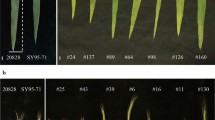
Abstract
Key message
QTLs and candidate gene markers associated with leaf morphological and color traits were identified in two immortalized populations of Brassica rapa, which will provide genetic information for marker-assisted breeding.
Abstract
Brassica rapa is an important leafy vegetable consumed worldwide and morphology is a key character for its breeding. To enhance genetic control, quantitative trait loci (QTLs) for leaf color and plant architecture were identified using two immortalized populations with replications of 2 and 4 years. Overall, 158 and 80 QTLs associated with 23 and 14 traits were detected in the DH and RIL populations, respectively. Among them, 23 common robust-QTLs belonging to 12 traits were detected in common loci over the replications. Through comparative analysis, five crucifer genetic blocks corresponding to morphology trait (R, J&U, F and E) and color trait (F, E) were identified in three major linkage groups (A2, A3 and A7). These might be key conserved genomic regions involved with the respective traits. Through synteny analysis with Arabidopsis, 64 candidate genes involved in chlorophyll biosynthesis, cell proliferation and elongation were co-localized within QTL intervals. Among them, SCO3, ABI3, FLU, HCF153, HEMB1, CAB3 were mapped within QTLs for leaf color; and CYCD3;1, CYCB2;4, AN3, ULT1 and ANT were co-localized in QTL regions for leaf size. These robust QTLs and their candidate genes provide useful information for further research into leaf architecture with crop breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chinese cabbage (Brassica rapa L. ssp. pekinensis; 2n = 20) is an inexpensive and nutritious vegetable that is consumed worldwide, especially in Asia. The leaves of B. rapa have diverse colors and sizes and come in various shapes. The greenish leaves are the main edible parts. However, in recent years, varieties with orange, yellow and purple leaves have become highly marketable for their appearance and health benefits. Leaf morphological features are important traits because consumers notice them. Therefore, many studies have attempted to identify genomic regions that influence leaf morphological traits for breeding (Kennard et al. 1994; Mocoeur et al. 2015; Veldboom et al. 1994).
Genetic information from genetic mapping and quantitative trait loci (QTLs) approaches provides clues for understanding complex traits. A precondition for QTL mapping studies is the availability of genetic markers. In recent decades, various molecular markers have been developed for B. rapa, such as amplified fragment length polymorphisms (AFLPs) (Choi et al. 2007; Panigrahi et al. 2009), simple sequence repeats (SSRs) (Kapoor et al. 2009; Kim et al. 2009; Wang et al. 2011) and single-nucleotide polymorphisms (SNPs) (Chung et al. 2014; Li et al. 2009). Subsequently, QTLs for traits controlling flowering time (Li et al. 2009; Lou et al. 2007), head traits (Yu et al. 2013), root morphology (Lu et al. 2008), seed color traits (Kebede et al. 2012) and leaf morphology (Li et al. 2013; Lu et al. 2008) have been mapped and the associated regions in the genome were reported. In addition, reference genome sequencing of the B. rapa has been done by next-generation sequencing and opened to the public (Wang et al. 2011). High-throughput sequencing and genotyping technology have permitted researchers to determine the co-location of candidate genes through QTLs for various traits (Azmach et al. 2013; Bajaj et al. 2015; Chung et al. 2014; Poczai et al. 2013). Therefore, development of candidate gene-based markers associated with different traits, which can be helpful for breeders and geneticists to understand the genetic mechanism of various complex phenotypes and accelerate the progress of molecular breeding (Kim et al. 2014).
Most quantitative genetic analyses of crop plants have used segregating populations, such as F2 or backcross populations. Those populations have the benefit of easy and fast construction; however, they have limited usefulness for analyzing complex agronomic traits because of the lack of genetically uniform individuals. There might be chance to overlook detection of QTL in a single investigation. Thus, immortal populations are appropriate for QTL studies because they reduce the need to estimate ambiguous phenotypes from heterozygotes and environmental effects.
Doubled haploid (DH) and recombinant inbred line (RIL) mapping populations could compensate for the lack of segregating populations for the following reasons: (i) each line of the two population types is stable and can provide a consistent genetic background; (ii) the low heritability of the populations means that they can be better used for morphological traits or quantitative traits; and (iii) tightly linked markers that are closely associated with those traits can be identified or used to accumulate additional markers. Therefore, linkage maps based on DH or RIL populations are sustained for genetic studies, especially for QTL analysis.
Many studies have considered the leaf morphology of B. rapa (Li et al. 2013; Lou et al. 2007; Lu et al. 2008; Zhao et al. 2008); however, most of the QTL results were estimated using non-immortal populations with restricted field trials. Further, information on candidate genes involved in the traits has been limited to qualitative characteristics such as leaf hair and seed coat color (Zhang et al. 2009a). In this study, we report the generation of an RIL population derived from a bi-parental cross. One of the parents is ‘Chiifu’, which was used for reference genome sequencing as a representative of the A genome. We also used 2998 SSRs designed from Chinese cabbage bacterial artificial chromosome (BAC)-end sequencing to construct the genetic map and analyze the QTLs.
Major factors involved in leaf morphology, such as leaf size, shape and number of leaves, are important to the study of plant architecture and breeding. Thirty-two traits were investigated in two different immortal populations, RIL and DH, and the linkage maps and QTLs were compared between these two permanent populations. Furthermore, using synteny analysis with Arabidopsis, candidate genes known to have an influence on leaf architecture were mapped. These results provide valuable information for identifying genomic regions corresponding to the morphological traits.
Materials and methods
Plant materials
Two diverse Chinese cabbage inbred lines, ‘Chiifu-402-42’ and ‘Kenshin-40-43’, used to construct the different mapping populations in this study. One was a DH population derived from an another culture of the F1 generation, termed CKDH (Choi et al. 2007; Kim et al. 2009; Li et al. 2010; Ramchiary et al. 2011). The other was an RIL population termed CKRI. CKRI consisted of 130 lines, 8 generation from the F2 via bud pollination.
‘Chiifu-402-42’ is an elite inbred line that has been adopted widely in South Korea and its genome was sequenced as a reference genome of B. rapa (Wang et al. 2011). ‘Kenshin’ is an inbred line from Taiwan. Both parents are characteristic leafy vegetables that can distinguish visually. Chiifu typically has denser foliage than Kenshin. Their heading habits at the mature stage are also different: Chiifu heads curl inwards, whereas Kenshin forms overlapping leaves. Chiifu is a late maturing type, showing late bolting, leaf hairs. Kenshin is an early maturing type, showing early bolting, no leaf hair. Plant genomic DNA was isolated from fresh leaf material according to the procedure used by Guillemaut and Maréchal-Drouard (1992).
Marker development and genetic mapping
Some of the SSRs reported in Kim et al. (2009) were used in this study to construct the genetic map frame of the RIL population. These marker sets were used as anchor markers to compare between previous map and current map, constructed with RIL population. To saturate the map, we designed new SSRs from 200,017 BAC-end sequences that were available in the GenBank database (https://www.ncbi.nlm.nih.gov/). The methods and parameters used to identify SSRs, and the primers designed followed those of Kim et al. (2009). The locus nomenclature followed that used in previous reports based on Choi et al. (2007). New markers were named with the prefix BRPGM (Brassica rapa pekinensis genomic microsatellite). Markers that were reproducibly polymorphic between the parent lines were genotyped in the RIL population. Linkage analysis and map construction were performed using JoinMap version 4.0 (Stam 1993; Van Ooijen 2011). Linked loci were grouped using a logarithm of odds (LOD) grouping threshold range of 2.3–4.1, and linkage groups (LGs) were assigned to 10 LGs corresponding to the first reported map of this species (Choi et al. 2007). Locus order within the LOD grouping were generated for each LG using a recombination frequency below 0.45 and an LOD score above 0.5 for all marker pairs within each LG. A “ripple” procedure was performed after the addition of each marker, and the “jump” thresholds were set to five. Recombination frequencies were converted to centiMorgans (cM) using Kosambi’s method for map distance calculation (Kosambi 1943). The genetic map was drawn using MapChart (Voorrips 2002).
Trait and phenotype investigation
Parent plants and the CKDH populations containing 78 lines were evaluated in two environments: they were planted in districts of the Anseong area in autumn during two years (1998, 1999), and in the Daejeon area during 4 years (2000, 2001, 2002, 2004). Each field experiment was arranged in a randomized block design, with 10–12 plants replicated annually. Twenty-three morphological traits related to four categories were selected based on the ‘descriptors for Brassica and Raphanus’ of the International Board for Plant Genetic Resources and investigated over 2–6 years; these included plant, leaf, and stem colors, plant weight, and all trait characteristics, abbreviations and description were list in Table 1. To allow for easier comparison with previous results, we used the same abbreviations for the traits as those used in previous studies. For example, the trait ID such as ‘LN’, ‘LL’, ‘LW’, ‘PL’ were used by Li et al. (2009, 2013), Lou et al. (2007). One hundred and thirty RIL populations (CKRI) and parent plants were grown in the Daejeon area in autumn of 2006 with three replicates. Each field experiment followed a randomized block design. A total of 14 overlapping traits were evaluated using the methods described above, except for leaf color, which was measured by SPAD instead of visually.
Statistical analysis
Statistical analysis conducted using SPSS program 22 (SPSS, Inc., USA). Pearson correlation coefficient analysis conducted in each phenotypical data. Broad-sense heritability was performed with the lmer function in the lmer4 package by R software (R Development Core Team, 2013; version 3.1.3; https://www.r-project.org/), calculated according to the formula H 2 = σ 2g /(σ 2g + σ 2e /n), where σ 2g is the variance of the genotype and σ 2e is the variance of the error term, n is the number of environment.
QTL analysis and comparative map
Analysis of variance was conducted for the replicated field trials of CKDH and CKRI populations using windows QTL Cartographer v. 2.0 (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm). QTLs were detected using the composite interval mapping (CIM) procedure in WinQTL Cartographer v. 2.0 (Zeng 1994). To identify significant QTL, the LOD thresholds were determined by permutation analysis with 1000 repetitions in trait, respectively (Churchill and Doerge 1994).
For the QTL analysis in CKDH populations, we selected 207 markers from 719 markers as a set, based on genetic distance (1 marker every 5 cM). We selected the QTLs based on adjusted LOD values and a comparison of the QTL regions within the annual data and in different population types for all traits.
To compare the genetic map and QTLs, the markers that were used in both maps were set as anchor points. The QTL results were also compared with those of previous reports such as Lou et al. (2007), Li et al. (2009, 2010), Ge et al. (2012) and Yu et al. (2013) to identify further conserved QTLs in the Brassica genome. All sequence information, such as marker primer sequences, PCR amplicons and BAC-end sequences were aligned with the A. thaliana genome using the BLASTN program of the NCBI (http://www.ncbi.nlm.nih.gov) to identify homologous chromosomal regions between the A. thaliana and B. rapa genomes, and to allow us to use model plant information for genome comparison. Based on the homology results, the syntenic regions of B. rapa corresponding to the Arabidopsis genome were identified as crucifer genomic blocks, following the method of Schranz et al. (2006).
Identification of candidate genes and SNP validation
To identify the related candidate/orthologous genes in the QTLs region, we first searched the gene list contained in the syntenic blocks in the Arabidopsis genome in the TAIR database (http://www.arabidopsis.org). We selected the genes involved in the trait based on the functional annotation of the genes. Second, we searched the lists for orthologous genes in the BRAD database (http://brassicadb.org/brad). Third, we selected orthologous genes that were located in a QTL region to design primers as target genes of the trait. Lasergene software (DNASTAR, Madison, USA) was used for DNA sequence analysis to detect SNPs or InDel by sequence alignment. Then, a 96 well Light Scanner® System (Idaho Technology Inc.) was used to genotype for SNP validation and the associations between QTLs and candidate genes. Finally, only the markers that were mapped within QTL intervals were regarded as a candidate genes associated with the traits. Candidate gene-target primers for SNP validation was designed with an amplicon size between 100 to 200 bp by primer3 software. PCR amplication was performed with a volume of 10 µl containing 4 ng of genomic DNA as template, 0.3 U of Taq polymerase (Intron Bio, Daejeon, Korea), 2 pmol of each primers, 1× PCR buffer, 250 µM dNTP, 2 mM MgCl, and 0.5× LC Green Plus as a dye (Idaho Technology Inc.). The PCR conditions was initiated with a 4 min at 94 °C for denaturation step, followed by 45 cycles of 15 s at 94 °C, 20 s at 55–60 °C, and 20 s at 72 °C, with a final extension step of 7 min at 72 °C. Melting temperature detection of PCR amplicon was carried by Light Scanner software (Idaho Technology Inc.), followed the protocols described in De Koeyer et al. (2010). There are several advantages of the high resolution melting (HRM) technique over normal gel electrophoresis. First, HRM is able to detect sequence variation, amplicon length variation and SNPs via the melting temperature. Second, HRM analysis is performed by scanning a PCR amplicon directly after the PCR, without further handling, such as amplicon fractionation. Thus, in this study, the results of 96 assays were generated within 15–20 min to score the SNPs, and the genotype data were recorded automatically in the machine.
Results
Identification of SSR motifs in the BAC-end sequences of B. rapa
A total of 200,017 BAC-end sequences of B. rapa were downloaded from the GenBank nucleotide database. The sequences were checked for sequence quality and duplication, and low-quality regions were removed using SeqClean software. The remaining 177,823 BAC-end sequences were searched for repeat motifs. A total of 30,008 SSRs were identified, among which (A)n and (T)n mono-nucleotide repeats were the most common (33.62%), followed by tri-nucleotide (32.95%) and di-nucleotide (29.44%) repeats (Supplementary Table 1). Among the repeats, (A)n was the most abundant overall, (AT)n and (AG)n were the most abundant di-nucleotide repeats, and n was the most abundant tri-nucleotide motif (at frequencies of 31.89, 13.99 and 12.85%, and 10.82%, respectively). Primers were designed from 2206 repeat motifs, excluding the mono-nucleotide repeats. All primer sequences and BAC clone IDs used as sequence sources are shown in Supplementary Table 2.
Construction of a genetic linkage map using the CKRI population
Of the 2206 primer pairs designed from the BAC-end sequence, 1618 primer pairs were synthesized to survey polymorphisms between the parental lines. The PCR fragment amplification was successful for 1554 primer pairs (96%), and 1111 primer pairs amplified a single fragment (68.7%). Polymorphic bands were generated by 375 primer pairs (23.1%), with a single amplification being generated by 303 primer pairs (Table 2). SSRs that showed clear amplification products (359) were scored in the RIL population. The RIL population was generated from F1 hybrid between the two inbred lines, Chiifu and Kenshin, by self-crossing nine times. Finally, 332 SSR markers were assigned to 10 LGs (Table 3; Fig. 1), corresponding to the LGs of the BRGP reference map (Choi et al. 2007). The total length of the genetic map was 985.5 cM, and the average distance between two loci was 3 cM. The largest LG was A1, which spanned 121.3 cM. The smallest was A10, which spanned 62 cM. The number of markers assigned to the LGs varied from 65 (A3) to 11 (A4). Two gaps of greater than 20 cM were present in A5 and A8. The characteristic of this map is that all the markers were from BAC clone sequence; therefore, the markers mapped on the genetic map might cover a wide genomic region in contrast to short sequence-based markers, such as expressed sequence tags and SNPs.
Brassica rapa genetic linkage map and QTLs for morphological traits discovered in doubled haploid lines (DH), and recombinant inbred lines (RIL) population. Genetic distances (cM) are shown to the left side of linkage group as a scale. The linkage groups of different maps are aligned based on common markers. The common markers between two maps connected with black lines. The abbreviations of traits described in Table 1. Rectangular bars indicate QTLs those found out controlling the morphological traits in multiple experiments. QTL regions discovered over two times were emblemized rectangular shadow. QTLs conserved between homologous linkage groups represented by red color character. Syntenic regions of Arabidopsis thaliana shown beside of linkage group with different color rectangle and the crucifer building blocks are recording. The numbers to the side of colored bars indicated aligned positions on the Arabidopsis chromosomes in megabase pairs (Mb). Candidate genes identified in common QTLs are presented beside the linkage group
QTL analysis in CKRI/CKDH population
Fourteen traits were investigated in the CKRI population with three replications in 2006, and 23 traits were investigated in 78 lines of the CKDH population. The frequency of investigation was two to six replications per year, depending on plant growth conditions. All traits were grouped into four categories: color, leaf morphology, plant weight and stem (Table 1).
Statistics of the mean value of morphological characteristics measured in parental lines and populations and broad sense heritability (H 2) summarized in Supplementary Table 3. Phenotypic variation represented transgressive segregations in the population in most traits. The heritability of each trait ranged from 0.06 to 0.89 in DH, and 0.04–0.89 in RIL population. Leaf blade length, leaf blade width, total no. of leaves traits had high heritability, ranged from 0.74–0.89, in both populations. The heritability showed tendency higher in DH population than RIL population.
In the 10 LGs, 80 and 158 QTLs were detected in the RIL and DH populations, respectively (Supplementary Tables 4, 5; Fig. 1). To identify whether the same or different QTL positions were detected in the two populations, the genetic maps were compared using the common markers as anchors.
Leaf color
Thirty-six QTLs were detected in the 10 LGs in the DH population for four color-related traits (CL, CIH, CP and COH) and six QTLs were detected for four LGs (A3, A4, A7 and A9) with two traits (CL, CIH) in the RIL population. The variation explained by each QTL ranged from 7.96 to 16.85% for the RIL population and from 4.20 to 35.40% for the DH population. Two major QTLs for the leaf color trait were detected in both RIL (qCL1/3, qCL2/2) and DH (qCL2/00, qCL7/00, qCL8/01) populations. This QTL co-located with marker cnu_m482 on the lower middle of A3 (F block). In the DH population the petiole color trait (qCP7/00) explained the largest amount of variation (72.60%); however, there was no reproducible result in this QTL region.
Leaf morphology related traits
Sixteen and nine leaf morphological traits, including leaf size, numbers, shapes and leaf hair, were investigated in different growing years. One hundred and twelve, and fifty tree QTLs were detected in the DH and RIL populations, respectively. In the RIL population, the explained variation per QTL was generally lower (3.97–25.16%) than in the DH population (6.40–33.60%). QTLs associated with leaf hair were detected consistently in all 6 years. A high percentage of phenotypic variance (14.50–81.40%) was explained by ‘qLT_13’ on A06 in the DH population. A minor region related to this trait was detected on A9, which explained 22.6% of the phenotypic variations. This QTL was investigated only in the DH population and detected in all growing conditions. One common QTL, which was conserved between homologous LGs in both populations, was detected in five traits (LNib, PLc, PW, LBS, and PT), and distributed on LGs A2 (qLNi4, qPL13), A4 (qPW15), and A7 (qLBS12, qPT14), respectively. Two common QTLs, associated with traits LNc, were detected on LGs A2 (qLN5), and A9 (qLN6). Three common QTLs were detected for trait LNob (qLNo1, qLNo2, qLNo3 on LGs A2, A5, A6), LLc (qLL7, qLL8, qLL9 on LGs A3, A7), LWc (qLW10, qLW11, qLW12 on LGs A3, A7) (Table 4; Supplementary Tables 4, 5). QTL regions that were detected more than twice were labeled with a rectangular shadow, and QTL names that were conserved between homologous LGs are indicated in red in Fig. 1. Additionally, there was a strong overlap on the top region (R block) of LG A02 for traits LNob, LNib, LNc, LWc and PLc, which were detected in both populations. The common markers ‘cnu_mBBSRC143’ and ‘cnu_mBRPGM112’ were included in this overlap region. Another co-location region was in the bottom of the LG A7 (E block), which was associated with traits LLc, LWc, LBS, PT and SL. Eight common SSR markers were co-mapped in this interval.
Whole plant weight
Five and ten QTLs associated with GW were located on four LGs (DH: A1, A3, A6 and A7) and on six LG (RIL: A1, A2, A3, A4, A7, A9), and individual variance ranged from 11.9 to 49.2% (DH), and from 7.8 to 11.6% (RIL). QTLs involved in plant weight located on LGs A1, A3 and A7 were detected consistently in both populations and in different years for a single population. Major QTLs ‘qGW2/98’ in DH and ‘qGW1/3’ in RIL were located in LG A1, explaining 49.2% of the phenotypic variation.
Stem trait
Two stem traits were used for QTL analysis: stem length (SL) and leaf diameter (LD). Two and eight QTLs were identified for SL in DH and RIL, respectively, and one common locus in A07 was detected in the E block. Two QTLs of SD trait were identified in each DH and RIL, but no common regions were detected.
Discussion
Genome coverage and robustness of marker order
A new genetic map of RIL populations was constructed using 332 SSR markers derived from a BAC clone sequence and compared with the previous genetic map generated using a DH population (Kim et al. 2009). The grouping of markers based on similarity of recombination values was exactly matched in the results of both populations. A few markers on LGs A3 and A5 showed inverted marker orders. Generally, most of the marker orders were collinear in both maps. In terms of marker number, the RIL map had fewer markers than the DH map, and the total map length was relatively shorter than the previous map. However, the previous map was constructed using non-sequence informative markers (31%), which are less useful for further applications, such as AFLPs and random amplified polymorphic DNA (RAPD). By contrast, the present map was constructed using SSRs exclusively, which were anchored on BAC clones and had long sequence fragments. Additionally, the lengths of the linkage group were enlarged at the top (A06, A07, A09) and at the bottom (A02, A3, A05, A08, A10) by inserting 1–11 markers. A03 gained most new markers at both ends by inserting 9 and 11 markers, respectively. Considering genetic loci and physical distance covers almost 80.1% (227.87 Mb) of the reference genome. The linkage group that showed the largest expansion of genome coverage was A02, increasing from 8.24 Mb (29.6%) to 24.16 Mb (86.7%). Based on the physical length of B. rapa, 529 Mb, 1 cM was estimated to be around 537 kb.
QTLs for morphological traits identified in this study were located in the same intervals
As a leafy vegetable, plant morphology in terms of leaf color, leaf shape, leaf number and individual lamina size are important in Chinese cabbage breeding to achieve more attractive plant architecture according to customers’ demands. Chiifu is a more leafy type than Kenshin in terms of the number of leaves, plant size, and hairiness. Plants show considerable diversity in size and shape; therefore, there have been many studies that attempted to understand plant architecture. However, plant architecture was determined by complex genetic regulation and interactions with the environment.
In this study, we used three to six replications of the experiments in different seasons and locations. We believe that this number of replications permitted accurate estimations of the genetic component of variance represented by the QTLs and allowed identification of the interactions between genes and the environment. In addition, we used two immortal populations, DH and RIL, to reduce the number of false-positive QTLs, which can result from heterozygotes and interactions between genes and the environment. DH populations are also easier to use in genetic mapping studies than segregating populations because they are derived from single meiotic events, with conserved gene combinations, and enhanced identification of genetic factors controlling quantitative traits (Burroughs et al. 1988; Kole et al. 1997). By comparing two different populations with many replications (over several years), consistent QTLs were detected that are highly likely to be related to leaf morphology in B. rapa. Twenty-two key genomic regions represented by robust QTLs controlling plant architecture were identified as consensus intervals on eight LGs that affected three categories (color, leaf architecture, and plant weight) in this study (Table 4).
Three major QTLs (qCL1, qCL2, qCIH1) which associated with leaf color trait were detected in both populations (Table 4; Fig. 2; Supplementary Tables 4, 5). Moreover, these QTLs co-located with same markers cnu_m482, cnu_m477 near the middle of LG A03 (F block), marker cnu_m553 on the bottom of LG A07 (E block), and marker cnu_m377 on LG A09 (I block).
Amplicon analysis of mapping population by HRM using two representative SNP markers designed from candidate genes, Bra016270 located in A07 and Bra024219 located in A03. Plot a represents the curve-shifted plot, and b represents a difference plot showing ‘Chiifu’ type in red color, ‘Kenshin’ type in grey color
Chlorophyll content is associated with crop yield (Rawson and Constable 1980) and biomass production (Wang et al. 2008), and is a multi-allelic system in sunflower, rice, barley and wheat (Guo et al. 2008; Zhang et al. 2009b). Ge et al. (2012) reported eight chlorophyll-associated QTLs distributed on seven LGs in B. rapa. Among them, four QTLs located on LG A3 (qClaA3b), A7 (qClbA7) and A9 (qClaA9a, qClaA9b) were detected in same genomic regions as our robust QTLs (qCL1, qCL2 and qCIH1) (Supplementary Tables 4, 5).
QTLs involved in number of outer leaves (qLNo1, qLNo2, qLNo3), number of inner leaves (qLNi4), and total number of leaves (qLN5, qLN6) were detected in three (A02, A05 and A06), one (A02) and two (A02, A09) LGs in both populations. Common QTLs for leaf number were detected on LG A2, which belongs to block R, and were consistent with those reported in previous studies in B. rapa (Li et al. 2010; Lou et al. 2007) and R. sativus (Yu et al. 2016). By contrast, the QTLs for leaf number on LG A05, belonging to block F, were newly identified in this study. This indicated that these QTLs might be a potentially new set of alleles representing the diversity of this genetic background in Chinese cabbage (Fig. 3).
Comparative QTL mapping between A. thalina and crop Brassicacea species revealed conservation of crucifer building blocks governing morphological and color traits. QTLs discovered in identical block shown inside of dotted line box. Common robust-QTLs, conserved DH and RIL population, were represented outside of dotted line box. QTL regions detected over two times were emblemized rectangular shadow. QTLs name that conserved between homologous linkage groups are indicated by red color character. QTLs results were compared in other previous results from Lou et al. (2007), Ge et al. (2012), and Yu et al. (2016)
For two traits involved with leaf size, QTLs for leaf blade length (qLL7, qLL8 and qLL9) and leaf blade width (qLW10, qLW11 and qLW12), detected in same genomic region of two LGs (A03, A07). Lou et al. (2007) reported QTLs controlling leaf size, such as leaf length and leaf width in seven LG. These results indicated that the leaf traits, breeders focus on, tend to be inherited quantitatively and are controlled by many genes and not single ones. Furthermore, phenotypic values show continuous distributions in nature, resulting from the combined allelic effects of many genes and interactions of the genotype with the environment. This suggests that many genes encode proteins that affect a trait; however, the effects of individual genes can be hard to distinguish. To solve this type of problem, we performed yearly replications and determined major genomic regions controlling phenotype in A03 and A07, respectively.
In this study, robust QTLs (qLW10, qLW11, and qLW12) for leaf width were detected with the common markers cnu_m241, cnu_m384, and cnu_m310 on LGs A3 (U, J), and A07 (E). Key genomic regions, controlling leaf length and width traits, was completely co-localized at the both ends of LG A3 in the two different populations. However, the Arabidopsis blocks containing the QTL region were different: U and J on A3. This difference of genetic block might suggest that leaf size controlled by more than one factor. QTLs affecting of all four categories were detected at the bottom of LG A7 with moderate to strong positive correlations among traits by Pearson’s correlation coefficient analysis (Supplementary Table 6). Interestingly, the leaf width trait represented a highly significant positive correlation with the leaf length trait based on Pearson correlation coefficient analysis in DH (r = 0.795–0.982), and RI (r = 0.718-0.931). This coincided exactly with a previous report by Li et al. (2010), and was detected partially by Lou et al. in B. rapa (2007) and by Yu et al. in R. sativus (2016), respectively. The other three robust QTLs (qLW10, qLW11, qLW12) co-segregated with ‘qLL8’, ‘qLL7’ and ‘qLL9’ on LGs A3 (U, J blocks) and A7 (E) in this study. Lou et al. (2007) reported QTLs that matched exactly in the same regions, respectively. This consistency in the QTL regions identified by different studies indicates that the locations of alleles for the morphological traits are conserved in different genetic and environmental backgrounds.
Three robust-QTLs (qLL8, qLW11, qLW12) were detected within several replications, as well as in different populations. These regions corresponded to crucifer genome blocks U, J and E. We hypothesized that these regions might contain the genes that control these traits. Gene specific primers were designed using Brassica orthologous genes and confirmed the co-segregation on the map. ‘qLW12’ on LG A7 was considered to be closely associated with leaf size trait even though no sequence variation was observed among the orthologous genes between the parents. Thus, we assume that either this gene effect is too weak to be detected or epigenomic effects play a role.
Functional markers associated with QTLs and marker-assisted selection
The parent line Chiifu was used as the reference genome sequence of B. rapa (A genome, The Brassica database: http://brassicadb.org/brad), which helped us to identify trait related genes and provided evidence for the gene function study. To develop gene-base markers for marker-assisted breeding, 35 functional genes (such as chloroplast morphology, chlorophyll biosynthesis genes and cell proliferation and elongation) were selected from Arabidopsis, which were located on QTL intervals and searched for orthologous genes in BRAD DB. Then we did comparison between the reference genome and the Kenshin genome to identify these candidate genes harboring SNP mutations. In total, 64 gene-based markers were developed in B. rapa corresponding to 35 Arabidopsis genes, with 1–3 copies in different genome segments, respectively (Table 5). The co-localization between mapped QTLs and candidate gene-base markers for morphological traits in B. rapa were validated through HRM approach. The genotyping scanning analysis of mapping population for two representative SNP markers were shown in Fig. 2. To further understands, the molecular mechanism for plant size and chlorophyll biosynthesis, twelve candidate genes involving leaf color and size were co-located in six robust QTLs. For leaf color, Bra-SCO3 was found to co-localize with the peaks of the QTL ‘qCL1/02’ in CKDH. The mutation of snowy cotyledon3 (sco3) in Arabidopsis impaired chloroplast and etioplast development in Arabidopsis seedlings (Albrecht et al. 2010). The gene Br-ABI3, which is positioned on chromosome 3, is located in the peaks of the QTL ‘qCL2/00’ in CKDH and ‘qCL1/3’ in CKRI, which interacts genetically with both FUS3 and LEC1 and is involved in controlling accumulation of chlorophyll and anthocyanins (Dekkers et al. 2016). HEMB1 encodes a putative 5-aminolevulinate dehydratase that is involved in chlorophyll biosynthesis, and is required for chlorophyll biosynthesis and plant growth (Wang et al. 2016), a homolog of Bra-HEMB1 was mapped in the clustered QTL (robust QTL, ‘qCL_3’) for ‘qCL7/00’ and ‘qCL8/01’ in CKDH and ‘qCL/2’ in CKRI of chromosome 7.
Many studies have attempted to understand leaf development and morphogenesis based from a genetic or epigenetic standpoint, and A. thaliana leaves are known to grow along three directions—that of the dorsiventral axis, proximodistal axis, and mediolateral axis (Barkoulas et al. 2007; Izhaki and Bowman 2007; Li et al. 2005; Shi et al. 2007; Siegfried et al. 1999). For leaf blade size, which was investigated by leaf length and width, cell proliferation and cell expansion are the basic factors that directly influence leaf growth processes.
ROUNIFOLIA3 (ROT3) and CYCD3;1 were found to co-localize with the peaks of the robust QTL ‘qLL_7’ (qLL2/00, qLL3/01 in CKDH, qLL3/3, qLL5/01 in CKRIL) on LGA3, According to Kim et al. (1998), ROT3 regulates longitudinal leaf growth in Arabidopsis. Mutant rot3 has a phenotype with reduced lengthwise cell expansion, round leaf blades and short petioles. Overexpression of ROT3 resulted in increased organ length, with no alteration in width (Kim et al. 1999). CYCD3;1, which regulates cell division in leaf expansion and promotes the cell cycle, encodes a cyclin D-type protein involved in the switch from cell proliferation to the final stages of differentiation. The gene is regulated transcriptionally by cytokinins and brassinosteroids (Dewitte et al. 2003; Shen et al. 2013). CYCD1 contains AS2 domain, is involved in correct abaxial/adaxial patterning (Horiguchi et al. 2005, 2011). Blast analysis identified a homolog in B. rapa, Bra007901. CYCD3;3 is important for determining cell number in developing lateral organs, cause of mediating cytokinin effects in apical growth and development (Dewitte et al. 2007; Mehrnia et al. 2013).
The robust QTL ‘qLW10’ region (qLW5/00 in DH, qLW4/1 in RIL) contained the candidate genes AINTEGUMENTA (ANT) and ULT1 on LG A03. ANT is required to control cell proliferation and encodes a putative transcriptional regulator similar to AP2. The mutant ant showed reduced leaf size, while overexpression of ANT increased the size of organs by augmenting cell numbers and cell proliferation, causing an increased period of cell growth and longer proliferation than normal (Mizukami and Fischer 2000). The Br-AN3 gene is positioned on chromosome 2 and located in the peaks of the QTL ‘qLW2/01’ in CKDH and ‘qLW1/1’ in CKRI. ANGUSTIFOLIA 3 (AN3) has prominent roles in regulating cell divisions in the leaf primordia and interacts with members of the growth-regulating factor family of transcription factors.
In conclusion, we detected numerous QTLs for various morphological traits using 3–6 year replications. The genomic regions controlling the morphological traits were confirmed at several time points and in different populations. To explain the QTLs, we identified the candidate genes related to the function of the trait and verified co-segregation based on their physical position. This robust QTL information will assist with further research on leaf morphology in Chinese cabbage. This study revealed robust QTLs and their candidate genes related to leaf morphology in Chinese cabbage. These findings could stimulate further research on leaf architecture in crops breeding.
Author contribution statement
SRC and XY designed the study, carried out the marker development, analyzed and interpreted all data, and drafted the manuscript. VD carried out primer designing, and the comparative anlaysis, and drafted the manuscript. XL performed the candidate gene identification, ZW, SYL, SHO, and WP were participated in marker survey and genotyping and genetic mapping. NR, CPH, SP carried out the SSR analysis and the comparative analysis of traits. ZP and HRK participated in its designing and coordination of this study. YPL conceived the study, participated as a director, and helped to draft the manuscript. All authors read and approved the final manuscript.
References
Albrecht V, Simkova K, Carrie C, Delannoy E, Giraud E, Whelan J, Small ID, Apel K, Badger MR, Pogson BJ (2010) The cytoskeleton and the peroxisomal-targeted snowy cotyledon3 protein are required for chloroplast development in Arabidopsis. Plant Cell 22:3423–3438
Azmach G, Gedil M, Menkir A, Spillane C (2013) Marker-trait association analysis of functional gene markers for provitamin A levels across diverse tropical yellow maize inbred lines. BMC Plant Biol 13:227
Bajaj D, Upadhyaya HD, Khan Y, Das S, Badoni S, Shree T, Kumar V, Tripathi S, Gowda CL, Singh S, Sharma S, Tyagi AK, Chattopdhyay D, Parida SK (2015) A combinatorial approach of comprehensive QTL-based comparative genome mapping and transcript profiling identified a seed weight-regulating candidate gene in chickpea. Sci Rep 5:9264
Barkoulas M, Galinha C, Grigg SP, Tsiantis M (2007) From genes to shape: regulatory interactions in leaf development. Curr Opin Plant Biol 10:660–666
Burroughs SF, Devine DV, Browne G, Kaplan ME (1988) The population of paroxysmal nocturnal hemoglobinuria neutrophils deficient in decay-accelerating factor is also deficient in alkaline phosphatase. Blood 71:1086–1089
Choi SR, Teakle GR, Plaha P, Kim JH, Allender CJ, Beynon E, Piao ZY, Soengas P, Han TH, King GJ, Barker GC, Hand P, Lydiate DJ, Batley J, Edwards D, Koo DH, Bang JW, Park BS, Lim YP (2007) The reference genetic linkage map for the multinational Brassica rapa genome sequencing project. Theor Appl Genet 115:777–792
Chung H, Jeong YM, Mun JH, Lee SS, Chung WH, Yu HJ (2014) Construction of a genetic map based on high-throughput SNP genotyping and genetic mapping of a TuMV resistance locus in Brassica rapa. Mol Genet Genomics 289:149–160
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
De Koeyer D, Douglass K, Murphy A, Whitney S, Nolan L, Song Y, De Jong W (2010) Application of high-resolution DNA melting for genotyping and variant scanning of diploid and autotetraploid potato. Mol Breed 25:67–90
Dekkers BJ, He H, Hanson J, Willems LA, Jamar DC, Cueff G, Rajjou L, Hilhorst HW, Bentsink L (2016) The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J 85(4):451–465
Dewitte W, Riou-Khamlichi C, Scofield S, Healy JM, Jacqmard A, Kilby NJ, Murray JA (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15:79–92
Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V, Murray JA (2007) Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci 104:14537–14542
Ge Y, Wang T, Wang N, Wang Z, Liang C, Ramchiary N, Choi SR, Lim YP, Piao ZY (2012) Genetic mapping and localization of quantitative trait loci for chlorophyll content in Chinese cabbage (Brassica rapa ssp. pekinensis). Sci Hortic 147:42–48
Guillemaut P, Maréchal-Drouard L (1992) Isolation of plant DNA: a fast, inexpensive and reliable method. Plant Mol Biol Rep 10:60–65
Guo PG, Baum M, Varshney RK, Graner A, Grando S, Ceccarelli S (2008) QTLs for chlorophyll and chlorophyll fluorescence parameters in barley under post-flowering drought. Euphytica 163:203–214
Horiguchi G, Kim GT, Tsukaya H (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43:68–78
Horiguchi G, Nakayama H, Ishikawa N, Kubo M, Demura T, Fukuda H, Tsukaya H (2011) ANGUSTIFOLIA3 plays roles in adaxial/abaxial patterning and growth in leaf morphogenesis. Plant Cell Physiol 52:112–124
Izhaki A, Bowman JL (2007) KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19:495–508
Kapoor R, Banga SS, Banga SK (2009) A microsatellite (SSR) based linkage map of Brassica rapa. New Biotechnol 26:239–243
Kebede B, Cheema K, Greenshields DL, Li C, Selvaraj G, Rahman H (2012) Construction of genetic linkage map and mapping of QTL for seed color in Brassica rapa. Genome 55:813–823
Kennard WC, Slocum MK, Figdore SS, Osborn TC (1994) Genetic analysis of morphological variation in Brassica oleracea using molecular markers. Theor Appl Genet 87:721–732
Kim GT, Tsukaya H, Uchimiya H (1998) The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev 12:2381–2391
Kim GT, Tsukaya H, Saito Y, Uchimiya H (1999) Changes in the shapes of leaves and flowers upon overexpression of cytochrome P450 in Arabidopsis. Proc Natl Acad Sci 96:9433–9437
Kim H, Choi SR, Bae J, Hong CP, Lee SY, Hossain MJ, Van Nguyen D, Jin M, Park BS, Bang JW, Bancroft I, Lim YP (2009) Sequenced BAC anchored reference genetic map that reconciles the ten individual chromosomes of Brassica rapa. BMC Genomics 10:432
Kim SM, Suh JP, Lee CK, Lee JH, Kim YG, Jena KK (2014) QTL mapping and development of candidate gene-derived DNA markers associated with seedling cold tolerance in rice (Oryza sativa L.). Mol Gen Genet 289:333–343
Kole C, Kole P, Vogelzang R, Osborn TC (1997) Genetic linkage map of a Brassica rapa recombinant inbred population. J Hered 88:553–557
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Li H, Xu L, Wang H, Yuan Z, Cao X, Yang Z, Zhang D, Xu Y, Huang H (2005) The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. Plant Cell 17:2157–2171
Li F, Kitashiba H, Inaba K, Nishio T (2009) A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res 16:311–323
Li X, Ramchiary N, Choi SR, Van Nguyen D, Hossain MJ, Yang HK, Lim YP (2010) Development of a high density integrated reference genetic linkage map for the multinational Brassica rapa Genome Sequencing Project. Genome 53:939–947
Li X, Ramchiary N, Dhandapani V, Choi SR, Hur Y, Nou IS, Yoon MK, Lim YP (2013) Quantitative trait loci mapping in Brassica rapa revealed the structural and functional conservation of genetic loci governing morphological and yield component traits in the A, B, and C subgenomes of Brassica species. DNA Res 20:1–16
Lou P, Zhao J, Kim JS, Shen S, Del Carpio DP, Song X, Jin M, Vreugdenhil D, Wang X, Koornneef M, Bonnema G (2007) Quantitative trait loci for flowering time and morphological traits in multiple populations of Brassica rapa. J Exp Bot 58:4005–4016
Lu G, Cao J, Yu X, Xiang X, Chen H (2008) Mapping QTLs for root morphological traits in Brassica rapa L. based on AFLP and RAPD markers. J Appl Genet 49:23–31
Mehrnia M, Balazadeh S, Zanor MI, Mueller-Roeber B (2013) EBE, an AP2/ERF transcription factor highly expressed in proliferating cells, affects shoot architecture in Arabidopsis. Plant Physiol 162:842–857
Mizukami Y, Fischer RL (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci 97:942–947
Mocoeur A, Zhang YM, Liu ZQ, Shen X, Zhang LM, Rasmussen SK, Jing HC (2015) Stability and genetic control of morphological, biomass and biofuel traits under temperate maritime and continental conditions in sweet sorghum (Sorghum bicolour). Theor Appl Genet 128:1685–1701
Panigrahi J, Patnaik A, Kole P, Koleb C (2009) Addition of restriction fragment length polymorphism markers to the genetic linkage map of Brassica rapa L. (syn. campestris). Z. Naturforschung C 64:882–890
Poczai P, Varga I, Laos M, Cseh A, Bell N, Valkonen JP, Hyvonen J (2013) Advances in plant gene-targeted and functional markers: a review. Plant Methods 9:6
Ramchiary N, Nguyen VD, Li X, Hong CP, Dhandapani V, Choi SR, Yu G, Piao ZY, Lim YP (2011) Genic microsatellite markers in Brassica rapa: development, characterization, mapping, and their utility in other cultivated and wild Brassica relatives. DNA Res 18:305–320
Rawson H, Constable G (1980) Carbon production of sunflower cultivars in field and controlled environments. I. Photosynthesis and transpiration of leaves, stems and heads. Funct Plant Biol 7:555–573
Schranz ME, Lysak MA, Mitchell-Olds T (2006) The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11:535–542
Shen Q, Wang YT, Tian H, Guo FQ (2013) Nitric oxide mediates cytokinin functions in cell proliferation and meristem maintenance in Arabidopsis. Mol Plant 6:1214–1225
Shi ZY, Wang J, Wan XS, Shen GZ, Wang XQ, Zhang JL (2007) Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 226:99–108
Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL (1999) Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126:4117–4128
Stam P (1993) Construction of integrated genetic linkage maps by means of a new computer package: join map. Plant J 3:739–744
Van Ooijen J (2011) Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet Res 93:343–349
Veldboom LR, Lee M, Woodman WL (1994) Molecular marker-facilitated studies in an elite maize population: I. Linkage analysis and determination of QTL for morphological traits. Theor Appl Genet 88:7–16
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang FH, Wang GX, Li XY, Huang JL, Zheng JK (2008) Heredity, physiology and mapping of a chlorophyll content gene of rice (Oryza sativa L.). J Plant Physiol 165:324–330
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, Huang S, Li X, Hua W, Wang J, Wang X, Freeling M, Pires JC, Paterson AH, Chalhoub B, Wang B, Hayward A, Sharpe AG, Park BS, Weisshaar B, Liu B, Li B, Liu B, Tong C, Song C, Duran C, Peng C, Geng C, Koh C, Lin C, Edwards D, Mu D, Shen D, Soumpourou E, Li F, Fraser F, Conant G, Lassalle G, King GJ, Bonnema G, Tang H, Wang H, Belcram H, Zhou H, Hirakawa H, Abe H, Guo H, Wang H, Jin H, Parkin IA, Batley J, Kim JS, Just J, Li J, Xu J, Deng J, Kim JA, Li J, Yu J, Meng J, Wang J, Min J, Poulain J, Wang J, Hatakeyama K, Wu K, Wang L, Fang L, Trick M, Links MG, Zhao M, Jin M, Ramchiary N, Drou N, Berkman PJ, Cai Q, Huang Q, Li R, Tabata S, Cheng S, Zhang S, Zhang S, Huang S, Sato S, Sun S, Kwon SJ, Choi SR, Lee TH, Fan W, Zhao X, Tan X, Xu X, Wang Y, Qiu Y, Yin Y, Li Y, Du Y, Liao Y, Lim Y, Narusaka Y, Wang Y, Wang Z, Li Z, Wang Z, Xiong Z, Zhang Z, Brassica rapa Genome Sequencing Project C (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Gen 43:1035–1039
Wang W, Tang W, Ma T, Niu D, Jin JB, Wang H, Lin R (2016) A pair of light signaling factors FHY3 and FAR1 regulates plant immunity by modulating chlorophyll biosynthesis. J Integr Plant Biol 58:91–103
Yu X, Wang H, Zhong W, Bai J, Liu P, He Y (2013) QTL mapping of leafy heads by genome resequencing in the RIL population of Brassica rapa. PLoS ONE 8:e76059
Yu X, Choi SR, Dhandapani V, Rameneni JJ, Li X, Pang W, Lee JY, Lim YP (2016) Quantitative trait loci for morphological traits and their association with functional genes in Raphanus sativus. Front Plant Sci 7:255
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Zhang J, Lu Y, Yuan Y, Zhang X, Geng J, Chen Y, Cloutier S, McVetty PB, Li G (2009a) Map-based cloning and characterization of a gene controlling hairiness and seed coat color traits in Brassica rapa. Plant Mol Biol 69:553–563
Zhang KP, Fang ZJ, Liang Y, Tian JC (2009b) Genetic dissection of chlorophyll content at different growth stages in common wheat. J Genet 88:183–189
Zhao J, Jamar DC, Lou P, Wang Y, Wu J, Wang X, Bonnema G, Koornneef M, Vreugdenhil D (2008) Quantitative trait loci analysis of phytate and phosphate concentrations in seeds and leaves of Brassica rapa. Plant Cell Environ 31:887–900
Acknowledgments
This research was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through Golden Seed Project, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (213006-05-1-SB110).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Communicated by Lixi Jiang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Choi, S.R., Yu, X., Dhandapani, V. et al. Integrated analysis of leaf morphological and color traits in different populations of Chinese cabbage (Brassica rapa ssp. pekinensis). Theor Appl Genet 130, 1617–1634 (2017). https://doi.org/10.1007/s00122-017-2914-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2914-4