:246–269 (in German)
De Vries AP (1971) Flowering biology of wheat, particularly in view of hybrid seed production—a review. Euphytica 20:152–170
Endelman JB (2011) Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 4:250–255
Endelman JB, Jannink J-L (2012) Shrinkage estimation of the realized relationship matrix. G3 Genes Genomes. Genet 2:1405–1413
Geiger HH, Miedaner T (1999) Hybrid rye and heterosis. In: Coors JG, Pandey S (eds) Genetics and exploitation of heterosis in crops. Crop Science Society of America, Madison, WI, pp 439–450
Gilmour AR, Gogel BG, Cullis BR, Thompson R (2009) ASReml user guide release 3.0. VSN International Ltd, Hertfordshire. http://www.vsni.co.uk
Gowda M, Kling CK, Würschum T, Liu W, Maurer HP, Hahn V, Reif JC (2010) Hybrid breeding in durum wheat: heterosis and combining ability. Crop Sci 50:2224–2230
IWGSC et al (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191
Langer SM, Longin CFH, Würschum T (2014) Phenotypic evaluation of floral and flowering traits with relevance for hybrid breeding in wheat (Triticum aestivum L.). Plant Breed 133:433–441
Longin CFH, Mühleisen J, Maurer HP, Zhang H, Gowda M, Reif JC (2012) Hybrid breeding in autogamous cereals. Theor Appl Genet 125:1087–1096
Longin CFH, Gowda M, Mühleisen J, Ebmeyer E, Kazman E, Schachschneider R, Schacht J, Kirchhoff M, Zhao Y, Reif JC (2013) Hybrid wheat: quantitative genetic parameters and consequences for the design of breeding programs. Theor Appl Genet 126:2791–2801
Meuwissen T, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Miedaner T, Schulthess AW, Gowda M, Reif JC, Longin CFH (2016) High accuracy of predicting hybrid performance of Fusarium head blight resistance by mid-parent values in wheat. Theor Appl Genet 130:461–470
Money D, Gardner K, Migicovsky Z, Schwaninger H, Zhong G, Myles S (2015) LinkImpute: fast and accurate genotype imputation for non-model organisms. G3 Genes Genomes. Genet 5:2383–2390
Muqaddasi QH, Brassac J, Börner A, Pillen K, Röder MS (2017) Genetic architecture of anther extrusion in spring and winter wheat. Front Plant Sci 8:754
Perenzin M, Corbellini M, Accerbi M, Vaccion P, Borghi B (1998) Bread wheat: F1 hybrid performance and parental diversity estimates using molecular markers. Euphytica 100:273–279
Piepho H-P, Möhring J (2007) Computing heritability and selection response from unbalanced plant breeding trials. Genetics 177:1881–1888
R Core Team (2016) R: a language and environment for statistical computing. http://www.R-project.org/
Rapp M, Lein V, Lacourdre F, Lafferty J, Müller E, Vida G, Bozhanova V, Ibraliu A, Thorwarth P, Piepho HP, Leiser WL, Würschum T, Longin CFH (2018) Simultaneous improvement of grain yield and protein content in durum wheat by different phenotypic indices and genomic selection. Theor Appl Genet 131:1315–1329
Spindel JE, Begum H, Akdemir D, Collard B, Redoña E, Jannink J, McCouch S (2016) Genome-wide prediction models that incorporate de novo GWAS are a powerful new tool for tropical rice improvement. Heredity 116:395–408
Thorwarth P, Piepho HP, Zhao Y, Ebmeyer E, Schacht J, Schachschneider R, Kazman E, Reif JC, Longin CFH (2018) Higher gain yield and higher grain protein deviation underline the potential of hybrid wheat for a sustainable agriculture. Plant Breed 137:326–337
Utz HF, Melchinger AE, Schön CC (2000) Bias and sampling error of the estimated proportion of genotypic variance explained by quantitative trait loci determined from experimental data in maize using cross validation and validation with independent samples. Genetics 154:1839–1849
Waines JG, Hegde SG (2003) Intraspecific gene flow in bread wheat as affected by reproductive biology and pollination ecology of wheat flowers. Crop Sci 43:451–463
Whittaker JC, Thompson R, Denham MC (2000) Marker-assisted selection using ridge regression. Genet Res 75:249–252
Williams E, Piepho HP, Whitaker D (2011) Augmented p-rep designs. Biom J 53:19–27
Würschum T, Reif JC, Kraft T, Janssen G, Zhao Y (2013) Genomic selection in sugar beet breeding populations. BMC Genet 14:85
Würschum T, Abel S, Zhao Y (2014) Potential of genomic selection in rapeseed (Brassica napus L.) breeding. Plant Breed 133:45–51
Würschum T, Langer SM, Longin CFH (2015) Genetic control of plant height in European winter wheat cultivars. Theor Appl Genet 128:865–874
Würschum T, Leiser WL, Weissmann S, Maurer HP (2017a) Genetic architecture of male fertility restoration of Triticum timopheevii cytoplasm and fine-mapping of the major restorer locus Rf3 on chromosome 1B. Theor Appl Genet 130:1253–1266
Würschum T, Leiser WL, Longin CFH (2017b) Molecular genetic characterization and association mapping in spelt wheat. Plant Breed 136:214–223
Würschum T, Liu G, Boeven PHG, Longin CFH, Mirdita V, Kazman E, Zhao Y, Reif JC (2018) Exploiting the Rht portfolio for hybrid wheat breeding. Theor Appl Genet 131:1433–1442
Yu J, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Zhao Y, Mette MF, Gowda M, Longin CFH, Reif JC (2014) Bridging the gap between marker-assisted and genomic selection of heading time and plant height in hybrid wheat. Heredity 112:638–645
Acknowledgements
W. Akel was financially supported by the Deutsche Forschungsgemeinschaft (DFG LO 1816/4-1). We thank the three anonymous reviewer, whose input considerably improved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
The authors declare that the experiments comply with the current laws of Germany.
Additional information
Communicated by Heiko C. Becker.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Akel, W., Rapp, M., Thorwarth, P. et al. Hybrid durum wheat: heterosis of grain yield and quality traits and genetic architecture of anther extrusion. Theor Appl Genet 132, 921–932 (2019). https://doi.org/10.1007/s00122-018-3248-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-018-3248-6