Abstract
Key message
Wheat lines with shortened Th. ponticum chromatin carrying Fhb7 and molecular markers linked to Fhb7 will accelerate the transfer of Fhb7 to breeding lines and provide an important resource for future map-based cloning of this gene.
Abstract
Fusarium head blight is a major wheat disease globally. A major FHB resistance gene, designated as Fhb7, derived from Thinopyrum ponticum, was earlier transferred to common wheat, but was not used in wheat breeding due to linkage drag. The aims of this study were to (1) saturate this FHB resistance gene region; (2) develop and characterize secondary translocation lines with shortened Thinopyrum segments carrying Fhb7 using ph1b; (3) pyramid Fhb7 and Fhb1 by marker-assisted selection. Fhb7 was mapped in a 1.7 cM interval that was flanked by molecular markers XsdauK66 and Xcfa2240 with SSR, diversity arrays technology, EST-derived and conserved markers. KS24-2 carrying Fhb7 was analyzed with molecular markers and genomic in situ hybridization, confirming it was a 7DS.7el2L Robertsonian translocation. To reduce the Thinopyrum chromatin segments carrying Fhb7, a BC1F2 population (Chinese Spring ph1bph1b*2/KS24-2) was developed and genotyped with the markers linked to Fhb7. Two new translocation lines (SDAU1881 and SDAU1886) carrying Fhb7 on shortened alien segments (approximately 16.1 and 17.3 % of the translocation chromosome, respectively) were developed. Furthermore, four wheat lines (SDAU1902, SDAU1903, SDAU1904, and SDAU1906) with the pyramided markers flanking Fhb1 and Fhb7 were developed and the FHB responses indicated lines with mean NDS ranging from 1.3 to 1.6 had successfully combined Fhb7 and Fhb1. Three new molecular markers associated with Fhb7 were identified and validated in 35 common wheat varieties. The translocation lines with shortened alien segments carrying Fhb7 (and Fhb1) and the markers closely linked to Fhb7 will be useful for improving wheat scab resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat (Triticum aestivum L., 2n = 6X = 42, genome AABBDD) scab, also called Fusarium head blight (FHB), is predominantly caused by the fungus Fusarium graminearum Schwabe [telomorph, Gibberella zeae (Schw.) Petch] and is one of the major diseases of wheat in many areas of the world (Bai and Shaner 2004; Goswami and Kistler 2004; West et al. 2012). It causes significant yield losses as well as reduced grain quality and functionality due to contamination with mycotoxins, such as deoxynivalenol (DON) and nivalenol (NIV) (D’mello et al. 1999; Cetin and Bullerman 2005). Climate change is predicted to increase the incidence of FHB on wheat (Chakraborty and Newton 2011; West et al. 2012). No single strategy has proven effective in mitigating the effects of FHB, but one promising avenue is the development of more resistant wheat cultivars, limiting both fungal growth and accumulation of mycotoxins.
Sources of FHB resistance used in current wheat breeding programs can be traced to very few parents, including Sumai 3 and its derivatives, Wangshuibai and Wuhan 1 (Bai and Shaner 2004; Buerstmayr et al. 2009). Development of only one or a few sources of resistance over large crop production areas poses vulnerability to resistance breakdown and disease epidemics. Resistance to FHB in wheat is a quantitative trait that is governed by multiple genes and modulated by environmental conditions. Numerous quantitative trait loci (QTL) have been reported as associated with FHB resistance (reviewed by Buerstmayr et al. 2009). A few of these FHB resistance-associated loci have been designated with a gene name: i.e., Fhb1 derived from T. aestivum cv. Sumai 3, Fhb2 derived from T. aestivum cv. Sumai 3, Fhb3 derived from Leymus racemosus, Fhb4 derived from T. aestivum cv. Wangshuibai, Fhb5 derived from T. aestivum cv. Wangshuibai, and Fhb6 derived from Elymus tsukushiensis have been mapped and designated previously (Cuthbert et al. 2006, 2007; Liu et al. 2006; Qi et al. 2008; Xue et al. 2010, 2011; Cainong et al. 2015). In addition, it is rare to find wheat cultivars with high levels of resistance to FHB, and no source of immunity has been identified (Bai and Shaner 2004; Yu et al. 2008; He et al. 2014). Therefore, discovery, development, and characterization of new resistance sources will provide breeders with a wider choice of germplasm.
Kim et al. (1993) described a wheat line carrying a wheatgrass, Thinopyrum ponticum (Podp.) Barkworth & D.R. Dewey (2n = 10X = 70) chromosome 7el2, substituted for wheat chromosome 7D. This highly resistant line carrying a FHB resistance gene, previously named as Fhblop (and designated as Fhb7 in the present study) and closely linked to molecular markers Xcfa2240 and XBE445653, was mapped on the long arm of chromosome 7el2. It explained 15.0–32.5 % of the phenotypic variation in FHB response (Shen et al. 2004; Shen and Ohm 2007; Zhang et al. 2011). However, due to linkage drag, the original translocation line, KS24-2, carrying a pair of Robertsonian translocation chromosomes identified as 7DS.7el2L (Kim et al. 1993), has not been used for FHB resistance breeding. In addition, the DNA marker density of the Fhb7 region is far lower than that required for positional cloning. Therefore, development of new molecular markers is needed for saturation mapping of Fhb7.
Diversity arrays technology (DArT), based on the use of restriction enzymes to digest genomic DNA to reduce genome complexity and enrich low copy sequences for marker development, has been widely used in plant genetic studies, genetic map construction, and selection of molecular markers linked to genes controlling agronomic traits in wheat, barley, sorghum, and other crop species (Wenzl et al. 2004; Akbari et al. 2006; Semagn et al. 2006). The large amount of crop genomic sequences, such as rice (Goff et al. 2002), Brachypodium distachyon (Vogel et al. 2010) and sorghum (Paterson et al. 2009), which are now publicly available, are rich resources for molecular marker development and comparative genomics analysis. Recently, based on synteny between wheat and rice, B. distachyon or Sorghum, comparative mapping has been widely used for construction of high-resolution genetic maps of interesting wheat genes or positional cloning of these genes (Liu and Anderson 2003; Wang et al. 2014a).
The objectives of the present study were to (1) saturate this FHB resistance gene region using SSR, DArT, and conserved markers; (2) develop and characterize small segment translocation lines carrying Fhb7 by reducing the amount of Th. ponticum chromatin using ph1b-induced homoeologous chromosome pairing (Qi et al. 2007; Marais et al. 2010; Niu et al. 2011); and (3) pyramid Fhb7 with Fhb1 by marker-assisted selection.
Plant materials and methods
Plant materials
A set of 156 F8:9 recombinatn inbred lines (RILs) derived from a cross between the FHB susceptible line K11463 (7el1[7D]) and the FHB resistant line K2620 (7el2[7D]) were utilized to saturate the 7el genetic linkage map of Fhb7 (Zhang et al. 2011). K11463 and K2620 were two wheat-Th. ponticum substitution lines, in which wheat chromosome 7D was substituted by Th. ponticum chromosome 7el1 and 7el2, respectively. Wheat variety Ning 7840 was used as the resistant control.
Wheat-Th. ponticum translocation line KS24-2 carrying FHB resistance gene Fhb7 on Th. ponticum chromatin in common wheat cv. Thatcher was used as the donor of the FHB resistance gene. This translocation was identified as 7DS.7el2L (Kim et al. 1993). The Chinese Spring Ph1b mutant and wheat cv. Jimai 22 were used as crossing and backcrossing parents. Chinese Spring (CS), Thatcher, and Jimai 22 were used as susceptible checks and Ning 7840 was used as the resistant check. CS nulli-tetrasomic lines N7AT7B, N7BT7D, and N7DT7A, and CS deletion lines 7AL16, 7AL1, 7DL3, 7BL2, 7BL7, 7BL10, 7DL2, and 7DL5 were used in molecular marker analyses. Thirty-five wheat cultivars were used for validation of newly developed markers along with the SSR marker Xcfa2240 (Table S1).
Comparative genomics analysis and conserved primer development
The FHB resistance gene Fhb7 was previously mapped on the long arm of the Th. ponticum chromosome 7el (Shen et al. 2004; Shen and Ohm 2007; Zhang et al. 2011). STS markers derived from wheat ESTs were developed and mapped to the long arm of the Th. ponticum chromosome 7el by Shen and Ohm (2007). The corresponding EST sequences of the polymorphic EST markers flanking the FHB resistance gene Fhb7 were used to perform a BLAST search against the genome sequence databases of rice (http://rice.plantbiology.msu.edu/), B. distachyon (http://www.Brachypodium.org/) and sorghum (http://www.plantgdb.org/SbGDB/cgi-bin/blastGDB.pl) to identify orthologous genomic regions in rice, B. distachyon and sorghum with cutoff parameters of E value ≦ 1E −10, identity ≧70 % and a minimum of a 100-bp match length. When several significant hits were found, only the best hit was adopted.
Coding sequences (CDS) of the putative genes in the regions were used as queries to search the NCBI database (http://www.ncbi.nlm.nih.gov) with cutoff parameter of E value ≦ 1E −10 for homologous Triticeae ESTs to develop new polymorphic markers linked to Fhb7. The conserved primers were designed with the online software version (http://probes.pw.usda.gov/cgi-bin/ConservedPrimers/ConservedPrimers.cgi) as described by You et al. (2009). The primer design parameters were as follows: amplification product size of 800–1500 bp with an optimum size 1000 bp, primer length of 18–24 bp, Tm of 55–65 °C, and GC content of 30–70 %.
SSR marker development
Fourteen scaffolds, i.e., Scaffold48913, scaffold89301, scaffold35216, scaffold38207, scaffold72072, scaffold70783, scaffold5083, scaffold11459, scaffold21010, scaffold23, scaffold76033, scaffold21011, scaffold92632, and scaffold32834, in the distal region of Ae. tauschii 7DL were downloaded and used for SSR marker development (Jia et al. 2013). SSR Finder (http://www.fresnostate.edu/ssrfinder/) was used to identify simple sequence repeat regions in the scaffolds, and sequences flanking the SSR region were selected to design SSR primers using Primer3 Input Version 0.4.0 (http://bioinfo.ut.ee/primer3-0.4.0/) under general settings.
The developed SSR and conserved primers were then tested on CS, CS nulli-tetra lines N7AT7B, N7BT7D, and N7DT7A and CS deletion lines 7AL16, 7AL1, 7DL3, 7DL2, and 7DL5, and screened for polymorphisms among wheat lines K11695, K11463, K2620, and KS24-2. Polymorphic molecular markers were used to genotype the 156 RILs. PCR products were separated on 8 % non-denaturing polyacrylamide gels (acrylamide:bisacrylamide = 39:1) and visualized with silver staining (Kong et al. 2008).
DArT marker development and genotyping
DArT service was provided by DArT Pty Ltd (DArT P/L) (Canberra, Australia; http://www.diversityarrays.com/), with genome profiling service. A bread wheat array was adopted for the two parental lines, K11463 and K2260 and its derived 156 RILs and the data were analyzed, as described by Akbari et al. (2006).
Genomic DNA isolation
Total genomic DNA was isolated from young leaves using the CTAB protocol (Zhang et al. 2011) with minor modifications (Supporting information). The concentration of each DNA sample was adjusted to 50–100 ng/μl after quantification on a ND-1000 Spectrophotometer V3.5 (NanoDrop Technologies, Inc., USA).
Molecular marker analysis
PCR amplifications were performed in 15 μl total volume containing 1× PCR buffer, 1.5 mM MgCl2, 200 μM dNTP, 0.7 μM of each primer, 1 unit of Taq polymerase, and 100 ng of template DNA. The amplification profile consisted of 1 cycle at 94 °C for 5 min, followed by 35 cycles of 50 s at 94 °C, 30 s at 50–60 °C (annealing temperature depending on specific primers) and 50 s at 72 °C, with a final extension of 10 min at 72 °C. PCR products were separated in 8 % non-denaturing polyacrylamide gels. Gels were silver stained and photographed (Zhang et al. 2011).
Chromosome manipulation
Wheat plants homozygous ph1bph1b and monosomic for wheat chromosome 7D and the translocated chromosome 7DS.7el2L were developed to enable meiotic recombination between 7D and 7DS.7el2L (Fig. 1). These plants were produced by crossing the CS Ph1 mutant (CS ph1b) to KS24-2 and backcrossing the F1 plants to the ph1bph1b line. BC1F1 plants were genotyped using the ph1b-specific molecular markers Xpsr128, Xpsr574, and XAWJL3 (Roberts et al. 1999) and SSR markers Xcfa2240 and Xgwm333, which were specific for chromosome 7el2 (Shen and Ohm 2007; Zhang et al. 2011). A BC1F2 population derived from BC1F1 plants that were monosomic for chromosomes 7D and 7DS. 7el2L was developed and screened by Fhb7-linked molecular markers (Xcfa2240, Xgwm333, Xswes130, XsdauK66, and Xmag1759) (Fig. S1) and genomic in situ hybridization (GISH). The overall procedure is summarized in Fig. 1. The progenies of the plants with homozygous putatively shortened Th. ponticum chromatin segments were examined by GISH and evaluated for FHB response. The sizes of the Th. ponticum chromatin segments in the selected lines were expressed as average percentages of the length of Th. ponticum chromatin segment/total length of the translocation chromosome, measured in at least 20 cells with good-quality mitotic metaphases.
Genomic in situ hybridization (GISH)
Seed germination and collection and the pretreatment and fixation of root tips were performed as described by Han et al. (2009) with minor modifications (Supporting information). Total genomic DNA of Th. ponticum (accession RM001128, from the Chinese Crop Germplasm Resources Information System) isolated from young leaves, was labeled with DIG-11-dUTP by nick translation and used as a probe. Sheared genomic DNA from Chinese Spring was used for blocking. Preparation of chromosome spreads and visualization of probe hybridization were performed as described by Han et al. (2009).
Validation of new molecular markers linked to Fhb7 on shortened Th. ponticum chromosome segments
Genomic DNA for identification and validation of molecular markers linked to Fhb7 on the shortened Th. ponticum segments in the new lines was extracted from seedling plants. Molecular marker and GISH analysis localized the shortened Th. ponticum chromatin segment on the distal region of chromosome 7DL in the new wheat lines. Then molecular markers located in the distal region of chromosome 7elL were selected and used to screen for polymorphism among Thatcher, CS, KS24-2, K2620 and the newly developed introgression lines with shortened Th. ponticum segments carrying Fhb7.
To validate molecular markers linked to Fhb7 on the shortened Th. ponticum chromosome segments, polymorphic molecular markers that were identified following the above procedures were used to genotype 35 common wheat cultivars (Table S1). Marker analyses were performed as described above.
Pyramiding of FHB resistance genes Fhb1 and Fhb7
Common wheat cv. Ning 7840 carrying Fhb1 was crossed to the new introgression lines. Markers Xgwm493 and Xgwm533 (Cuthbert et al. 2006; Buerstmayr et al. 2009), which are closely linked to Fhb1 and the SSR markers XsdauK66 and Xcfa2240, which are closely linked to Fhb7 were used to select homozygous F2 plants. These homozygous plants were evaluated for FHB response in the greenhouse (Fig. S2A). PCR amplifications were performed as described by Cuthbert et al. (2006). Amplification products were then separated in 2 % agarose gels and visualized by ethidium bromide staining.
Introgression of Fhb7 into Chinese major winter wheat cultivar Jimai 22
Jimai 22 was crossed to the new translocation lines and the hybrids were backcrossed to Jimai 22 for three generations. BC3F1 plants with the shortened Th. ponticum chromatin segments were selected by the markers flanking Fhb7 and then self-fertilized. BC3F2 plants homozygous for the marker were evaluated for FHB response in the greenhouse (Fig. S2B).
Determination of FHB response
Type II FHB resistance, defined as inhibition of disease spread from a point infection, was determined in a greenhouse at Shandong Agricultural University over four seasons, 2012 Winter (2012 W), 2013 Winter (2013 W), 2014 Spring (2014 S), and 2014 Winter (2014 W). All experimental lines, that is KS24-2, Thatcher, CS ph1b, new Th. ponticum derivatives, lines combining markers flanking Fhb1 and Fhb7, Jimai 22, Jimai 22 backcross derivatives homozygous for markers flanking Fhb7, and Ning 7840, were screened. Eight to ten spikes of each line were inoculated when each spike reached 50 % anthesis. One basal flower in the 4th or 5th spikelet from the tip of the spike was inoculated by injecting 10 μl micro-conidial suspension (50,000 spores/ml) between the lemma and palea of a floret with a syringe. Inoculated spikes were covered for 72 h with a plastic bag to maintain high humidity. The number of diseased spikelets (NDS), including the inoculated spikelet, were counted at 21 days post-inoculation. Data were analyzed with SAS software version 9.0. The mean values of putatively resistant lines were compared with Thatcher, Chinese Spring ph1bph1b, Ning 7840 and Jimai 22. Duncan’s multiple range test was used to test for significant differences.
Genetic linkage map construction
All DArT, SSR, and conserved markers polymorphic between K11463 and K2620 were mapped in the K11463/K2620 mapping population of 156 RILs (Zhang et al. 2011). The linkage map was constructed using the software Joinmap version 4.0 (Van Ooijen 2006) with an initial theshold of LOD ≧ 10.0. After a final order of markers was reached, a representative marker from each group of cosegregating markers (termed as a recombination block) was selected and a map was constructed with LOD ≧ 3.0 by using the Kosambi mapping function (Kosambi 1943). The computer program QTL Icimapping V4.0 (Wang et al. 2014b) was used for mapping the FHB resistance QTL with a LOD threshold 3.0 using the data collected by Zhang et al. (2011).
Results
Identification of genomic region in rice, B. distachyon, and Sorghum with Th. ponticum chromosome 7el containing Fhb7 and comparative genomic analysis
The FHB resistance gene Fhb7 was previously mapped on the long arm of Th. ponticum chromosome 7el and was closely associated with 10 EST-derived STS markers (XBE404744, XBM137749, XBE637476, XBE445506, XBF483039, XBG607810, XB145935, XBE445653, Xmag1932 [CD880294], and XHX2166 [CJ579028]) (Shen and Ohm 2007; Zhang et al. 2011). Based on the corresponding EST sequences of these EST-STS markers, the orthologous genomic regions were identified in rice, B. distachyon, and sorghum. The Th. ponticum genomic region containing Fhb7 was orthologous to a distal region of rice chromosome 6 (Fig. 2). Although this gene was not mapped between two EST-derived markers, this orthologous region in rice contained only two PAC clones (AP005750 and AP006616).
Based on the above information and blast analysis, the Th. ponticum genomic region containing Fhb7 was presumably orthologous to rice chromosome 6 (Os06g51330 and Os06g51570), B. distachyon chromosome 1 (Bradi1g29130 and Bradi1g29510), and sorghum chromosome 10 (Sb10g031110 and Sb10g031310, Fig. 2; Table 1). Three genomic regions spanning 154.1, 366.2, and 222.1 kb in rice, B. distachyon and sorghum, respectively, appeared to be the orthologous genomic regions to Th. ponticum harboring the FHB resistance gene Fhb7. Detailed comparative analysis revealed that 17 of 25 predicted rice genes on chromosome 6 were orthologous to 15 of 39 predicted B. distachyon genes and 14 of 20 predicted sorghum genes in the corresponding genomic regions, which showed high levels of genomic collinearity among these three species (Fig. 2; Table 1).
However, very common phenomena of gene insertion or deletion and gene order inversions or chromosomal rearrangements were also observed in the three species. A segmental inversion, Bradi1g29390-Bradi1g29510 was present in B. distachyon compared to rice and sorghum in the corresponding orthologous genomic region (Fig. 3). Another inversion, Sb10g031240-Sb10g031265, was present in sorghum compared to rice and B. distachyon. Finally, some specific genomic regions were also observed. A segment Os06g51520-Os06g51570 was specific to rice and was not present in B. distachyon or sorghum. Five segments Bradi1g29140-Bradi1g29160, Bradi1g29200-Bradi1g29230, Bradi1g29260-29280, Bradi1g29330-Brai1g29380, and Bradi1g29434-Brai1g29450 were specific to B. distachyon and not present in the corresponding genomic region of rice and sorghum. Two segments Sb10g031160-Sb10g031170 and Sb10g031200-Sb10g031210 were specific to sorghum, which were not present in rice and B. distachyon (Table 1; Fig. 3).
Structure variations of the Fhb7 orthologous genomic regions in rice, B. distachyon, and sorghum. Green regions indicate the same gene order among rice, B. distachyon, and sorghum. Orange region represents B. distachyon inversion in gene order compared to rice and sorghum. Purple region and black region represent sorghum inversion in gene order compared to rice and Brachypodium, respectively. Light blue regions represent rice-specific regions compared to B. distachyon and sorghum. Light green regions represent B. distachyon-specific regions compared to rice and sorghum. Blue regions represent sorghum-specific regions compared to rice and B. distachyon. Superscript letter a represents the start position of putatively syntenic regions in rice B. distachyon and sorghum corresponding to Th. ponticum chromosome 7el regions harboring Fhb7. Superscript letter b represents the end position of putatively syntenic regions in rice B. distachyon and sorghum corresponding to Th. ponticum chromosome 7el regions harboring Fhb7 (color figure online)
A refined genetic map of the region of Th. ponticum chromosome 7el2 surrounding the FHB resistance gene Fhb7
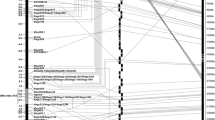
The predicted genes of rice, B. distachyon, and sorghum in the corresponding orthologous region of Th. ponticum chromosome 7el were used to develop molecular markers linked to the FHB resistance gene Fhb7. The putative orthologous or non-orthologous genes between Os06g51330 and Os06g51570 in rice, Bradi1g29130 and Bradi1g29510 in B. distachyon, and Sb10g031110 and Sb10g031310 in sorghum were selected to develop markers to narrow the collinearity regions among Th. ponticum, rice, B. distachyon, and sorghum. The CDS of these genes were used as queries to search for orthologous wheat (T. aestivum, T. turgidum, and T. monococcum), barley and wheatgrass (Elymus, Leymus, and Secale) ESTs. The numbers of orthologs identified in wheat, barley, and wheatgrass were different. Among the 25 rice putative genes investigated, 72 orthologs, 44 orthologs, 5 orthologs, 4 orthologs, and 1 orthologs were identified in T. aestivum, H. vulgare, Leymus, E. spicatus, and T. turgidum, respectively. None were identified in T. monococcum and S. cereale (Table 1; Excel S1). When the 39 B. distachyon putative genes were queried, 191 orthologs, 71 orthologs, 18 orthologs, 15 orthologs, 15 orthologs, and 2 orthologs were identified in T. aestivum, H. vulgare, Leymus, T. turgidum, E. spicatus, and T. monococcum, respectively. However, none were identified in S. cereale (Table 1; Excel S1). With the 20 sorghum putative genes as queries, 90 orthologs, 51 orthologs, 7 orthologs, 16 orthologs, 7 orthologs, 1 orthologs, and 1 orthologs were identified in T. aestivum, H. vulgare, Leymus, T. turgidum, E. spicatus, S. cereale, and T. monococcum, respectively (Table 1; Excel S1). Then, 10 rice, 15 B. distachyon, and 4 sorghum putative genes were used to develop conserved markers. Finally, 26 conserved markers were produced by the software ConservedPrimer 2.0 (Table S2). Out of them, 6 conserved markers, XsdauK13, XsdauK60, XsdauK66, XsdauK71, XsdauK130, and XsdauK144 were polymorphic between the two parents, K11463 and K2620. DArT was also used to produce polymorphic markers between K11463 and K2620 and 499 polymorphic DArT markers were produced. In order to produce more markers surrounding Fhb7, fourteen scaffolds, i.e., Scaffold48913, scaffold89301, scaffold35216, scaffold38207, scaffold72072, scaffold70783, scaffold5083, scaffold11459, scaffold21010, scaffold23, scaffold76033, scaffold21011, scaffold92632, and scaffold32834, in the distal region of Ae. tauschii 7DL (Jia et al. 2013) were downloaded and used for SSR marker development. Finally, 14 SSR markers were developed (Table S2) and one SSR marker, XsdauK352, showed polymorphisms between K11463 and K2620. In total, 506 polymorphic markers, including 1 SSR, 499 DArT and 6 conserved markers, were used to construct a linkage map of Th. ponticum chromosome 7el combining the map of Zhang et al. (2011) with the present results. After elimination of unlinked markers and other small linkage groups, the high-density genetic map contained 167 markers covering 158.97 cM, with an average density of one marker per 0.95 cM (Excel S2). Of the six conserved markers identified above, XsdauK60 and XsdauK130 were assigned to deletion bin 7BL10-0.78-1.0, and XsdauK13 and XsdauK66 were assigned to deletion bin 7DL10-0.82-1.0. These markers permitted mapping of Fhb7 within a 1.7 cM interval flanked by molecular markers XsdauK66 and Xcfa2240 (Fig. 4). Thus, Fhb7 was most closely linked to Os06g51490 and Os06g51500 in rice representing a 18.8 kb genomic region and corresponding to a 37.1 kb genomic region in B. distachyon and a 27.0 kb genomic region in sorghum.
A high-density genetic map of Thinopyrum chromosome 7el constructed with DArT, SSR, EST-derived, and conserved markers and genetic detection of FHB resistance gene Fhb7 derived from Th. ponticum. FHB response data from the greenhouse (2005 USA) at Purdue University are presented in red. FHB response data from the greenhouse (2008 China) at Shandong Agricultural University are presented in green. The mean response data from both locations are presented in blue. Composite interval mapping was performed with Icimapping V4.0. The DArT markers are painted red; SSR markers are painted blue; EST-derived markers are painted green; conserved markers are painted pink; and the rest are painted black (color figure online)
Development and characterization of Fhb7-carrying wheat lines with reduced Th. ponticum chromatin segments induced by ph1b with marker-assisted introgression and genomic in situ hybridization
For characterization of the original translocation line KS24-2, all molecular markers except DArT markers mapped on the 7el chromosome (Excel S2) were screened among Chinese Spring, KS24-2, K2620, Thatcher, and Th. ponticum and 12 markers were polymorphic among them (Table S4). Finally, markers Xcfa2240 and XsdauK66 mapped in wheat chromosome bin 7DL3-0.82-1.0 and closely linked to Fhb7 as described above, Xgwm333 previously mapped in wheat chromosome bin 7BL2-0.33-7BL7-0.63 Sourdille et al. (2004), Xmag1759 mapped in wheat chromosome bin 7BL7-0.63-7BL10-0.78, and Xswes130 showing polymorphic between KS24-2 and wheat checks Thatcher and Chinese Spring were selected for further analysis (Figs S1, 4; Table S4). The SSR marker Xcfa2240 mapped to the long arms of chromosomes 7A and 7D with CS 7L-deletion lines (7AL1, 7AL16, 7DL2, 7DL3, and 7DL5) (Fig. 5d). GISH results confirmed that the interchanged chromosome in line KS24-2 is a 7DS.7el2L Robertsonian translocation (Fig. 6b).
Characterization of two new wheat lines (SDAU1881 and SDAU1886) carrying Fhb7 in small segments of Thinopyrum chromatin. a, b, and c Gel images of the PCR products of three co-dominant markers Xcfa2240, XsdauK352, and XsdauK66 run on 8 % non-denaturing polyacrylamide gels, respectively. The numbers at the top of the gels are lane numbers: M, 100 bp ladder; 1, Th. ponticum; 2, 7el2(7D); 3, KS24-2; 4, SDAU1881; 5, SDAU1886; 6, Chinese Spring (CS); 7, Thatcher; 8, K11463; 9, K11695. Arrows indicate Thinopyrum-specific bands linked to Fhb7. d Gel image of the PCR products of Xcfa2240 run on 8 % non-denaturing polyacrylamide gels. The numbers at the top of the gels are lane numbers: M, 100 bp ladder; 1, CS; 2, N7AT7B; 3, N7BT7D; 4, N7DT7A; 5, 7AL16; 6, 7AL1; 7, 7DL5; 8, 7DL2; 9, 7DL3. Arrows indicate 7D-specific bands in bin 7DL3-0.82-1.0. Arrowhead indicates 7A-specific bands in bin 7AL16-0.86-1.0. e FHB reactions
Images from GISH analysis of wheat-Thinopyrum translocation lines carrying FHB resistance gene Fhb7. GISH was performed using Th. ponticum genomic DNA as the probe and Chinese Spring (CS) genomic DNA as the blocker on a CS b KS24-2 c SDAU1881 d SDAU1886. Bars 10 μm. Thinopyrum chromatin is painted green and indicated arrows. Wheat chromosomes are painted blue (color figure online)
When the T7DS.7el2L × CS ph1bph1b F1 was used as the female parent in crossing with CS ph1bph1b, 61 plants had the translocation chromosome (96.8 %) and two did not (3.2 %). However, in the reciprocal cross 32 plants (53.3 %) had the translocation chromosome and 28 (46.7 %) lacked it. Clearly, there was preferential transmission of the T7DS.7el2L translocation chromosome by female gametes whereas male transmission was normal.
To identify introgression lines carrying Fhb7 on smaller alien segments, 208 BC1F2 individuals were genotyped for five markers; i.e., Xcfa2240, Xgwm333, Xswes130, XsdauK66, and Xmag1759 (Figs. S1, 1; Table S4); 34 carried wheat CS ph1b alleles at all five SSR loci (Plant type 8; Table 2), 15 carried Th. ponticum alleles at 1 or 2 SSR loci (Plant types 4, 5, 7, and 9) and the remaining 159 plants retained Th. ponticum alleles at three or more marker loci (Plant types 1, 2, 3, 6, and 10). Fhb7 was located on chromosome 7el2L and was flanked by molecular markers Xcfa2240 and XsdauK66 as described above. GISH was utilized to examine the two BC1F2 plants with only the Th. ponticum alleles at the Xcfa2240 and XsdauK66 locus. Lines 338-29 and 338-63 (Type 4, Table 2) carrying much smaller translocated Th. ponticum segments were presumed to have the structure T7DS.7DL-7el2L (Fig. 6). In addition, six molecular markers (Xbarc76, Xwmc273, XBM137749, XHX2166, XsdauK352, and Xpsp3003) between Xmag1759 and Xcfa2240 were selected to screen the above two short lines identified (Table S5). Our results indicated the 7elL-7DL translocation might be between Xwmc273 and XBM137749.
The two lines with putatively shortened segments and homozygous for the Th. ponticum Xcfa2240 and XsdauK66 alleles were evaluated for FHB response using the parental lines and the resistant and susceptible varieties as controls. Plants were classified as resistant or susceptible. FHB response differences represented by the NDS between the parental lines KS24-2 and CS ph1b were significant at p = 0.01. The two new lines with terminal 7el2L fragments, designated as SDAU1881 and SDAU1886, were resistant (Table 3; Figs. 5d, 6b, c), with mean NDS of 1.4 and 1.5, respectively, similar to KS24-2. In contrast, three other derivatives; i.e., SDAU1885, SDAU1893, and SDAU1894 (all belonging to plant type 8), had mean NDS of 5.8, 5.2, and 5.7, respectively. The FHB responses were not significantly different from CS ph1b (Table 3)
Based on the FHB responses, GISH, and molecular maker analysis, the shortened Th. ponticum chromatin segments in SDAU1881 and SDAU1886 were located in the distal region of chromosome 7DL (Table 3; Figs. 5d, 6c, d). The sizes of the Th. ponticum chromatin segments in the entire chromosomes were 16.1 % (25 cells) for SDAU1881 and 17.3 % (30 cells) for SDAU1886 compared to 49.0 % (20 cells) for line KS24-2. Therefore, approximately 65 % of the Th. ponticum chromatin proximal to Fhb7 was removed in the two new translocation lines after ph1b-induced homoeologous recombination.
Validation of new molecular markers linked to Fhb7 on the shortened Th. ponticum chromosome segments
The GISH patterns and physical position of the SSR marker Xcfa2240 on chromosome 7D indicated that Fhb7 was located in the deletion bin 7DL3-0.82-1.0 (Fig. 5d). The three co-dominant markers; i.e., XsdauK352, Xcfa2240, and XsdauK66, were selected and validated with 35 wheat accessions (Table S1). The co-dominant marker XsdauK352 derived from the scaffold76033 amplified two fragments of approximately 210 and 483 bp in KS24-2, 7el2(7D), SDAU1881 and SDAU1886, and amplified one 490 bp fragment in Thatcher and CS ph1b (Figs. 4, 5b, Table S1). The SSR marker Xcfa2240 amplified two fragments of 240 and 280 bp in KS24-2, 7el2(7D), SDAU1881 and SDAU1886, and two fragments of 245 and 280 bp in Thatcher and CS ph1b (Figs. 4, 5a; Table S1). The conserved marker, XsdauK66, derived from the rice gene Os06g51500 (Fig. 4), amplified three fragments of approximately 358, 360, and 420 bp in KS24-2, 7el2(7D), SDAU1881, and SDAU1886, and amplified one 360 bp fragment in Thatcher and CS ph1b (Figs. 4, 5c, Table S1).
Confirmation of Fhb7 in the Chinese major wheat cultivar Jimai 22 and its pyramiding with Fhb1
To confirm the FHB resistance conferred by Fhb7 and its flanking markers, a BC3F2 population totaling 200 plants was produced by crossing SDAU1881 to the susceptible cv. Jimai 22 and then backcrossing to Jimai 22 for three generations. Two BC3F2:3 lines that were putatively homozygous for Fhb7 and four non-Fhb7 lines were selected by marker-assisted selection using markers XsdauK66 and Xcfa2240 flanking Fhb7, and were then evaluated for FHB response in the greenhouse. The four non-Fhb7 lines determined with the above markers, SDAUI081, SDAUI082, SDAUI083, and SDAUI084, had mean NDS of 6.2, 6.2, 5.5, and 5.0, respectively, which was similar to Jimai 22. The two lines that were agronomically attractive predicted to carry Fhb7 had means NDS of 1.5 and 1.6, which were not significantly different from the donor parent SDAU1881 (Table 4; Fig. S6). The two newly improved lines that were homozygous to Fhb7 were designated as SDAU2003 and SDAU2028.
For pyramiding Fhb7 and Fhb1, a population totaling 300 F2 plants was generated from crosses of the new wheat-Th. ponticum derivatives and cv. Ning 7840. Four lines homozygous for markers Xcfa2240 and XsdauK66 flanking Fhb7, and Xgwm493 and Xgwm533 flanking Fhb1, were selected (Fig. 2). The FHB responses indicated that four F2:3 lines were resistant to FHB with mean NDS ranging from 1.3 to 1.6 and not significantly different from KS24-2 or Ning 7840 (Table 3; Fig. S6). The four pyramided lines putatively homozygous for both Fhb7 and Fhb1 were designated as SDAU1902, SDAU1903, SDAU1904, and SDAU1906.
Inheritance of FHB resistance gene Fhb7 derived from Th. ponticum
We evaluated 63 F2 progeny from a cross between Jimai 22 and SDAU1881 and grouped them based on the molecular markers XsdauK352, Xcfa2240, and XsdauK66, which were classified as homozygous for T7DS.7DL-7el2L, heterozygous for 7D/T7DS.7DL-7el2L, and homozygous for 7D. The ratio of the above three groups was 15:34:14, which fitted the expected 1:2:1 ratio (\(\chi_{ 1: 2: 1}^{2}\) = 0.4, p = 0.8). The mean NDS in plants with only one copy of T7DS.7DL-7el2L was not significantly different from lines without T7DS.7DL-7el2L (LSD0.05 = 1.6), while plants homozygous for T7DS.7DL-7el2L had significantly lower NDS (Table 5).
Discussion
Thinopyrum species, as members of the tertiary gene pool for wheat, harbor many biotic and abiotic stress genes (Wang 2011). To date, at least 3 FHB resistance genes or QTLs have been identified in Thinopyrum derivatives. Among them, one gene located on chromosome 1E derived from Th. elongatum showed high FHB resistance in the greenhouse, but no visible effect was observed in the field (Jauhar 2014). Another gene Fhb7, located on chromosome 7el2 derived from Th. ponticum showed high FHB resistance in the greenhouse and was resistant in the field (Shen et al. 2004; Shen and Ohm 2007; Zhang et al. 2011). A third gene located on chromosome 7E derived from Th. elongatum also showed FHB resistance, but it was not used due to linkage drag (Shen et al. 2004; Fu et al. 2012). We are developing a backcrossed population between 7E(7D) and wheat the Ph1 mutant inducing homoeologous chromosome pairing in our ongoing research.
The FHB resistance gene Fhb7 was identified in the wheat-Th. ponticum substitution line K2620 and mapped closely linked to Xcfa2240 on the long arm of chromosome 7el (Shen and Ohm 2007; Zhang et al. 2011), but the DNA marker density of the Fhb7 region was far lower than that required for fine mapping and positional cloning. Therefore, development of new molecular markers is needed for saturation mapping of Fhb7. Comparative genomics approaches have been widely used to construct high-density genetic maps of important wheat genes based on synteny among wheat, rice, B. distachyon, and sorghum. Based on synteny among these species, the FHB resistance gene Fhb1 and the powdery mildew resistance gene Pm41 were narrowed to sub-centiMorgan intervals (Liu and Anderson 2003; Cuthbert et al. 2006; Liu et al. 2006; Wang et al. 2014b). In this study, synteny among Th. ponticum, rice, B. distachyon, and sorghum was established. Subsequently, conserved primers were developed and used to map the FHB resistance gene Fhb7, which was flanked by XsdauK66 and XsdauK60, corresponding to 18.8, 37.1, and 27.0 kb genomic regions in rice, B. distachyon, and sorghum, respectively. Of the 6 polymorphic markers, 3 were derived from the orthologous genes presented in the three species, which indicated the orthologous genes of the three species were more useful for marker development than others. However, the genes specific for one species or two species could also be useful for high-density genetic linkage map construction (Figs. 2, 4). Previous studies showed that Thinopyrum species were closely related to wheat (Dvořák 1980; Dewey 1983; Wang and Lu 2014). We expect that the newly published genome sequences of T. uratu (Ling et al. 2013), Aegilops tauschii (Jia et al. 2013), and T. aestivum (Mayer et al. 2014) will be more useful for marker development and mapping of important genes in Thinopyrum being transferred to wheat.
Creation and development of crops with stable or high disease resistance play important roles in sustainable crop production (Duveiller et al. 2007). Both the spectrum of protection against different races of pathogens, and the degree of resistance, can be increased by combining, or pyramiding resistance genes in the same line. This process is greatly aided by the availability of genetic markers because the required genotypes cannot be recognized by disease response alone. In this study, the introgression lines SDAU1881 and SDAU1886 with shortened Th. ponticum segments carrying Fhb7 were identified and developed using the CS ph1b mutant, molecular markers, and GISH. Both lines were characterized as T7DS.7DL-7el2L translocations and showed resistance to FHB, with outcomes consistent with the genetic mapping data (Fig. 4). The alien segments in these two lines were shortened by approximately 65 % compared to the original line KS24-2 (49 %), which meant that the distal 35 % of the long arm of chromosome 7el2L or 7DL is common in these two lines, i.e., SDAU1881 and SDAU1886. In addition, co-dominant markers Xcfa2240, XsdauK352, and XsdauK66 closely linked to Fhb7 were identified and validated. MAS based on these markers will expedite transfer of Fhb7 to other genotypes and the newly developed polymorphic markers would be helpful to select shorter 7el2L chromosome segment carrying Fhb7 through a second round of homeologous recombination. Two agronomically attractive Jimai 22 backcross derivatives (SDAU2003 and SDAU2028) developed by MAS using markers closely linked to Fhb7 were highly resistant to FHB. Four lines putatively carrying Fhb1 and Fhb7 (SDAU1902, SDAU1903, SDAU1904, and SDAU1906) were developed by MAS and all showed high resistance to FHB, although none was more resistant than the Fhb1 donor parent Ning 7840, or the new introgession lines SDAU1881 or SDAU1886. Nevertheless, we believe that the pursuit of resistance gene combinations in breeding is worthwhile because of the likelihood of more stable resistance to multiple pathogenic types and across environments and genetic backgrounds.
The FHB resistance conferred by translocation T7DS.7DL-7el2L in the CS background is novel and the level of resistance is similar to Ning 7840 (Table 3). However, our screening results of a population segregating for T7DS.7DL-7el2L showed that the resistance gene Fhb7 in 7DL-7el2L is likely homozygous effective (Table 5). We are presently transferring the 7DS.7DL-7el2L translocation to different commercial cultivars, which will allow us to evaluate the effect of 7DL-7el2L on FHB resistance and other agronomical traits in other genetic backgrounds.
In summary, new wheat lines with shortened Th. ponticum chromatin segments carrying Fhb7 were developed by molecular chromosome engineering using the Ph1 mutant. Identification of molecular markers closely linked to Fhb7 will accelerate the transfer of Fhb7 to breeding lines and also provide an important resource for future map-based cloning of this FHB resistance gene.
Author's contribution statement
LR Kong and Herbert W. Ohm conceived and designed the experiments. J Guo, XL Zhang, YR Hou, and XR Shen performed the experiments. J Guo, XL Zhang, YR Hou, JJ Cai, and LR Kong analyzed the data. TT Zhou, HH Xu, HW Wang, AF Li, FP Han, and HG Wang contributed reagents/materials/analysis tools. J Guo and LR Kong wrote the paper.
References
Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang S, Uszynski G, Mohler V, Lehmensiek A, Kuchel H, Hayden MJ, Howes N, Sharp P, Vaughan P, Rathmell B, Huttner E, Kilian A (2006) Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor Appl Genet 113:1409–1420
Bai G, Shaner G (2004) Management and resistance in wheat and barley to Fusarium head blight 1. Annu Rev Phytopathol 42:135–161
Buerstmayr H, Ban T, Anderson JA (2009) QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breed 128:1–26
Cainong JC, Bockus WW, Feng Y, Chen P, Qi L, Sehgal SK, Danilova TV, Koo D-H, Friebe B, Gill BS (2015) Chromosome engineering, mapping, and transferring of resistance to Fusarium head blight disease from Elymus tsukushiensis into wheat. Theor Appl Genet 128:1019–1027
Cetin Y, Bullerman LB (2005) Cytotoxicity of Fusarium mycotoxins to mammalian cell cultures as determined by the MTT bioassay. Food Chem Toxicol 43:755–764
Chakraborty S, Newton AC (2011) Climate change, plant diseases and food security: an overview. Plant Pathol 60:2–14
Cuthbert PA, Somers DJ, Thomas J, Cloutier S, Brulé-Babel A (2006) Fine mapping Fhb1, a major gene controlling Fusarium head blight resistance in bread wheat (Triticum aestivum L.). Theor Appl Genet 112:1465–1472
Cuthbert PA, Somers DJ, Brulé-Babel A (2007) Mapping of Fhb2 on chromosome 6BS: a gene controlling Fusarium head blight field resistance in bread wheat (Triticum aestivum L.). Theor Appl Genet 114:429–437
D’mello J, Placinta C, Macdonald A (1999) Fusarium mycotoxins: a review of global implications for animal health, welfare and productivity. Anim Feed Sci Technol 80:183–205
Dewey DR (1983) Historical and current taxonomic perspectives of Agropyron, Elymus, and related genera. Crop Sci 23:637–642
Duveiller E, Singh RP, Nicol JM (2007) The challenges of maintaining wheat productivity: pests, diseases, and potential epidemics. Euphytica 157:417–430
Dvořák J (1980) Homoeology between Agropyron elongatum chromosomes and Triticum aestivum chromosomes. Can J Genet Cytol 22:237–259
Fu S, Lv Z, Qi B, Guo X, Li J, Liu B, Han F (2012) Molecular cytogenetic characterization of wheat—Thinopyrum elongatum addition, substitution and translocation lines with a novel source of resistance to wheat Fusarium head blight. J Genet Genomics 39:103–110
Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100
Goswami RS, Kistler HC (2004) Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol 5:515–525
Han F, Gao Z, Birchler JA (2009) Reactivation of an inactive centromere reveals epigenetic and structural components for centromere specification in maize. Plant Cell 21:1929–1939
He X, Singh PK, Schlang N, Duveiller E, Dreisigacker S, Payne T, He Z (2014) Characterization of Chinese wheat germplasm for resistance to Fusarium head blight at CIMMYT, Mexico. Euphytica 195:383–395
Jauhar PP (2014) Durum wheat genetic stocks involving chromosome 1E of diploid wheatgrass: resistance to Fusarium head blight. Nucleus 57:19–23
Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, Appels R, Pfeifer M, Tao Y, Zhang X, Jing R, Zhang C, Ma Y, Gao L, Gao C, Spannagl M, Mayer K, Li D, Pan S, Zheng F, Hu Q, Xia X, Li J, Liang Q, Chen J, Wicker T, Gou C, Kuang H, He G, Luo Y, Keller B, Xia Q, Lu P, Wang J, Zou H, Zhang R, Xu J, Gao J, Middleton C, Quan Z, Liu G, Wang J, International Wheat Genome Sequencing Consortium, Yang H, Liu X, He Z, Mao L, Wang J (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496:91–95
Kim NS, Armstrong K, Knott D (1993) Molecular detection of Lophopyrum chromatin in wheat-Lophopyrum recombinants and their use in the physical mapping of chromosome 7D. Theor Appl Genet 85:561–567
Kong L, Cambron SE, Ohm HW (2008) Hessian fly resistance genes H16 and H17 are mapped to a resistance gene cluster in the distal region of chromosome 1AS in wheat. Mol Breed 21:183–194
Kosambi D (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Ling HQ, Zhao S, Liu D, Wang J, Sun H, Zhang C, Fan H, Li D, Dong L, Tao Y (2013) Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496:87–90
Liu S, Anderson JA (2003) Targeted molecular mapping of a major wheat QTL for Fusarium head blight resistance using wheat ESTs and synteny with rice. Genome 46:817–823
Liu S, Zhang X, Pumphrey MO, Stack RW, Gill BS, Anderson JA (2006) Complex microcolinearity among wheat, rice, and barley revealed by fine mapping of the genomic region harboring a major QTL for resistance to Fusarium head blight in wheat. Funct Integr Genomics 6:83–89
Marais G, Kotze L, Eksteen A (2010) Allosyndetic recombinants of the Aegilops peregrina-derived Lr59 translocation in common wheat. Plant Breed. doi:10.1111/j.1439-0523.2009.01713.x
Mayer KF, Rogers J, Doležel J, Pozniak C, Eversole K, Feuillet C, Gill B, Friebe B, Lukaszewski AJ, Sourdille P (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788
Niu Z, Klindworth DL, Friesen TL, Chao S, Jin Y, Cai X, Xu SS (2011) Targeted introgression of a wheat stem rust resistance gene by DNA marker-assisted chromosome engineering. Genetics 187:1011–1021
Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Qi L, Friebe B, Zhang P, Gill BS (2007) Homoeologous recombination, chromosome engineering and crop improvement. Chromosome Res 15:3–19
Qi L, Pumphrey M, Friebe B, Chen P, Gill B (2008) Molecular cytogenetic characterization of alien introgressions with gene Fhb3 for resistance to Fusarium head blight disease of wheat. Theor Appl Genet 117:1155–1166
Roberts MA, Reader SM, Dalgliesh C, Miller TE, Foote TN, Fish LJ, Snape JW, Moore G (1999) Induction and characterization of Ph1 wheat mutants. Genetics 153:1909–1918
Semagn K, Bjørnstad Å, Skinnes H, Marøy AG, Tarkegne Y, William M (2006) Distribution of DArT, AFLP, and SSR markers in a genetic linkage map of a doubled-haploid hexaploid wheat population. Genome 49:545–555
Shen X, Ohm H (2007) Molecular mapping of Thinopyrum-derived Fusarium head blight resistance in common wheat. Mol Breed 20:131–140
Shen X, Kong L, Ohm H (2004) Fusarium head blight resistance in hexaploid wheat (Triticum aestivum)-Lophopyrum genetic lines and tagging of the alien chromatin by PCR markers. Theor Appl Genet 108:808–813
Sourdille P, Singh S, Cadalen T, Brown-Guedira GL, Gay G, Qi L, Gill BS, Dufour P, Murigneux A, Bernard M (2004) Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct Integr Genomics 4:12–25
Van Ooijen JW (2006) JoinMap® 4.0: software for the calculation of genetic linkage maps in experimental population. Kyazma BV, Wageningen, p. 33
Vogel JP, Garvin DF, Mockler TC, Schmutz J, Rokhsar D, Bevan MW, Barry K, Lucas S, Harmon-Smith M, Lail K (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463:763–768
Wang RR (2011) Agropyron and Psathyrostachys. In: Kole C (ed) Wild crop relatives: genomic and breeding resources, cereals, 1st edn. Springer, Berlin, pp 77–108
Wang RR, Lu B (2014) Biosystematics and evolutionary relationships of perennial Triticeae species revealed by genomic analyses. J Syst Evol 52:697–705
Wang J, Li H, Zhang L, Meng L (2014a) Users’ Manual of QTL IciMapping. The Quantitative Genetics Group, Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS), Beijing 100081, China, and Genetic Resources Program, International Maize and Wheat Improvement Center (CIMMYT), Apdo. Postal 6-641, 06600 Mexico, D.F., Mexico
Wang Z, Cui Y, Chen Y, Zhang D, Liang Y, Zhang D, Wu Q, Xie J, Ouyang S, Li D (2014b) Comparative genetic mapping and genomic region collinearity analysis of the powdery mildew resistance gene Pm41. Theor Appl Genet 127:1741–1751
Wenzl P, Carling J, Kudrna D, Jaccoud D, Huttner E, Kleinhofs A, Kilian A (2004) Diversity arrays technology (DArT) for whole-genome profiling of barley. Proc Natl Acad Sci USA 101:9915–9920
West JS, Holdgate S, Townsend JA, Edwards SG, Jennings P, Fitt BD (2012) Impacts of changing climate and agronomic factors on Fusarium ear blight of wheat in the UK. Fungal Ecol 5:53–61
Xue S, Li G, Jia H, Xu F, Lin F, Tang M, Wang Y, An X, Xu H, Zhang L (2010) Fine mapping Fhb4, a major QTL conditioning resistance to Fusarium infection in bread wheat (Triticum aestivum L.). Theor Appl Genet 121:147–156
Xue S, Xu F, Tang M, Zhou Y, Li G, An X, Lin F, Xu H, Jia H, Zhang L (2011) Precise mapping Fhb5, a major QTL conditioning resistance to Fusarium infection in bread wheat (Triticum aestivum L.). Theor Appl Genet 123:1055–1063
You FM, Huo N, Gu YQ, Lazo GR, Dvorak J, Anderson OD (2009) ConservedPrimers 2.0: a high-throughput pipeline for comparative genome referenced intron-flanking PCR primer design and its application in wheat SNP discovery. BMC Bioinformatics 10:331
Yu J, Bai G, Cai S, Dong Y, Ban T (2008) New Fusarium head blight-resistant sources from Asian wheat germplasm. Crop Sci 48:1090–1097
Zhang X, Shen X, Hao Y, Cai J, Ohm HW, Kong L (2011) A genetic map of Lophopyrum ponticum chromosome 7E, harboring resistance genes to Fusarium head blight and leaf rust. Theor Appl Genet 122:263–270
Acknowledgments
The authors thank Robert McIntosh, University of Sydney Plant Breeding Institute–Cobbitty, PB4011, Narellan, NSW 2567, Australia, for review of the manuscript before submission. We thank Dr. B. S. Gill, Kansas state university for providing seeds of wheat nulli-tetra and deletion stocks of group 7. We acknowledge financial supports by the National High-Tech R&D Program of China (2011AA100102 and 2012AA101105), the NSF of China (Grant nos. 31171553 and 31471488), Shandong Seed Engineering Project (2015-2019), and the International Collaboration Program (948 Project, 2013-S19).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Communicated by H. Buerstmayr.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Guo, J., Zhang, X., Hou, Y. et al. High-density mapping of the major FHB resistance gene Fhb7 derived from Thinopyrum ponticum and its pyramiding with Fhb1 by marker-assisted selection. Theor Appl Genet 128, 2301–2316 (2015). https://doi.org/10.1007/s00122-015-2586-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2586-x